Highlights
► We detected antibodies to HCoV-OC43 NP in over 90% of tested cord blood samples. ► Three patterns for the immunoreactivities of the structural regions of HCoV-OC43 NP. ► The central-linker region of the NP showed highly immunoreactive in human serum.
Abbreviations: RNP, ribonucleoprotein; HCoV-OC43, human coronavirus OC43; NP, nucleocapsid protein; IPTG, β-d-1-thiogalactopyranoside; S, spike; M, matrix; E, envelope; SPR, surface plasmon resonance
Keywords: Human coronavirus, OC43 strain, Nucleocapsid protein, Antigenicity, Polyclonal antibody, Virus infection
Abstract
Previous studies have reported that a prokaryotic-expressed recombinant nucleocapsid protein (NP) is a suitable reagent for the epidemiological screening of coronavirus infection. In this study, soluble recombinant human coronavirus OC43 (HCoV-OC43) NP was produced to examine the antigenicity of the HCoV-OC43 NP of betacoronavirus. Using the purified recombinant NP as an antigen, a polyclonal antibody from rabbit serum with specificity for HCoV-OC43 NP was generated; this antibody reacts specifically with HCoV-OC43 NP and does not cross-react with other human CoV NPs (including those of SARS-CoV and HCoV-229E) by Western blot. Sera from 26 young adults, 17 middle-aged and elderly patients with respiratory infection, and 15 cord blood samples were also tested. Strong reactivity to the NPs of HCoV-OC43 was observed in 96%, 82%, and 93% of the serum samples from the young adults, respiratory patients, and cord blood samples, respectively. To identify the immunoreactivities of the three structural regions of the NP that are recognised by the rabbit polyclonal antibody and human serum, the antigenicities of three protein fragments, including the N-terminal domain (aa 1-173), the central-linker region (aa 174-300), and the C-terminal domain (aa 301-448), were evaluated by Western blot. The rabbit polyclonal antibody demonstrated greater immunoreactivity to the central-linker region and the C-terminal domain than to the N-terminal domain. Three different patterns for the immunoreactivities of the three structural regions of HCoV-OC43 NP were observed in human serum, suggesting variability in the immune responses that occur during HCoV-OC43 infection in humans. The central-linker region of the NP appeared to be the most highly immunoreactive region for all three patterns observed. The goal of this study was to offer insight into the design of diagnostic tools for HCoV infection.
1. Introduction
HCoV-OC43 was identified in the 1960s and is responsible for the majority of “common colds” in humans (St-Jean et al., 2004, Vabret et al., 2003). Although HCoV-OC43 infections are generally mild, more severe upper and lower respiratory tract infections such as bronchiolitis and pneumonia, which are particularly severe in infants, elderly individuals, and immunocompromised patients, have been documented (El-Sahly et al., 2000, Gagneur et al., 2002, St-Jean et al., 2004). There have also been reports of clusters of HCoV-OC43 infections that cause pneumonia in adults (Vabret et al., 2003, Wenzel et al., 1974). In addition, a previous study has reported that the neurotropism and neuroinvasion of HCoV are associated with multiple sclerosis (Arbour et al., 2000). In recent years, several emerging human coronaviruses have been discovered (Skowronski et al., 2005, Vabret et al., 2005, Vabret et al., 2006), and between 2003 and 2004, the SARS-CoV outbreak caused a worldwide epidemic that had a significant economic impact in countries where the disease outbreak occurred (Skowronski et al., 2005). Phylogenetic analyses have shown that SARS-CoV contains sequences that are closely related to sequences found in the betacoronaviruses. In 2004, another alphacoronavirus, HCoV-NL63, which was isolated from a 7-month old child suffering from bronchiolitis and conjunctivitis, was reported in the Netherlands (Vabret et al., 2005). Woo et al. (2005) described a novel betacoronavirus, HKU1, which was found in patients with respiratory tract infections (Woo et al., 2005).
The RNA genomes of coronaviruses include the genes encoding the structural proteins S (spike), M (matrix), E (envelope), and N (nucleocapsid). Additionally, some coronaviruses encode a third glycoprotein, HE (hemagglutinin-esterase), which is present in most of the betacoronaviruses (Lai and Cavanagh, 1997). The primary function of CoV NP is to recognise a stretch of RNA that serves as a packaging signal, leading to the formation of the ribonucleoprotein (RNP) complex during viral assembly (Huang et al., 2004, Lai, 2003, Navas-Martin and Weiss, 2004, Nelson et al., 2000). Previous studies have shown that the NPs are the immunodominant domain in hosts infected with several coronaviruses (Chan et al., 2005, Che et al., 2005, Lau et al., 2004). Additionally, it has been shown that NPs can accumulate intracellularly before being packaged in mature viruses (Garoff et al., 1998). Together, these characteristics make the NP a suitable candidate for early diagnosis of coronavirus infection (Chan et al., 2005, Mourez et al., 2007).
In this study, a purified soluble full-length HCoV-OC43 NP was produced and characterised using highly specific rabbit polyclonal antibody. Sera from young healthy adults, respiratory infection patients, and cord blood samples were also analysed using Western blot assays, using the purified recombinant NP as an antigen. To identify the immunodominant regions of the HCoV-OC43 NP, the antigenicities of three structural regions, aa 1-173, aa 174-300, and aa 301-448, were analysed. These results, which defined the immunoreactivity patterns of the three structural regions recognised by the rabbit polyclonal antibody and human serum, may foster the development of a diagnostic test for CoV infection.
2. Materials and methods
Drugs and reagents were purchased from Sigma (St. Louis, MO, USA). All oligoribonucleotides (or oligodeoxyribonucleotides) were synthesised using an automated DNA synthesiser and purified by gel electrophoresis.
2.1. Expression and purification of recombinant NPs
The templates for the HCoV-OC43 NP were kindly provided by the Institute of Biological Chemistry, Academia Sinica (Taipei, Taiwan). To generate recombinant NPs, the NP gene was amplified by the polymerase chain reaction (PCR) from the plasmid pGENT using various primers. The PCR products were digested with NdeI and XhoI, and the DNA fragments were cloned into pET28a. The recombinant plasmid was transformed into Escherichia coli BL21-RIL cells using the heat-shock method, and the cells were grown at 37 °C in Luria-Bertani medium. Protein expression was induced at an OD600 of 0.6 by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 10 °C for 24 h. After harvesting the bacteria by centrifugation (3500 × g, 30 min, 4 °C), the bacterial pellets were lysed with lysis buffer (50 mM Tris-buffered solution, pH 7.3, 150 mM NaCl, and 15 mM imidazole). The soluble proteins were collected from the supernatant following centrifugation (27,200 × g, 30 min, 4 °C) to remove the precipitate. The methods for protein purification have been described previously (Chen et al., 2010, Hung et al., 2012). Full-length and truncated HCoV-OC43 NPs have a His6-tag at the N-terminus and the C-terminus, and the expressed proteins were purified using a Ni-NTA column (Novagen) with an elution gradient ranging from 15 to 300 mM imidazole. The pure fractions were collected and dialysed against a low-salt buffer. The protein concentrations were determined using the Bradford method with Bio-Rad protein assay reagents. The SARS-CoV and HCoV-229E NPs were expressed in E. coli and purified as described previously (Tang et al., 2005).
2.2. Immunisation of rabbits
Soluble and purified recombinant NPs prepared as described above were used as antigens for immunisation and the immunoassays. All animal procedures described were approved by the Animal Care Use Committee of National Chung Hsing University. Rabbits (New Zealand White strain) weighing approximately 3 kg were immunised by an intrasplenic injection with 400 μg of recombinant HCoV-OC43 NP per immunisation. The antigen was administered together with an equal amount of Gold TiterMax adjuvant (CytRx, Norcross, GA, USA). Additional booster immunisations were administered to obtain a high titre of the antisera, and high-titre polyclonal antibodies were obtained in 6–8 weeks. The rabbit antisera were used without purification for the majority of the subsequent experiments. The titre of rabbit sera was determined for the NP antigen by Western blot.
2.3. Serum collections
Twenty-six human serum specimens, denoted H1 to H26, from young healthy adults approximately 18–26 years of age were collected at random from Shih-Chien University. Seventeen serum samples, denoted R1 to R17, were collected from patients that were approximately 50-80 years of age who had reported to the Emergency Department of the Medicine Chang Gung Memorial hospital with symptoms of respiratory tract infections. Fifteen cord blood samples, denoted C1 to C15, were collected from the Department of Obstetrics and Gynaecology, Chung Shan Medical University. The criteria used for the selection of human serum samples from the middle-aged and elderly patients with respiratory infections were largely based on clinical symptoms. No symptoms indicative of respiratory infection were noted by physicians in the young healthy adults or the newborns from when cord blood was obtained. All sera were stored at −20 °C. All of the human serum collections were approved by Institutional Review Boards.
2.4. Western blot immunoassay using the rabbit polyclonal antibody
The NPs were solubilised in sample buffer containing 50 mM Tris–HCl, pH 7.5, 0.1% CHAPS, 10% glycerol, 150 mM NaCl and 10 mM dithiothreitol and boiled for 5 min. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a discontinuous buffer system, and the polypeptide bands were revealed by staining the gel with Coomassie Brilliant Blue G-250. For immunoblotting, polypeptides separated by SDS-PAGE were electrotransferred onto a PVDF membrane with transfer buffer containing 50 mM Tris, 384 mM glycine, and 20% (v/v) methanol, pH 8.3. Electrotransfer was conducted at 65 V for 1 h. The PVDF membrane was then incubated for 1 h in PBS buffer containing 0.05% Tween 20 (PBS-T). After washing three times in PBS-T, the membrane was incubated for 2 h at room temperature with rabbit antisera. The three washes were followed by the addition of horseradish peroxidase-conjugated goat anti-rabbit IgG at a dilution of 1:10,000. After a 2-h incubation at room temperature, the membrane was washed three times and covered with the peroxidase substrate 3,3′-diaminobenzidine (DAB). The blot was allowed to develop, and the reaction was stopped by washing the membrane in distilled water. Quantitation of the protein bands on the gel was achieved using the Uniphoto Band Tool software.
2.5. Western blot immunoassay using human serum
An identical protocol to that described above was used to test human sera by Western blotting. Each well contained 50 ng of recombinant NP, and human sera were diluted 200-fold in blocking solution. The secondary antibody, horseradish peroxidase-conjugated goat anti-human antibody IgG + IgM, was used and detected as described above.
3. Results
3.1. Purification of recombinant HCoV-OC43 NP
To express soluble HCoV-OC43 NP as a set of fusion proteins in E. coli, the NP gene was cloned into the pET-28a expression vector. CHAPS, which demonstrates low light absorbance in the ultraviolet region of the electromagnetic spectrum, facilitates the characterisation of protein properties when using CD and UV–VIS spectra. To solve the problem of the solubility of the NP, buffers containing CHAPS were used to obtain NP-His6 in a soluble form for subsequent analysis. Fig. 1A shows that NP-His6 was soluble in Tris-buffer in the presence of 0.1, 0.2, and 0.4% CHAPS. Large-scale expression and purification of the His-tag NP in the presence of 0.1% CHAPS from E. coli was then performed. Fig. 1B shows that the His-tag NP was purified from the soluble fraction using Ni-NTA column chromatography, resulting in a single band of the expected mass of 55 kDa. The final yield of NP-His6 was approximately 10 mg/l from the E. coli culture. The NP is responsible for recognising a stretch of RNA that serves as a viral packaging signal, leading to the formation of the ribonucleoprotein (RNP) complex during viral assembly (Lai and Cavanagh, 1997). Previous surface plasmon resonance (SPR) data showed that purified recombinant HCoV-OC43 NP can bind single-stranded RNA, suggesting that recombinant HCoV-OC43 NP is folded properly (Huang et al., 2009).
Fig. 1.
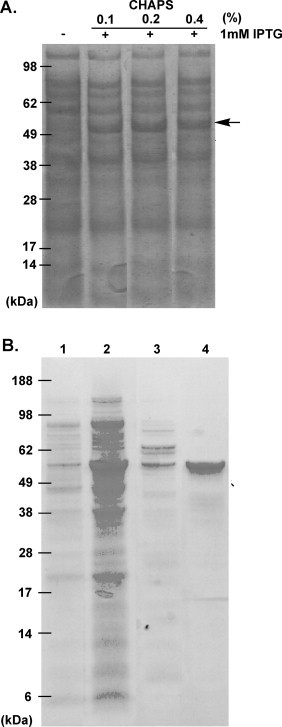
(A) Soluble protein fraction of recombinant HCoV-OC43 NP from IPTG-induced cells prepared using different concentrations of CHAPS in 50 mM Tris–HCl (pH 7.3) and 150 mM NaCl buffer. (B) Expression and purification of OC43 NP. Extract of un-induced cells (lane 1), extract of induced cells (lane 2), protein eluted with 15 mM imidazole (lane 3), and purified NP (lane 4).
3.2. Production and antigenicity analysis of rabbit polyclonal antibodies against HCoV-OC43 NP
Rabbits were immunised with soluble recombinant full-length HCoV-OC43 NP to produce a high-quality polyclonal antibody. Following three booster immunisations, the titre of antibody against full-length HCoV-OC43 NP in the rabbit serum was analysed using Western blotting. Western blot data showed that 50 ng NP were clearly detected at dilutions of at least 1:1,900,000 (Fig. 2A). The specificity and cross-reactivity of the rabbit polyclonal antibody against HCoV-229E and SARS-CoV NPs were also determined by Western blot analysis. Despite the presence of several conserved motifs in the NPs from these coronaviruses, the specificity of the anti-HCoV-OC43 NP polyclonal antibody was excellent, and the antibody did not cross-react with other human CoV NPs, including SARS-CoV and HCoV-229E at 1:150,000 dilutions (Fig. 2B). Previous studies have suggested that the NPs of CoVs share the same modular organisation (Fig. 3A) (Huang et al., 2009). The PONDR program, a predictor of naturally disordered regions, predicts three structural regions in HCoV-OC43 NP: aa 1-173 (the N-terminal domain), aa 174-300 (the central-linker region), and aa 301-448 (the C-terminal domain) (Romero et al., 2004) (Fig. 3A). To analyse the antigenicity of these three structural regions in the NP, fragments encoding aa 1-173, aa 174-300, and aa 301-448, which together span the entire coding region, were cloned, expressed, and successfully purified as single bands on SDS-PAGE gels (Fig. 3B). The results demonstrated that the rabbit polyclonal antibody reacted strongly with the regions aa 174-300 and aa 301-448, whereas the aa 1-173 region showed low reactivity with the antibody at dilutions of 1:50,000 and 1:450,000 (Fig. 3B–D). In Fig. 3C, a minor band slightly below the major band is apparent, indicating that aa 174-300 was partially degraded.
Fig. 2.
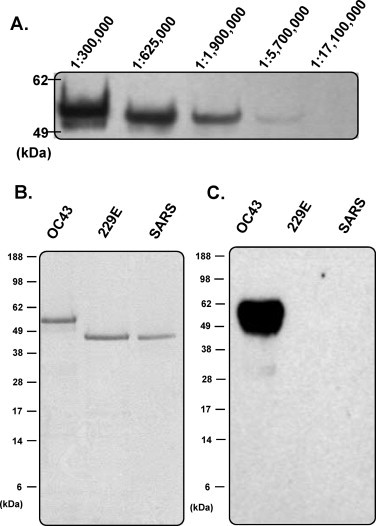
(A) Titre analysis of the rabbit polyclonal antibody against HCoV-OC43 NP using Western blotting. (B) SDS-PAGE analysis of the NPs from HCoV-OC43, HCoV-229E, and SARS-CoV purified by Ni-NTA chromatography. (C) Specificity analysis of the rabbit polyclonal antibody against HCoV-OC43, HCoV-229E, and SARS-CoV NPs by Western blot.
Fig. 3.
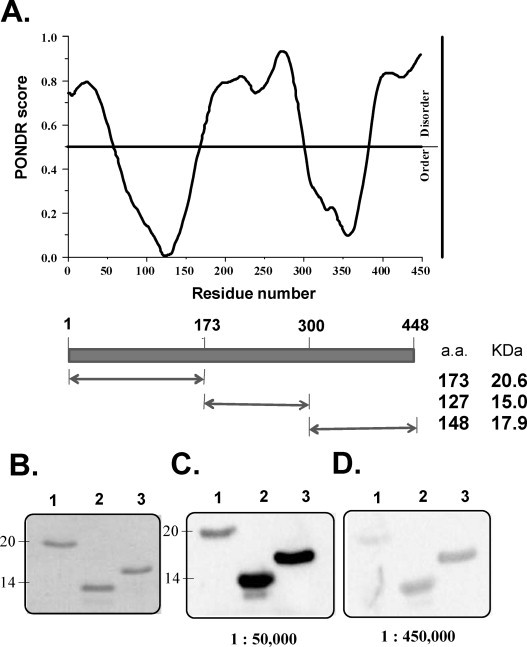
(A) Conformational analysis of HCoV-OC43 NP by the VL3-BA predictor of the PONDR program. (B) SDS-PAGE analysis of three truncated fragments of HCoV-OC43 NP, aa 1–173, aa 174–300, and aa 301–448, purified by Ni-NTA chromatography. Antigenicity analysis of three truncated fragments of HCoV-OC43 NP, aa 1–173, aa 174–300, and aa 301–448, against the rabbit polyclonal antibody at dilutions of (C) 1:50,000 and (D) 1:450,000.
3.3. Serological assay and antigenicity analysis of the NP antibody in humans
The recombinant protein-based Western blot assay was used to screen human serum from 26 young adults, 17 middle-aged and elderly patients with respiratory infection symptoms, and 15 cord blood samples (Fig. 4A). The serum samples from young adults, middle-aged and elderly patients with respiratory infection, and cord blood samples were 92.3%, 82.3%, and 93.3% seropositive for HCoV-OC43, respectively (Table 1 ). None of the samples reacted with the SARS-CoV NP. Interestingly, approximately 81% of the serum samples reacted to the HCoV-229E NP. Thirty-eight human sera that tested positive for HCoV-OC43 NP in the recombinant protein Western blot assay were chosen for further analysis in order to examine the antigenicity of the three following structural regions of NP: regions aa 1-173, aa 174-300, and aa 301-448. The different immunoreactivity patterns that were observed in human serum were classified into three types: I, II, and III. One case selected from each type is shown (Fig. 4B). In 16 of 38 persons that had Type I immunoreactivity, which is similar to the immunoreactivity of rabbit serum, the highly immunoreactive regions are located in the aa 174-300 and aa 301-448 regions (Table 1). For Type II immunoreactivities (11 people), the results showed that the aa 174-300 region was the most immunodominant region among the three structural regions of the NP. For Type III immunoreactivities (11 persons), all three of the NP regions demonstrated strong antigenicity.
Fig. 4.
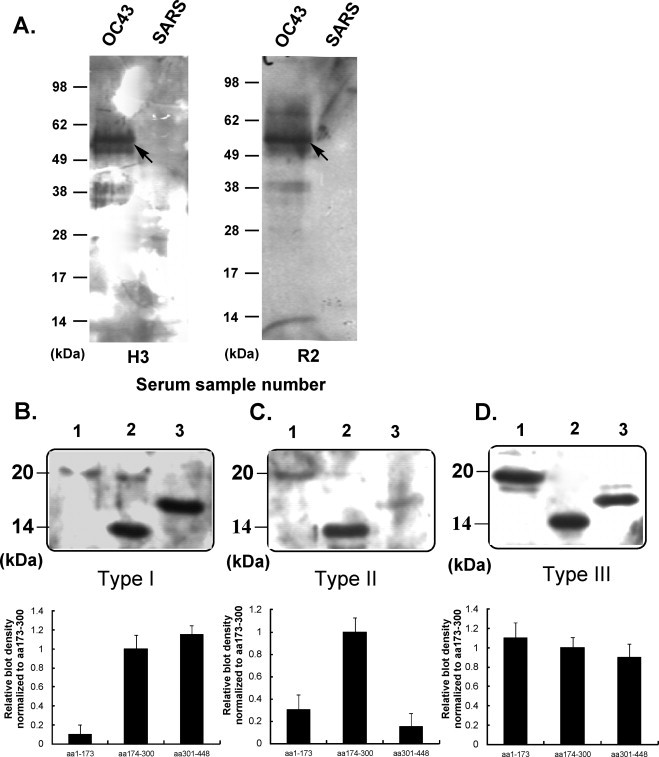
(A) Western blot analysis of the individual human sera obtained from young healthy adults and patients with respiratory infection symptoms tested against HCoV-OC43 and SARS-CoV NPs (denoted by H3 and R2, respectively) at an antibody dilution of 1:1000. (B) Type I, (C) Type II, and (D) Type III immunoreactivity patterns of three structural regions of HCoV-OC43 NP, aa 1–173, aa 174–300, and aa 301–448, against human antisera at a 1:1000 dilution determined by Western blotting (above). The bar diagram represents the relative density of each of the three truncated fragments, determined by immunoreactivity analysis for human sera; aa 174–300 was used as a control (below).
Table 1.
Screening of human sera for HCoV-OC43 NP antibodies.
| Group | Total number of individuals screened | HCoV-OC43 positive (%) | HCoV-229E positive (%) | SARS-CoV positive (%) | Immunoreactivity patterns for the three structural regions of HCoV-OC43 NP |
||
|---|---|---|---|---|---|---|---|
| Type I | Type II | Type III | |||||
| 18–25 years | 26 | 24 (92.3) | 21 (80.7) | 0 | 9 | 8 | 7 |
| 50–80 yearsa | 17 | 14 (82.3) | 13 (76.4) | 0 | 7 | 3 | 4 |
| Cord blood | 15 | 14 (93.3)c | 13 (86.6) | 0 | –b | – | – |
| Total | 58 | 52 (89.7) | 47 (81.0) | 0 | 16 | 11 | 11 |
These specimens were taken from patients with respiratory tract infection symptoms.
Not determined, due to the limited quantities of cord blood collected per sample.
Sera from cord blood samples were analysed using purified NP as an antigen.
Because the central-linker region of the HCoV-OC43 NP was the most highly immunoreactive region in human serum in these experiments, the central-linker region was used as an antigen to test the antibody titre in the serum of young adults and middle-aged and elderly patients with respiratory infection symptoms. To titrate the sera, two-fold dilutions of serum in the range of 1:2000–1:64,000 were examined. All titres were in the range of 1:8000–1:64,000, and most of the sera displayed titres of 1:32,000, showing no significant differences in antibody titres between young healthy adults and patients with respiratory infection symptoms (Figure S1).
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jviromet.2012.11.009.
Titers of antibodies specific for the central-linker region of HCoV-OC43 NP in serum samples from 24 young healthy adults (H) or 14 middle-aged and elderly patients with respiratory infection symptoms (R).
4. Discussion
Coronaviruses cause colds of mild to moderate severity and are transmitted by aerosols of respiratory secretions, the faecal-oral route, and mechanical transmission (Cavanagh and Brown, 1990). The most common symptoms of coronavirus infection are nasal catarrh and a sore throat, and the illness typically lasts approximately 6–8 days (Kaye et al., 1971). The early diagnosis of common colds caused by coronavirus is an important step in preventing the recurrence of a global outbreak (Carattoli et al., 2005, Das et al., 2010, Wang et al., 2004). HCoVs are difficult to detect, and the current diagnosis of coronaviral infection is based on reverse-transcription polymerase chain reaction (RT-PCR) (Huang et al., 2005, Ng and Lo, 2006). The NP is involved in the pathological reaction to human coronavirus and is a key antigen for the development of a sensitive diagnostic assay (Di et al., 2005, He et al., 2005). Che et al. (2004) reported that NP in the serum samples of SARS patients can be detected as early as 1 day after disease onset.
Prokaryotic-expressed NPs have been used successfully as antigens in ELISA-based assays for the detection of antibodies specific to many viruses, including SARS-CoV and several animal coronaviruses (Blanchard et al., 2011, Loa et al., 2004, Timani et al., 2004, Wu et al., 2004). In the current study, large quantities of soluble recombinant HCoV-OC43 NP from E. coli were expressed and purified in the presence of 0.1% CHAPS. The yield from 1 l of bacterial culture was as much as 10 mg of pure NP after extraction and column chromatography. The ability of the recombinant HCoV-OC43 protein to bind the intergenic sequence of the RNA genomic leader by SPR was examined (Huang et al., 2009). These results, which showed efficient binding of HCoV-OC43 to the RNA, suggest that the purified recombinant NP was folded properly. The prokaryotic expression system used in this study is high-yield, inexpensive, and highly efficient, does not require viral cultures, and is nontoxic. Despite these advantages, viral proteins expressed in prokaryotic cells lack post-translational modifications that are present in proteins expressed in baculovirus expression systems (Mourez et al., 2007).
The production of the polyclonal antibody against coronavirus NP generated in this study could be used to develop a rapid, easy, and specific diagnostic tool for the detection of HCoV-OC43 infections by immunofluorescence or ELISA-based tests (Carattoli et al., 2005). Whereas cross-reactivity between the SARS-CoV NP and the anti-HCoV-229E antibody or the anti-HCoV-OC43 antibody has been reported (Che et al., 2005), high-titre anti-HCoV-OC43 NP polyclonal antibody did not cross-react with other human CoV NPs, including those of SARS-CoV and HCoV-229E, despite the presence of highly conserved motifs in these coronavirus NPs. Previous studies also showed that the anti-SARS CoV NP and anti-HCoV-229E NP polyclonal antibodies did not cross-react with other human CoV NPs (Che et al., 2005, Lee et al., 2010, Lehmann et al., 2008).
HCoV-OC43 is distributed worldwide (Patrick et al., 2006). Indeed, the results presented in this study showed that approximately 90% of serum samples from young adults and middle-aged and elderly patients with respiratory infections reacted strongly to the HCoV-OC43 NPs, indicating prior exposure to this disease. This finding is consistent with previous epidemiological surveys that concluded that seroprevalence increases rapidly during childhood, attaining a seroprevalence rate of up to 90% in adults (Lehmann et al., 2008, Mourez et al., 2007). Additionally, antibodies against HCoV-OC43 NP were detected in over 90% of cord blood samples tested. Maternally acquired antibodies may help protect a newborn baby from HCoV-OC43 infection, although this protection appears to wane by 4–5 months of age. In addition, 81% of serum samples tested in this study were seropositive for the HCo-229E NP. The reactivity of the serum samples to the HCoV-229E NP is most likely due to a prior infection with HCoV-229E or the cross-reactivity of the serum to the conserved region within the NPs (Che et al., 2005). In 2004, a new coronavirus, HCoV-NL63, belonging to the betacoronaviruses, was identified (Pyrc et al., 2004, Vabret et al., 2005). HCoV-NL63 is a coronavirus that is also identified frequently in respiratory tract infections and is reported most frequently during the winter season. The epidemiology and characteristics of infection for this virus do not differ significantly from those described for HCoV-OC43. Cross-reactivity between the OC43 and NL63 HCoVs could not be excluded in the current study, due to the lack of a modified, culturable strain in our laboratory.
CoV NPs contain multiple immunodominant epitopes and antigenic sites (Yu et al., 2008, Zhao et al., 2007). To compare the immunoreactivity of the three structural regions of HCoV-OC43 NP, three truncated recombinant fragments (aa 1-173 (the N-terminal domain), aa 174-300 (the central region), and aa 301-448 (the C-terminal domain)), were produced in E. coli; these regions were chosen based on PONDR predictions. The reactivities of a specific rabbit polyclonal antibody and human serum against these fragments were determined by Western blotting. It is commonly accepted that Western blot is the most effective method for unequivocally locating linear or continuous immunodominant epitopes (Mourez et al., 2007, Pohl-Koppe et al., 1995). The rabbit polyclonal antibody generated in this study reacted strongly with the central region and the C-terminal domain of the NP, whereas the N-terminal domain demonstrated low reactivity with the antibody. According to protein sequence alignment, the NP of HCoV-OC43 contains 27% to 30% sequence identity with various other animal coronaviruses. To minimise possible cross-reactions between the NPs of HCoV-OC43 and other human coronaviruses, a minimally derived region encoding the NP that could be detected efficiently by all human sera was sought. Three types of immunoreactivity patterns had been found in human sera, suggesting that there was variability in the immune responses occurring in humans during HCoV-OC43 infection. In the Type I immunoreactivity pattern, which had a similar reactivity as the rabbit serum, the immunodominant region was located in the aa 174-300 and aa 301-448 regions. This result was consistent with previous studies conducted with sera from SARS patients, which revealed that the immunodominant regions were located in the middle and C-terminal regions of the SARS-CoV NP (Lee et al., 2008, Yu et al., 2008). In the Type II immunoreactivity pattern, the immunodominant region was located in the aa 174-300 region, while in the Type III immunoreactivity pattern, all of the three structural regions showed strong antigenicity. According to previous circular dichroism and SPR studies, the central region is necessary for the disorder conformation and has been shown to have RNA-binding activity (Huang et al., 2009).
In conclusion, prokaryotic expression of recombinant HCoV-OC43 NP is a suitable reagent for high-sensitivity, highly specific antibody production and can be used for epidemiological screening of HCoV-OC43 infection. The strong immunogenicity of the central region of HCoV-OC43 NP suggests that this protein could serve as an HCoV-OC43-specific antigen that could be used for the diagnosis of CoV infection.
Acknowledgement
This work was supported by NSC grant 97-2311-B-005-003-MY3 to M.-H. Hou.
References
- Arbour N., Day R., Newcombe J., Talbot P.J. Neuroinvasion by human respiratory coronaviruses. J. Virol. 2000;74:8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E.G., Miao C., Haupt T.E., Anderson L.J., Haynes L.M. Development of a recombinant truncated nucleocapsid protein based immunoassay for detection of antibodies against human coronavirus OC43. J. Virol. Methods. 2011;177:100–106. doi: 10.1016/j.jviromet.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Di Bonito P., Grasso F., Giorgi C., Blasi F., Niedrig M., Cassone A. Recombinant protein-based ELISA and immuno-cytochemical assay for the diagnosis of SARS. J. Med. Virol. 2005;76:137–142. doi: 10.1002/jmv.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Brown T.D.K. Plenum Press; New York: 1990. Coronaviruses and their Diseases. [Google Scholar]
- Chan P.K., Liu E.Y., Leung D.T., Cheung J.L., Ma C.H., Tam F.C., Hui M., Tam J.S., Lim P.L. Evaluation of a recombinant nucleocapsid protein-based assay for anti-SARS-CoV IgG detection. J. Med. Virol. 2005;75:181–184. doi: 10.1002/jmv.20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Hao W., Wang Y., Di B., Yin K., Xu Y.C., Feng C.S., Wan Z.Y., Cheng V.C., Yuen K.Y. Nucleocapsid protein as early diagnostic marker for SARS. Emerg. Infect. Dis. 2004;10:1947–1949. doi: 10.3201/eid1011.040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Qiu L.W., Liao Z.Y., Wang Y.D., Wen K., Pan Y.X., Hao W., Mei Y.B., Cheng V.C., Yuen K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.J., Chou C.C., Liu C.L., Lee C.C., Kan L.S., Hou M.H. Crystallization and preliminary X-ray diffraction analysis of the N-terminal domain of human coronavirus OC43 nucleocapsid protein. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2010;66:815–818. doi: 10.1107/S1744309110017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Kammila S., Suresh M.R. Development, characterization, and application of monoclonal antibodies against severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Vaccine Immunol.: CVI. 2010;17:2033–2036. doi: 10.1128/CVI.00293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di B., Hao W., Gao Y., Wang M., Wang Y.D., Qiu L.W., Wen K., Zhou D.H., Wu X.W., Lu E.J., Liao Z.Y., Mei Y.B., Zheng B.J., Che X.Y. Monoclonal antibody-based antigen capture enzyme-linked immunosorbent assay reveals high sensitivity of the nucleocapsid protein in acute-phase sera of severe acute respiratory syndrome patients. Clin. Diagn. Lab. Immunol. 2005;12:135–140. doi: 10.1128/CDLI.12.1.135-140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sahly H.M., Atmar R.L., Glezen W.P., Greenberg S.B. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 2000;31:96–100. doi: 10.1086/313937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneur A., Sizun J., Vallet S., Legr M.C., Picard B., Talbot P.J. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J. Hosp. Infect. 2002;51:59–64. doi: 10.1053/jhin.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Hewson R., Opstelten D.J. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Du Q., Lau S., Manopo I., Lu L., Chan S.W., Fenner B.J., Kwang J. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods. 2005;127:46–53. doi: 10.1016/j.jviromet.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Hsu Y.L., Chiang W.L., Hou M.H. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci. 2009;18:2209–2218. doi: 10.1002/pro.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.L., Lin H.T., Wang Y.M., Yeh Y.C., Peck K., Lin B.L., Liu H.W., Chen A., Lin C.S. Rapid and sensitive detection of multiple genes from the SARS-coronavirus using quantitative RT-PCR with dual systems. J. Med. Virol. 2005;77:151–158. doi: 10.1002/jmv.20432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Hung H.C., Liu C.L., Hsu J.T., Horng J.T., Fang M.Y., Wu S.Y., Ueng S.H., Wang M.Y., Yaw C.W., Hou M.H. Development of an anti-influenza drug screening assay targeting nucleoproteins with tryptophan fluorescence quenching. Anal. Chem. 2012;84(84):6391–6399. doi: 10.1021/ac2022426. [DOI] [PubMed] [Google Scholar]
- Kaye H.S., Marsh H.B., Dowdle W.R. Seroepidemiologic survey of coronavirus (strain OC43) related infections in a children's population. Am. J. Epidemiol. 1971;94:43–49. doi: 10.1093/oxfordjournals.aje.a121293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. SARS virus: the beginning of the unraveling of a new coronavirus. J. Biomed. Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Wong B.H., Tsoi H.W., Woo G.K., Poon R.W., Chan K.H., Wei W.I., Peiris J.S., Yuen K.Y. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in sars patients by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2004;42:2884–2889. doi: 10.1128/JCM.42.7.2884-2889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lee B.H., Dutta N.K., Seok S.H., Baek M.W., Lee H.Y., Kim D.J., Na Y.R., Noh K.J., Park S.H., Kariwa H., Nakauchi M., Mai le Q., Heo S.J., Park J.H. Detection of antibodies against SARS-Coronavirus using recombinant truncated nucleocapsid proteins by ELISA. J. Microbiol. Biotechnol. 2008;18:1717–1721. [PubMed] [Google Scholar]
- Lee H.K., Lee B.H., Seok S.H., Baek M.W., Lee H.Y., Kim D.J., Na Y.R., Noh K.J., Park S.H., Kumar D.N., Kariwa H., Nakauchi M., Heo S.J., Park J.H. Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J. Vet. Sci. 2010;11:165–167. doi: 10.4142/jvs.2010.11.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann C., Wolf H., Xu J., Zhao Q., Shao Y., Motz M., Lindner P. A line immunoassay utilizing recombinant nucleocapsid proteins for detection of antibodies to human coronaviruses. Diagn. Microbiol. Infect. Dis. 2008;61:40–48. doi: 10.1016/j.diagmicrobio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Hooper T., Schrader D. Expression and purification of turkey coronavirus nucleocapsid protein in Escherichia coli. J. Virol. Methods. 2004;116:161–167. doi: 10.1016/j.jviromet.2003.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourez T., Vabret A., Han Y., Dina J., Legrand L., Corbet S., Freymuth F. Baculovirus expression of HCoV-OC43 nucleocapsid protein and development of a Western blot assay for detection of human antibodies against HCoV-OC43. J. Virol. Methods. 2007;139:175–180. doi: 10.1016/j.jviromet.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Martin S.R., Weiss S. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J. Neurovirol. 2004;10:75–85. doi: 10.1080/13550280490280292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- Ng E.K., Lo Y.M. Molecular diagnosis of severe acute respiratory syndrome. Methods Mol. Biol. 2006;336:163–175. doi: 10.1385/1-59745-074-X:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect. Dis. Med. Microbiol. 2006;17:330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl-Koppe A., Raabe T., Siddell S.G., ter Meulen V. Detection of human coronavirus 229E-specific antibodies using recombinant fusion proteins. J. Virol. Methods. 1995;55:175–183. doi: 10.1016/0166-0934(95)00041-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Jebbink M.F., Berkhout B., van der Hoek L. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Dunker A.K. Natively disordered proteins: functions and predictions. Appl. Bioinformatics. 2004;3:105–113. doi: 10.2165/00822942-200403020-00005. [DOI] [PubMed] [Google Scholar]
- Skowronski D.M., Astell C., Brunham R.C., Low D.E., Petric M., Roper R.L., Talbot P.J., Tam T., Babiuk L. Severe acute respiratory syndrome (SARS): a year in review. Annu. Rev. Med. 2005;56:357–381. doi: 10.1146/annurev.med.56.091103.134135. [DOI] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.K., Wu M.P., Chen S.T., Hou M.H., Hong M.H., Pan F.M., Yu H.M., Chen J.H., Yao C.W., Wang A.H. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani K.A., Ye L., Zhu Y., Wu Z., Gong Z. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 2004;30:309–312. doi: 10.1016/j.jcv.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Dina J., Gouarin S., Petitjean J., Corbet S., Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 2006;42:634–639. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Dina J., van der Hoek L., Gouarin S., Petitjean J., Brouard J., Freymuth F. Human coronavirus NL63, France. Emerg. Infect. Dis. 2005;11:1225–1229. doi: 10.3201/eid1108.050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.K., Chen S.Y., Liu I.J., Chen Y.C., Chen H.L., Yang C.F., Chen P.J., Yeh S.H., Kao C.L., Huang L.M., Hsueh P.R., Wang J.T., Sheng W.H., Fang C.T., Hung C.C., Hsieh S.M., Su C.P., Chiang W.C., Yang J.Y., Lin J.H., Hsieh S.C., Hu H.P., Chiang Y.P., Yang P.C., Chang S.C. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg. Infect. Dis. 2004;10:1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel R.P., Hendley J.O., Davies J.A., Gwaltney J.M., Jr. Coronavirus infections in military recruits. Three-year study with coronavirus strains OC43 and 229E. Am. Rev. Respir. Dis. 1974;109:621–624. doi: 10.1164/arrd.1974.109.6.621. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.S., Hsieh Y.C., Su I.J., Lin T.H., Chiu S.C., Hsu Y.F., Lin J.H., Wang M.C., Chen J.Y., Hsiao P.W., Chang G.D., Wang A.H., Ting H.W., Chou C.M., Huang C.J. Early detection of antibodies against various structural proteins of the SARS-associated coronavirus in SARS patients. J. Biomed. Sci. 2004;11:117–126. doi: 10.1007/BF02256554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Stevens V., Berry J.D., Crameri G., McEachern J., Tu C., Shi Z., Liang G., Weingartl H., Cardosa J., Eaton B.T., Wang L.F. Determination and application of immunodominant regions of SARS coronavirus spike and nucleocapsid proteins recognized by sera from different animal species. J. Immunol. Methods. 2008;331:1–12. doi: 10.1016/j.jim.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Titers of antibodies specific for the central-linker region of HCoV-OC43 NP in serum samples from 24 young healthy adults (H) or 14 middle-aged and elderly patients with respiratory infection symptoms (R).


