Abstract
Endoribonuclease-prepared siRNA (esiRNA) is an alternative tool to chemical synthetic siRNA for gene silencing. Since esiRNAs are directed against long target sequences, the genetic variations in the target sequences will have little influence on their effectiveness. The ability of esiRNAs to inhibit hepatitis B virus (HBV) gene expression and replication was tested. EsiRNAs targeting the coding region of HBV surface antigen (HBsAg) and the nucleocapsid (HBcAg) inhibited specifically the expression of HBsAg and HBcAg when cotransfected with the respective expression plasmids. Both esiRNAs reduced the HBV transcripts and replication intermediates in transient transfected cells and cells with HBV genomes integrated stably. Compared with synthetic siRNA, esiRNA targeting HBsAg was less effective than the selected synthetic siRNA in terms of the inhibition of HBV gene expression and replication. However, while the ability of synthetic siRNAs for specific gene silencing was impaired strongly by the nucleotide substitutions within the target sequences. The efficiency of gene silencing by esiRNAs was not influenced by sequence variation. The transfection of esiRNA did not induce interferon-stimulated genes (ISGs) like STAT1 and ISG15, indicating the absence of off-target effects. In general, esiRNAs strongly inhibited HBV gene expression and replication and may have an advantage against HBV strains which are variable genetically.
Keywords: RNA interference (RNAi), Endoribonuclease-prepared siRNA (esiRNA), Hepatitis B virus (HBV)
1. Introduction
Hepatitis B virus (HBV) causes acute and chronic infection in humans. Although effective vaccines are available to prevent the transmission of HBV, HBV infection remains a global health problem due to about 350 million of chronically HBV-infected people worldwide. These individuals have a relatively high risk of developing end-stage liver diseases, such as liver cirrhosis and hepatocellular carcinoma (Seeger and Mason, 2000). To date, treatment regiments for chronic hepatitis B are costly and have limited effectiveness. Only about one-third of the patients treated with alpha-interferon show a sustained response. Nucleoside analogues do not eliminate the virus completely and may select resistant viral variants (Marcellin, 2002). Thus, it is urgent to develop new antiviral drugs against HBV.
RNA interference (RNAi) is a process whereby double-stranded RNA (dsRNA) induces a sequence-specific degradation of homologous messenger RNA (mRNA) (Hannon, 2002). This process is mediated by small interfering RNAs (siRNAs) of the length of 21–23 nucleotides. In the natural RNAi pathway, siRNAs are derived from the processing of long dsRNAs by the nuclease dicer into discrete 21-mers. Using chemically synthesized or vector-expressed siRNAs, many clinically important viruses including human immunodeficiency virus (HIV), severe acute respiratory syndrome coronavirus, HBV, and hepatitis C virus (HCV) could be inhibited in vitro (Haasnoot et al., 2003, Li et al., 2005, Randall and Rice, 2004, Stevenson, 2003, Wu and Nandamuri, 2004). A number of recent studies have demonstrated that the HBV gene expression and viral replication could be inhibited (Giladi et al., 2003, Guo et al., 2005; Hamasaki et al., 2003, Klein et al., 2003, Konishi et al., 2003, McCaffrey et al., 2003, Morrissey et al., 2005a, Morrissey et al., 2005b, Shlomai and Shaul, 2003, Wu et al., 2005), or even cleared from liver of transgenic mice by siRNAs synthesized chemically or vector-expressed (Uprichard et al., 2005). In these studies, siRNAs targeting different region of the HBV genome were used.
The genetic variation of viral genomes may lead to escape from the silencing effect, as reported for poliovirus (Gitlin et al., 2005), HIV-1 (Boden et al., 2003, Das et al., 2004), and HCV (Wilson and Richardson, 2005). This problem could be overcome by targeting alternative sites on viral genomes as shown for HCV (Wilson and Richardson, 2005). Due to the genetic variability of HBV, a treatment with siRNAs faces the problem with naturally occurring or treatment-induced nucleotide substitutions in the HBV genomes. Thus, gene silencing strategies targeting multiple sites are warranted. Yang et al. (2002) showed that Escherichia coli RNase III can digest dsRNA efficiently into short pieces with the same end structures as siRNAs. These endoribonuclease-prepared siRNAs (esiRNAs) are able to target multiple sites within an mRNA, and have been verified to silence target mRNA efficiently and specifically (Calegari et al., 2002, Kittler et al., 2004, D. Yang et al., 2004, H. Yang et al., 2004, Zhu and Jiang, 2005). In the present study, the ability of esiRNA to inhibit the HBV gene expression and replication was investigated. The esiRNAs targeting the HBV S and C gene were prepared by in vitro transcription and RNase III digestion. The inhibition of the HBV gene expression and the replication by esiRNA were demonstrated in transient cotransfection experiments and in a cell line with a stably integrated HBV genome. The effect of esiRNA targeting HBV S gene was compared further with that of chemical synthetic siRNAs.
2. Methods
2.1. Preparation of esiRNAs targeting the coding region of HBV surface antigen (HBsAg) and nucleocapsid (HBcAg)
The coding regions for HBsAg (nt 129–842) and HBcAg (nt 1901–2348) were amplified from a HBV genome of subtype ayw (GenBank accession no. U95551) and cloned into pCR2.1 vector (Invitrogen, Karlsruhe, DE). The primers S6C, S7D, HcNCO, and Hc-149s used for PCR amplification are listed in Table 1 . Clones with the inserts in both orientations with regard to the T7 promoter were selected and sequenced to verify that the construction was correct. EsiRNAs targeting the coding region of HBsAg and HBcAg (SesiRNA and CesiRNA) were prepared by using Silencer siRNA Cocktail Kit (RNase III) (Ambion, Darmstadt, DE) according to the manufacturer's instructions. Briefly, single strand RNAs were transcribed from the plasmids using T7 polymerase and then annealed to form double strand RNA. EsiRNAs were generated finally by digestion of the purified dsRNA with RNase III at 37 °C for 1 h, and verified by gel electrophoresis on a 4% agarose gel. HCV-C5, a plasmid containing HCV core region (nt 631–900) (Lu et al., 1995) was used to prepare HCVesiRNA as a scramble esiRNA control.
Table 1.
Primers used for cloning of the coding region of HBsAg and HBcAg and RT-PCR
| Amplicon | Primer | Polarity | Nucleotide sequence | Position of the 5′-base |
|---|---|---|---|---|
| HBV S gene | S6C | Sense | 5′-gca cac gga att ccg agg act ggg gac cct g-3′ | 129a |
| S7D | Antisense | 5′-gac acc aag ctt ggt tag ggt tta aat gta tac c-3′ | 842a | |
| HBV C gene | HcNCO | Sense | 5′-acc atg gac att gac cct tat aaa g-3′ | 1901a |
| Hc-149s | Antisense | 5′-agg atc caa caa cag tag ttt ccg g-3′ | 2348a | |
| HBc-EV1 | Sense | 5′-gga tat cat gga cat cga ccc tta t-3′ | 1901a | |
| HBc-EV2 | Antisense | 5′-act cga gac att gag att ccc gag a-3′ | 2451a | |
| HBV preC/C | HpreC-EV1 | Sense | 5′-gga tat cat gca act ttt tca cct ctg-3′ | 1814a |
| HBc-EV2 | Antisense | 5′-act cga gac att gag att ccc gag a-3′ | 2451a | |
| STAT1 | STAT1-1 | Sense | 5′-cca tgg aaa tca gac agt acc tgg c-3′ | 431b |
| STAT1-3 | Antisense | 5′-cct tca cat ttc tga ctt tac tgt c-3′ | 802b | |
| ISG15 | ISG15k | Sense | 5′-gac ctg acg gtg aag atg ctg g-3′ | 84c |
| ISG15k-2 | Antisense | 5′-ttc ctc acc agg atg ctc aga gg-3′ | 337c | |
| β-Actin | A-1 | Sense | 5′-tgg aat cct gtg gca tcc atg aaa c-3′ | 879d |
| A-2 | Antisense | 5′-taa aac gca gct cag taa cag tcc g-3′ | 1224d | |
The italicized parts of primers S6C, S7D, HcNCO, Hc-149s, HBc-EV1, HpreC-EV1, and HBc-EV2 indicate the specific cleavage sites of restriction enzymes EcoRI, HindIII, NcoI, BamHI, EcoRV, and XhoI, respectively.
Numbering of the sequence is according to U95551.
Numbering of the sequence is according to NM_007315.2.
Numbering of the sequence is according to XM_010527.
Numbering of the sequence is according to NM_001101.2.
2.2. siRNAs
The sequences of 3 siRNAs targeting the coding region of S antigen of HBV adw strain were designed by using an online software from Ambion (www.ambion.com) and combined with the algorithm described by Reynolds et al. (2004). The sequences of the siRNAs are: siHBs1, 5′-GAA TCC TCA CAA TAC CGC A-3′ (nt 225–243); siHBs2, 5′-GGT ATG TTG CCC GTT TGT C-3′ (nt 458–496); siHBs3, 5′-GGA ACC TCT ATG TAT CCC T-3′ (nt 542–560). A siRNA against green fluorescent protein, siGFP (5′-CGG CAA GCT GAC CCT GAA GTT-3′), was used as control. All siRNAs were purchased from Qiagen.
2.3. Cell culture and transfection
BHK cells, HepG2 cells, and HepG2.2.15 cells (provided by Prof. G. Arc; HBV serotype ayw, genotype D; GenBank accession no. U95551) (Sells et al., 1987) were maintained in Eagle's minimal essential medium or RPMI 1640 medium, respectively, supplemented with 10% of fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were seeded in an 8-well chamber slide or 24-well plate at about 60% confluence. After 24 h, cells were transfected with lipofectamine 2000 (Invitrogen, Karlsruhe, DE) following the manufacturer's instructions. 0.2 μg or 0.5 μg of plasmid DNA and 0.5 μl or 2 μl of lipofectamine 2000 were placed in each well of an 8-well chamber slide or in a 24-well plate, respectively.
2.4. Construction of an HBV infectious clone and expression plasmids encoding HBsAg, HBeAg, and HBcAg
A replication-competent HBV construct pHY106 + wta was generated by inserting a full length wild type HBV genome from pSM2 (kindly provided by Prof. Hans Will, Genotype D, subtype ayw, GenBank accession no. V01460) into pHY106 vector (H. Yang et al., 2004). The pHY106 vector contains a cytomegalovirus (CMV) promoter upstream of a short, recombinant HBV sequence that allows the in-frame insertion of a full length HBV genome following SapI digestion. The CMV promoter upstream of the precore initiation site allows an efficient transcription of the 3.5-kb pregenomic RNA following transfection of liver cell lines. Two vectors pHBeex and pHBcex were constructed for the expression of HBeAg and HBcAg, respectively. The regions of the HBV genome (nt 1814–2451) and (nt 1901–2451) were amplified with primers HpreC-EV1, HBc-EV2 and HBc-EV1, HBc-EV2, respectively (Table 1), digested with EcoRV and XhoI, and inserted into the pcDNA3 vector via the EcoRV and XhoI sites. The expression plasmids encoding HBsAg 1056Sp (Genotype D, subtype ayw), HK188 (Genotype C, subtype adr), and 91-4696 (Genotype A, subtype adw) were described by Ireland et al. (2000).
2.5. HBsAg and HBeAg chemiluminescent microparticle immunoassay (CMIA)
Levels of HBsAg and HBeAg in cell supernatants were determined by using the Architect system and HBsAg and HBeAg CMIA kits (Abbott Laboratories, Wiesbaden-Delkenheim, DE) according to the manufacturer's instructions.
2.6. Immunofluorescence (IF) staining for hepatitis B core antigen (HBcAg)
Hepatitis B core antigen was visualized by IF staining with specific antibodies. Transfected cells were cultured for 24 h, then fixed with 50% of methanol in phosphate-buffered saline (PBS), and stained with polyclonal rabbit anti-HBc antibody (DAKO, Hamburg, DE). Goat anti-rabbit immunoglobulin G-fluorescein isothiocyanate (Sigma, Munich, DE) was used as a secondary antibody for the experiments. Staining was visualized under a fluorescence microscope (Nikon, Tokyo, JP) with an excitation wavelength of 490 nm.
2.7. Isolation and analysis of viral RNA
Total RNA was extracted from transfected cells with TRIzol reagent (Invitrogen, Karlsruhe, DE) according to the manufacturer's instruction. Northern blot analysis was carried out by agarose–formaldehyde method. Briefly, 5 μg of total RNA per sample were separated on 1% agarose–formaldehyde gel and blotted to a Hybond-N+ nylon membrane (Amersham, Buckinghamshire, GB). HBV transcripts were detected by using a 32P-labeled full length HBV probe. Hybridization signals were visualized and analyzed by a Phospho-Imager (Cyclon, Parkard Instrument).
2.8. Purification and analysis of HBV DNA from intracellular core particles
HBV replicative intermediates were purified from intracellular core particles according to the method described by Sterneck et al. (1998) with minor modification. Briefly, cells were washed in ice-cold PBS and lysed in 0.4 ml of lysis buffer containing 50 mM Tris–HCl, pH 7.4, 1 mM EDTA, 1% NP-40 at 4 °C for 15 min. Nuclei were pelleted by centrifugation. The supernatant was adjusted to 10 mM MgCl2 and treated with 100 μg/ml DNase I (Roche, Mannheim, DE) at 37 °C for 30 min. The reaction was stopped by the addition of EDTA to a final concentration of 25 mM. Proteins were then digested with 0.5 mg/ml proteinase K (Qiagen, Düsseldorf, DE) and 1% sodium dodecyl sulfate at 55 °C for 2 h. HBV nucleocapsid-associated DNA was purified by phenol/chloroform (1:1) extraction followed by isopropanol precipitation by adding 15 μg of tRNA and 1/10 volume of 3 M sodium acetate, pH 5.2. The isolated HBV DNA was subjected to agarose gel electrophoresis, followed by denaturation and Southern blotting. HBV DNA was detected by hybridization with a 32P-labeled full length HBV probe. Hybridization signals were visualized and analyzed by a Phospho-Imager (Cyclon, Parkard Instrument).
2.9. RT-PCR and real-time PCR
Two micrograms of total RNA per sample was reverse transcribed by using Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) (Promega, Mannheim, DE) and Oligo (dT) (Invitrogen, Karlsruhe, DE) as primer. The cDNA fragments of STAT1 and ISG15 were amplified by using the primers listed in Table 1. Cycle parameters were (i) 1 cycle: 94 °C, 4 min; (ii) 30 cycles: 94 °C, 30 s; 55 °C, 30 s; 72 °C, 30 s; and (iii) 1 cycle: 72 °C, 10 min. PCR products were subjected to agarose gel electrophoresis and visualized by ethidium bromide staining. Quantitative PCR was carried out using Platinum SYBR Green qPCR SuperMix UDG (Invitrogen, Karlsruhe, DE) in a Roche Lightcycler V.3. The PCR was performed with the following cycling parameters over 45 cycles: 95 °C for 5 s, 58 °C for 10 s, and 72 °C for 10 s. The specificity of the PCR products was verified by melting curve analysis and agarose gel electrophoresis.
3. Results
3.1. Inhibition of the HBV gene expression by specific esiRNAs
To examine the inhibitory effect of esiRNAs on the HBV gene expression, the expression plasmids encoding HBsAg and HBeAg were cotransfected with esiRNA into HepG2 cells. The expression levels of HBsAg and HBeAg in cell culture supernatants were determined 72 h later by CMIA. Cotransfection with SesiRNA and CesiRNA at the concentration of 100 nM reduced the expression levels of HBsAg and HBeAg to 20% and 10% of the control without transfection of esiRNA, respectively (Fig. 1a and b). The inhibition of the HBcAg expression by CesiRNA was verified by IF staining of cells transfected with the HBcAg expression plasmid with and without CesiRNA (Fig. 1c). The HBcAg expression in transfected cells was strongly reduced by CesiRNA.
Fig. 1.
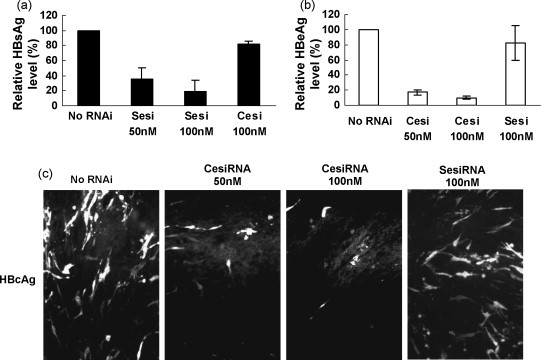
The effects of esiRNAs on HBV S and core antigen expression. HepG2 cells were cotransfected with 0.5 μg of the HBsAg or HBeAg expression plasmid and esiRNAs at concentrations of 50 nM or 100 nM. The HBsAg (a) and HBeAg (b) levels in cell culture media were measured by CMIA 72 h post-transfection. (c) BHK cells in 8-well chamber slide were cotransfected with 0.2 μg of the expression plasmid encoding HBcAg and CesiRNA at concentration of 50 nM or 100 nM, or 100 nM of SesiRNA as a control, and fixed 24 h post-transfection. HBcAg was stained with a polyclonal rabbit anti-HBc serum. Sesi, SesiRNA; Cesi, CesiRNA.
3.2. Inhibition of HBV replication in cell culture by specific esiRNAs
To determine whether esiRNAs are able to inhibit the HBV replication, HepG2 cells were transfected with pHY106 + wta in the presence of SesiRNA or CesiRNA. The expression of HBsAg and HBeAg in cell culture supernatants was reduced significantly by esiRNAs (Fig. 2a). A decrease of 76.0% and 68.1% of the HBsAg levels in supernatants was measured by cotransfection with SesiRNA and CesiRNA, respectively. Likewise, SesiRNA and CesiRNA suppressed the expression levels of HBeAg to 20% and 22% of the control, respectively (Fig. 2a). The amount of HBV replicative intermediates was reduced in HepG2 cells by both esiRNAs in a dose-dependent manner (Fig. 2b). Only 10% of HBV replication intermediates was detected in HepG2 cells treated with 100 nM of SesiRNA and CesiRNA. Northern blot analysis showed that HBV transcripts were reduced to a level of about 40% of the control (Fig. 2c).
Fig. 2.
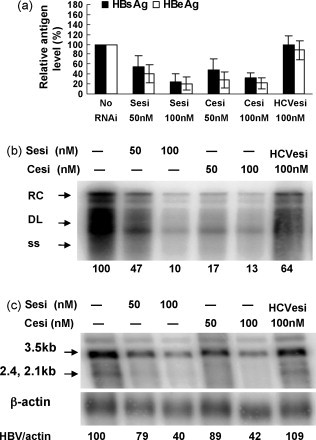
The effects of esiRNAs on HBV gene expression and replication. HepG2 cells were cotransfected with 0.5 μg of pHY106 + wta and esiRNAs at concentrations of 50 nM or 100 nM, or 100 nM of HCVesiRNA as a control. The HBsAg and HBeAg levels in cell culture supernatants (a), HBV replicative intermediates (b), and HBV transcripts (c) were analyzed 72 h post-transfection, by CMIA, Southern blot, and Northern blot analysis, respectively. The relative strength of signals detected in Southern blot and Northern blot was calculated by setting the untransfected control as 100% and given in (b) and (c). Sesi, SesiRNA; Cesi, CesiRNA; HCVesi, HCVesiRNA.
SesiRNA and CesiRNA were then examined in HepG2.2.15 cells with a stably integrated HBV dimer. This cell line produces HBV RNA and replication intermediates at a stable level. HepG2.2.15 cells were transfected with 50 nM or 100 nM of SesiRNA and CesiRNA and incubated further for 72 h. The treatment with esiRNAs did not reduce the HBsAg concentrations in culture supernatants (Fig. 3a). The HBeAg concentrations in culture supernatants were not changed by transfection with SesiRNA. CesiRNA was effective in suppressing the HBeAg level to about 50%. The HBV replication intermediates were reduced to 29% and 35% of the control by SesiRNA and CesiRNA at a concentration of 100 nM, respectively (Fig. 3b). Similarly, the level of HBV transcripts decreased to 33% and 49% of the control. Thus, the silencing of HBV transcripts in cells transfected stably by a single transfection with esiRNA had a limited effect.
Fig. 3.
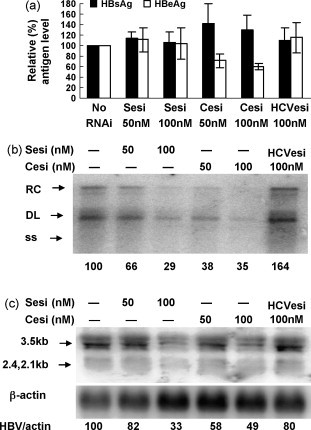
The effects of esiRNAs on HBV gene expression and replication in a stable HBV-producing cell line. HepG2.2.15 cells were transfected with SesiRNA, CesiRNA, or 100 nM of HCVesiRNA as a control. The HBsAg and HBeAg levels in cell culture medium (a), HBV replicative intermediates (b), and HBV transcripts (c) were analyzed 72 h post-transfection, by CMIA, Southern blot, and Northern blot analysis, respectively. The relative strength of signals detected in Southern blot and Northern blot was calculated by setting the control as 100% and given in (b) and (c). Sesi, SesiRNA; Cesi, CesiRNA; HCVesi, HCVesiRNA.
3.3. Comparison of esiRNAs with synthetic siRNAs
A number of specific siRNAs have been shown to knock down the corresponding HBV transcripts efficiently in vitro and in vivo. Therefore, the ability of esiRNA and synthetic siRNAs to inhibit HBV gene expression and viral replication was compared. Three synthetic siRNAs siHBs1–3 targeting the coding region of HBsAg inhibited the HBsAg expression with different effectiveness (Fig. 4a). However, the silencing of the HBsAg expression by synthetic siRNAs was impaired strongly by nucleotide variations (Fig. 4b). A G to A substitution at the position 17 in the target sequence reduced the ability of siHBs1 to inhibit the expression of HBsAg. Similarly, the two nucleotide substitutions in the target sequence of siHBs2 abolished the silencing effect completely. In contrast, these mutations did not affect the silencing effect of SesiRNA (data not shown, see below).
Fig. 4.
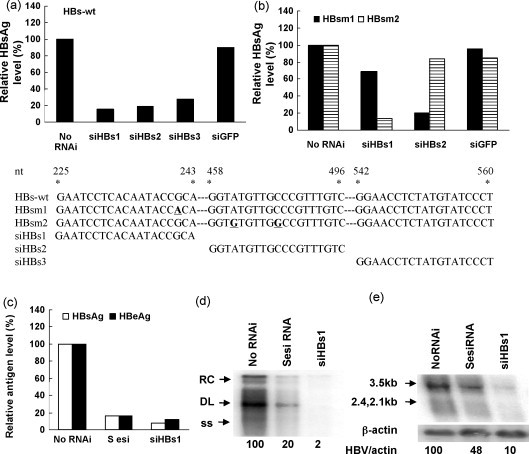
Comparison of esiRNA and siRNAs. HepG2 cells were cotransfected with 10 nM of siRNA targeting HBV S gene (siHBs1–3) and an expression plasmid encoding wild type HBsAg (HBs-wt) (a) or mutated HBsAg (HBsm1 and HBsm2) (b). To compare the gene silencing by synthetic siRNAs and esiRNAs, HepG2 cells were cotransfected with pHy106 + wta and SesiRNA or siHBs1 (c–e). The HBsAg and HBeAg levels in cell culture supernatants (c), HBV replicative intermediates (d), and HBV transcripts (e) were analyzed 72 h post-transfection by CMIA, Southern blot, and Northern blot analysis, respectively. The sequences of siRNA and their targets were shown. siRNA targeting green fluorescence protein (siGFP) was used as a scramble siRNA control. The relative strength of signals detected in Southern blot and Northern blot was calculated by setting the untransfected control as 100% and given in (d) and (e).
The ability of siHBs1 and SesiRNA to inhibit HBV gene expression and replication was compared in HepG2 cells by cotransfection with pHY106 + wta and 100 nM of SesiRNA or 12.5 nM of siHBs1. The production of HBsAg and HBeAg was suppressed strongly by both SesiRNA and siHBs1 (Fig. 4c). The level of HBV replicative intermediates was reduced to 20% of the control by SeiRNA vs. 2% by siHBs1 (Fig. 4d). Further, siHBs1 led to a significant decrease of the amounts of HBV transcripts to 10% of the control (Fig. 4e). Therefore, synthetic siRNAs may be more efficient for gene silencing than esiRNAs in the present use of synthetic siRNAs.
It is suggested that esiRNAs are directed to multiple sites of a target sequence. Therefore, the ability of SesiRNA to inhibit the expression of heterologous HBsAg sequences was examined. Three HBsAg expression plasmids encoding different HBsAg geno/subtypes were cotransfected with SesiRNA. The determination of the HBsAg expression level in supernatants showed that cotransfection with SesiRNA reduced equally the expression of HBsAg, regardless of the subtypes (Fig. 5 ).
Fig. 5.
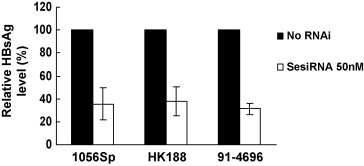
Effects of esiRNA on naturally occurred HBs mutants. HepG2 cells were cotransfected with SesiRNA and three expression plasmids encoding different types of HBsAg, 1056Sp (genotype D, ayw), HK188 (genotype C, adr), and 91-4696 (genotype A, adw). The HBsAg level in cell culture supernatants was determined by CMIA 72 h post-transfection.
3.4. EsiRNA did not induce the expression of interferon-stimulated genes (ISGs)
Large dsRNA with a length over 35 bp might induce the interferon response in cells. Thus, it is necessary to examine whether the use of esiRNAs induces ISGs in HepG2.2.15 cells to exclude the off-target effect. RT-PCR analysis was carried out to monitor the induction of STAT1 and ISG15 in cells. The expression of STAT1 and ISG15 in HepG2.2.15 cells was increased after a 24-h treatment with 100 units of IFN-α. The transfection of HepG2.2.15 cells with SesiRNA and CesiRNA did not increase the expression of STAT1 and ISG15 (Fig. 6 ). Thus, esiRNA was not able to activate the interferon response in hepatoma cells.
Fig. 6.
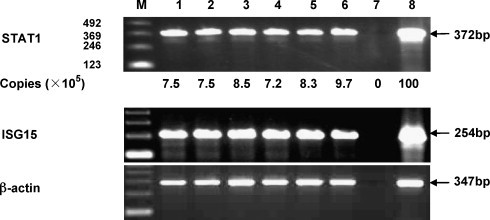
RT-PCR analysis of STAT1, ISG15, and β-actin mRNA in cells transfected with esiRNAs. Total RNAs were purified from HepG2.2.15 cells transfected with or without esiRNA. cDNAs were prepared by reverse transcription with Oligo (dT) primer. The cDNA fragments of STAT1, ISG15, and β-actin were amplified and examined by agarose gel electrophoresis. In addition, the relative amounts of STAT1 mRNAs were determined by real-time RT-PCR as shown in copies per microgram RNA. M, 123 bp DNA Ladder; lane 1, no RNAi; lane 2, SesiRNA 50 nM; lane 3, SesiRNA 100 nM; lane 4, CesiRNA 50 nM; lane 5, CesiRNA 100 nM; lane 6, HCVesiRNA 100 nM; lane 7, negative control; lane 8, positive control: total RNA purified from HepG2.2.15 cells stimulated with interferon-alpha at a concentration of 100 units/ml.
4. Discussion
In this study, the ability of specific esiRNAs to silence the HBV gene expression and replication was examined in detail. The esiRNA directed against the coding sequences of HBsAg and HBcAg was capable of inhibiting the HBV gene expression and viral replication both in transient cotransfection system and in HepG2.2.15 cells. The inhibition effect was specific and dose-dependent. Theoretically, the esiRNA targeted at HBsAg should be more effective than its counterpart targeted at HBcAg. Since the HBV S region is shared by the major viral transcripts, all these RNA species were suppressed by SesiRNA. However, CesiRNA was as effective as or even more effective than SesiRNA. CesiRNA inhibited the HBsAg expression likely acting indirectly through reduction of HBV replication or by other unknown mechanisms. Interestingly, both SesiRNA and CesiRNA effectively inhibited HBV replication in HepG2.2.15 cells while only HBeAg expression was reduced by CesiRNA. One explanation is that only a small amount of HBsAg transcripts is needed for the synthesis of HBsAg.
These data demonstrated that it is advantageous to use esiRNAs against genetically heterogeneous target sequences. The ability to use siRNAs targeted simultaneously at different regions of a viral genome may increase the efficiency of the treatment and, in addition, will prevent the appearance of resistant mutants. Eight genotypes A–H of HBV can be distinguished. Each genotype differs from the others by more than 8% at the nucleotide level. Phylogenetic analysis has shown that the eight genotypes can be subdivided further into genotypical subtypes (Schaefer, 2005). It is difficult to identify a highly effective siRNA target at a conserved 19–21 nt sequence in all these genotypes and genotypical subtypes. EsiRNAs generated from long dsRNA are mixtures of siRNAs with diverse specificities, and would be able to target the whole mRNA sequence and retain the ability for gene silencing. The results in the current study showed that esiRNA generated from HBV subtype ayw is applicable for different HBsAg subtypes tested. In contrast, single nucleotide mutation in the target sequence abrogates the silencing effect of chemically synthetic siRNAs. Similarly, Weinberg et al. (2007) used a vector to produce a dicer substrate that could generate multiple siRNAs. This approach would have the advantage of limiting escape and targeting a range of sequences found in different viral genotypes or quasispecies.
siRNA-specific features such as low G/C content, a bias towards low internal stability at the sense strand 3′-terminus, lack of inverted repeats, and sense strand base preference are likely to contribute to efficient RNAi process (Reynolds et al., 2004). According to these criteria, there would be only a limited number of optional sequences for RNAi targeted on a given mRNA. Processing of long dsRNA generates a variety of small dsRNA molecules with different degrees for RNA interference. Thus, it is understandable that esiRNAs would be less effective in comparison with a defined, optimized synthetic siRNA within the range of used concentrations. However, synthetic siRNAs may have different abilities for gene silencing, thus, some of them may be less efficient than esiRNAs (Xuan et al., 2006).
Recently, several in vivo studies based on the use of cationic liposomes, polyplexes, and chemical modified siRNAs showed improved effects of siRNA (Aigner, 2006, Morrissey et al., 2005a, Morrissey et al., 2005b). Using liver-specific apo A-I-mediated siRNA delivery method, Kim et al. (2007) showed that administration of synthetic siRNAs reduced significantly HBV protein expression with the advantages of effectiveness at low doses and long-term effect. This unique approach to siRNA delivery creates a foundation for the development of a new class of promising therapeutic method against hepatitis viruses. The in vivo use of esiRNA needs to be investigated further using a liver-specific siRNA delivery method.
Acknowledgements
We are thankful for critical reading of Delia Cosgrove. This work is supported by grants of Deutsche Forschungsgemeinschaft (Lu 669/2-1, GRK1045/1, and Lu 669/5-1).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jviromet.2008.02.008.
Appendix A. Supplementary data
References
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J. Biotechnol. 2006;124(1):12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calegari F., Haubensak W., Yang D., Huttner W.B., Buchholz F. Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14236–14240. doi: 10.1073/pnas.192559699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.T., Brummelkamp T.R., Westerhout E.M., Vink M., Madiredjo M., Bernards R., Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H., Ketzinel-Gilad M., Rivkin L., Felig Y., Nussbaum O., Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Gitlin L., Stone J.K., Andino R. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Guo H., Zhang L., Xie H., Zhao X., Wang F., Li Z., Wang Y., Ma S., Tao J., Wang W., Zhou Y., Yang W., Chen J. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J. Virol. 2005;79:14392–14403. doi: 10.1128/JVI.79.22.14392-14403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot P.C., Cupac D., Berkhout B. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 2003;10:607–616. doi: 10.1159/000073526. [DOI] [PubMed] [Google Scholar]
- Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ireland J.H., O’Donnell B., Basuni A.A., Kean J.D., Wallace L.A., Lau G.K., Carman W.F. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology. 2000;31:1176–1182. doi: 10.1053/he.2000.6407. [DOI] [PubMed] [Google Scholar]
- Kim S.I., Shin D., Choi T.H., Lee J.C., Cheon G.J., Kim K.Y., Park M., Kim M. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol. Ther. 2007;15(6):1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., Yaspo M.L., Himmelbauer H., Korn B., Neugebauer K., Pisabarro M.T., Buchholz F. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Klein C., Bock C.T., Wedemeyer H., Wüstefeld T., Locarnini S., Dienes H.P., Kubicka S., Manns M.P., Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- Konishi M., Wu C.H., Wu G.Y. Inhibition of HBV replication by siRNA in a stable HBV-producing cell line. Hepatology. 2003;38:842–850. doi: 10.1053/jhep.2003.50416. [DOI] [PubMed] [Google Scholar]
- Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C., Zhong N., Lu P.Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Funsch B., Wiese M., Roggendorf M. Analysis of hepatitis C virus quasispecies populations by temperature gradient gel electrophoresis. J. Gen. Virol. 1995;76:881–887. doi: 10.1099/0022-1317-76-4-881. [DOI] [PubMed] [Google Scholar]
- Marcellin P. Advances in therapy for chronic hepatitis B. Semin. Liver Dis. 2002;22:33–36. doi: 10.1055/s-2002-35698. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V., Blanchard K., Shaw L., Jensen K., Lockridge J.A., Dickinson B., McSwiggen J.A., Vargeese C., Bowman K., Shaffer C.S., Polisky B.A., Zinnen S. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., Vaish N., Zinnen S., Vargeese C., Bowman K., Shaffer C.S., Jeffs L.B., Judge A., MacLachlan I., Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Randall G., Rice C.M. Interfering with hepatitis C virus RNA replication. Virus Res. 2004;102:19–25. doi: 10.1016/j.virusres.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Schaefer S. Hepatitis B virus: significance of genotypes. J. Viral Hepat. 2005;12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells M.A., Chen M.L., Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. U.S.A. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A., Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37(4):764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Sterneck M., Kalinina T., Günther S., Fischer L., Santantonio T., Greten H., Will H. Functional analysis of HBV genomes from patients with fulminant hepatitis. Hepatology. 1998;28:1390–1397. doi: 10.1002/hep.510280530. [DOI] [PubMed] [Google Scholar]
- Stevenson M. Dissecting HIV-1 through RNA interference. Nat. Rev. Immunol. 2003;3:851–858. doi: 10.1038/nri1227. [DOI] [PubMed] [Google Scholar]
- Uprichard S.L., Boyd B., Althage A., Chisari F.V. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc. Natl. Acad. Sci. U.S.A. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg M.S., Ely A., Barichievy S., Crowther C., Mufamadi S., Carmona S., Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol. Ther. 2007;15(3):534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Richardson C.D. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J. Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.L., Huang L.R., Huang C.C., Lai H.L., Liu C.J., Huang Y.T., Hsu Y.W., Lu C.Y., Chen D.S., Chen P.J. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708–716. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Nandamuri K.M. Inhibition of hepatitis viral replication by siRNA. Expert Opin. Biol. Ther. 2004;4:1649–1659. doi: 10.1517/14712598.4.10.1649. [DOI] [PubMed] [Google Scholar]
- Xuan B., Qian Z., Hong J., Huang W. EsiRNAs inhibit hepatitis B virus replication in mice model more efficiently than synthesized siRNAs. Virus Res. 2006;118:150–155. doi: 10.1016/j.virusres.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Yang D., Buchholz F., Huang Z., Goga A., Chen C.Y., Brodsky F.M., Bishop J.M. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Goga A., Bishop J.M. RNA interference (RNAi) with RNase III-prepared siRNAs. Methods Mol. Biol. 2004;252:471–482. doi: 10.1385/1-59259-746-7:471. [DOI] [PubMed] [Google Scholar]
- Yang H., Westland C., Xiong S., Delaney W.E., IV In vitro antiviral susceptibility of full-length clinical hepatitis B virus isolates cloned with a novel expression vector. Antiviral Res. 2004;61:27–36. doi: 10.1016/j.antiviral.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Zhu C., Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


