Abstract
Toroviruses (ToVs) are a group of emerging viruses that cause gastroenteritis in domestic animals and humans. Currently, methods such as real-time reverse transcription-polymerase chain reaction (real-time RT-PCR) have not yet been developed for the rapid detection and quantitation of bovine (BToV) and porcine (PToV) toroviruses. Using BToV and PToV RNA standards generated by in vitro transcription, the detection limit of the SYBR Green real-time RT-PCR assay was 2.54 × 102 BToV and 2.17 × 103 PToV copies/reaction (correlation coefficiency = 0.99 and 0.97, respectively), whereas those of RT-PCR and nested PCR were 2.54 × 105 and 2.54 × 104 (BToV) and 2.17 × 107 and 2.17 × 105 (PToV) cRNA viral copies/reaction, respectively. Archived diarrhea specimens of calves (n = 121) and piglets (n = 86) were subjected to RT-PCR, nested PCR and SYBR Green real-time RT-PCR. By conventional RT-PCR, 1 (0.8%) bovine and 7 (8.1%) porcine samples tested positive to BToV and PToV, respectively. With nested PCR, 13 (10.7%) bovine and 17 (19.8%) porcine samples tested positive. SYBR Green real-time RT-PCR assay detected BToV and PToV in 22 of 121 (18.2%) bovine and 31 of 86 (36.0%) porcine samples. These results indicate that SYBR Green real-time RT-PCR (P < 0.05) is a more sensitive assay, which can be reproduced as a reliable, sensitive, and rapid tool for the detection and quantitation of toroviruses.
Keywords: BToV, PToV, SYBR Green real-time RT-PCR, Detection, Quantitation
1. Introduction
Toroviruses (ToVs) are enveloped, positive-stranded polyadenylated RNA viruses and are classified as the second genus in the family of Coronaviridae, order Nidovirales that contains many important pathogens. Toroviruses infect both animals and humans and are associated predominantly with enteric diseases (Koopmans and Horzinek, 1994). The first report of ToVs described the identification of the Berne virus in 1972 from a rectal swab of a horse with diarrhea in Berne, Switzerland (Weiss et al., 1983). A decade later, a virus that was related morphologically and antigenically to the Berne virus, which was designated the Breda virus (BToV) and Breda virus-like particles, was described in the stools of neonatal calves and children (Beards et al., 1984, Woode et al., 1982). A porcine torovirus (PToV) has been identified and characterized in the feces of swine (Kroneman et al., 1998). Toroviruses have been established as infectious gastrointestinal agents in cattle and as a predominant cause of acute enteric infections in piglets (Ito et al., 2007, Kroneman et al., 1998). Significantly, zoonosis was possibly implicated when ToV-like particles detected in patients with gastroenteritis exhibited similar morphology and cross-reacted with BToV, which supports the need for continuous monitoring of potential zoonotic infection with ToVs (Hoet and Saif, 2004).
Detection of BToVs and PToVs usually involves a cross-hybridization assay, electron microscopy (EM), enzyme-linked immunosorbent assay (ELISA) and reverse transcription-polymerase chain reaction (RT-PCR) (Durham et al., 1989, Koopmans and Horzinek, 1994, Matiz et al., 2002, Smits et al., 2003). Real-time RT-PCR is a more recent method that is more sensitive than EM and conventional RT-PCR for quantitating virus load, pathogenesis studies and identification of virus latency (Glass et al., 2000, Park et al., 2009). Real-time RT-PCR is relatively easy to perform and has a high-throughput capacity as reported with other viruses using SYBR Green chemistry and/or the TaqMan assay (Chun et al., 2010, Park et al., 2009). As far as we know, there is only one report describing the use of real-time RT-PCR for the detection of PToV (Pignatelli et al., 2010). There is no report of the use of real-time RT-PCR for BToV.
The present study developed, optimized and validated a SYBR Green real-time RT-PCR assay using a universal primer pair for the detection and quantitation of both BToVs and PToVs in archived stool samples. From the same samples evaluated previously by conventional RT-PCR and nested PCR assays, SYBR Green real-time RT-PCR proved to be a more specific and sensitive detection and quantitation method.
2. Materials and methods
2.1. Specimens
Bovine (n = 121) and porcine (n = 86) stool specimens were selected from archived fecal samples submitted to the Laboratory of Animal Diseases, College of Veterinary Medicine, Chonnam National University, by local veterinary clinicians in Korea during 2004–2005 and 2007. The stool specimens were stored at −80 °C until used.
2.2. RNA extraction, conventional RT-PCR and nested PCR
RNA was extracted using Trizol-LS (Gibco-BRL, Grand Island, NY) from a 200 μl starting volume of a centrifuged 10% fecal suspension of each sample. Total recovered RNA was suspended in 50 μl of RNase-free water and stored at −80 °C until analysis. The oligonucleotide primers, and conventional RT-PCR and nested PCR conditions for the detection of BToV and PToV (Table 1 ) were described previously (Park et al., 2008, Shin et al., 2010).
Table 1.
List of primers used for conventional RT-PCR, nested PCR and SYBR Green real-time RT-PCR assay for the detection and quantitation of bovine and porcine toroviruses in the fecal specimens from diarrheic calves and piglets.
| Primer name | Sequence (5′–3′)a | Annealing temperature (°C) | Product size (bp) | Gene location | Source or references |
|---|---|---|---|---|---|
| Conventional RT-PCRb | |||||
| BToV | F: TTCTTACTACACTTTTTGGA R: ACTCAAACTTAACACTAGAC |
43 | 603 | 25989–26005c 26438–26458c |
Park et al. (2008) |
| PToV | F: GCCTTTTCCAGACCAGGCCC R: GCAAACCATTGTCCATTAACAC |
55 | 555 | 21–40d 554–575d |
Shin et al. (2010) |
| Nested PCRb | |||||
| BToV | F: TATGTACTATGTTTCCAGCT R: CCAACACAAATCCGCAACGC |
43 | 409 | 25989–26005c 26298–26315c |
Park et al. (2008) |
| PToV | F: ATCTTTGGCAATTGCTTA R: ACCACGAATAGCAATT |
55 | 175 | 231–248d 390–405d |
Shin et al. (2010) |
| SYBR Green real-time RT-PCRe | |||||
| BToV | F: TTACTGGYTATTGGGCMYT R: AAAGGRGTGCAGTGWAGCTT |
48 | 187 | 26029–26047c 26196–26215c |
This study |
| PToV | 272–290d 439–458d |
||||
F: forward primer for conventional RT-PCR, nested PCR, and SYBR Green real-time RT-PCR; R: reverse primer for Conventional RT-PCR, nested PCR, and SYBR Green real-time RT-PCR.
All the procedure of RNA extraction, conventional RT-PCR and nested PCR were performed as described previously (Cho et al., 2001, Park et al., 2008).
Position as counted from the start codon of the complete genome of bovine Breda strain.
Position as counted from the start codon of the porcine torovirus Markelo strain M gene.
Universal primer pair for SYBR Green real-time RT-PCR is designed from the M gene of the bovine and porcine torovirus strains reported in Genbank database as follows; AY427798, AJ575377, DQ778043, AB285126, DQ778053, AJ575374, DQ778048, DQ778049, AJ575371, AJ575370, AJ575368, AJ575369, GU181240, GU181241, GU181244, GU181246, DQ778045.
2.3. Real-time RT-PCR using SYBR Green chemistry
A one-step real-time RT-PCR was developed based on the detection of SYBR Green. The primers were designed based on the published sequence of the membrane (M) gene (Table 1). All reactions were performed using a Corbett Research Rotor-Gene 6000 series Real-Time Amplification system (Corbett Research, Mortlake, Australia) and SensiMix one-step RT-PCR kit with SYBR Green (Quantace, London, UK). Optimization of primer concentration was achieved using RNA from positive fecal samples. Reactions were run using primer concentrations from 0.3 to 0.9 μM. A concentration of 0.5 μM of each primer was found to give the highest sensitivity. Real-time RT-PCR was performed with a final volume of 25 μl containing 5 μl of RNA template, 12.5 μl of SensiMix one-step mixture, 1 μl each of 0.5 μM forward and reverse primers (final concentration of each primer: 20 nM), 0.5 μl of 50× SYBR Green solution (final concentration: 1×), 0.5 μl of RNase inhibitor (final concentration: 10 units), 0.5 μl of MgCl2 (final concentration: 4.0 mM) and 4 μl of RNase-free water. Reverse transcription was carried out at 42 °C for 30 min, followed by the activation of the hot-start DNA polymerase at 95 °C for 10 min and 45 three-step cycles: 94 °C for 15 s, 50 °C for 45 s and 72 °C for 20 s. To perform the melting curve analysis, after 45 reaction cycles, the temperature ramp was programmed from 72 to 95 °C in increments of 1 °C, waiting 5 s before each acquisition. Samples were considered positive if both an exponential increase of fluorescence and a BToV- or PToV-specific melting peak were observed. Quantitative detection of ToVs was averaged from three independent runs with duplicate reactions.
2.4. In vitro transcription of complementary RNA (cRNA) standards
The targeted 603 bp M gene fragment in BToV and the 555 bp fragment in PToV obtained by RT-PCR were used as the source DNA for the preparation of in vitro RNA transcripts (Table 1). The amplicons were inserted into a yT&A cloning vector (Yeastern Biotech, iNtron Biotechnology, Taipei, Taiwan) and, after cloning, a clone was selected based on correct sequence of the insert. Plasmid DNA of the recombinant clone was digested with the restriction enzyme PstI, electrophoresed using a 1.2% agarose gel, and the purified digested plasmid was recovered from the gel using the QIAQuick gel-extraction kit (Qiagen, Valencia, CA). The gel-purified linearized DNA clone served as the template for the in vitro transcription. Reverse transcription reaction was performed using T7 RNA polymerase in a mMessage mMachine T7 kit (Ambion, Austin, TX). After 1 h of incubation at 30 °C, the DNA template was removed by digestion with DNase using the TURBO DNA Free kit (Ambion). cRNA was then purified with the RNeasy Mini kit (Qiagen). After dilution, the concentrations were calculated by measuring the absorbance at 260 nm with NanoDropND1000 (NanoDrop Technologies, Wilmington, DE). cRNA samples were stored at −80 °C until used. The regression lines between the logarithms of the input amounts of cRNAs and the corresponding mean threshold cycle (Ct) values were calculated using the Rotor-Gene software version 6.0.19 (Corbett Research).
2.5. Determination of the performance parameters
To check the reproducibility of the new real-time RT-PCR method, 10-fold serial dilutions of the obtained cRNA were assayed in triplicate by real-time RT-PCR. The intra-assay coefficient variation (CV) was computed from the results obtained with three replicates of each dilution sample tested at the same time. The inter-assay CV was computed for each nine diluted samples carried out by three independent assays on different days. Both CVs were calculated by dividing the standard deviation of each tested sample by its mean and multiplying that result by 100 (Pignatelli et al., 2010).
2.6. Statistical validation
Statistical analyses were performed by SPSS version 11.5.1 for Windows (SPSS, Chicago, IL). The two-tailed Fisher's exact test was used to assess the statistical significance of the detection rate between real-time RT-PCR, conventional RT-PCR and nested PCR. A P-value of <0.05 was considered significant statistically.
3. Results
3.1. Validation of SYBR Green real-time RT-PCR assay for sensitivity, reproducibility, and specificity using in vitro transcripts of BToV and PToV RNA
Real-time RT-PCR was standardized using in vitro transcribed cRNA of BToV and PToV. The standard RNA was diluted in a 10-fold dilution series ranging from 1.0 × 100 to 1.0 × 10−9, and was tested for each run within the SYBR Green real-time RT-PCR assay. Quantities used for each dilution corresponded to 2.54 × 102–2.54 × 1011 and 2.17 × 102–2.17 × 1011 viral copies per reaction of BToV and PToV, respectively. The SYBR Green real-time RT-PCR assay detected as little as 2.54 × 102 copies of BToV cRNA and 2.17 × 103 copies of PToV cRNA per reaction, and displayed linearity over a wide dynamic range of copy numbers (Fig. 1A and D). Amplicons of the expected size by SYBR Green real-time RT-PCR were visualized by gel electrophoresis (Fig. 1C and F). Standard curves with a higher correlation coefficient (R 2 > 0.97) and slope values of 3.42 and 3.33 (Fig. 1B and E) were generated using a serial dilution of in vitro BToV and PToV RNA transcripts, respectively. The standard RNA subjected to SYBR Green real-time RT-PCR presented a specific fluorescence signal and Ct values between 3.7 and 34.6 for BToV and between 6.8 and 39.8 for PToV (data not shown). The intra-assay reproducibility showed that at the highest dilution where cRNA was detected, both replicates were found positive for BToV or PToV. At the 10−1 dilution containing cRNA copies, 100% reproducibility was achieved for the two viruses. The intra-assay coefficient variation of Ct values ranged from 0.02% to 0.23% and 0.001% to 0.24% for standard dilutions from 100 to 10−9 BToV and PToV cRNA copies per reaction, respectively. The inter-assay coefficient variation of Ct values ranged from 0.03% to 0.15% and 0.04% to 0.34% for standard dilutions from 100 to 10−9 BToV and PToV cRNA copies per reaction, respectively. These results indicate that real-time RT-PCR was successful in detecting and quantifying cRNA for BToV and PToV.
Fig. 1.
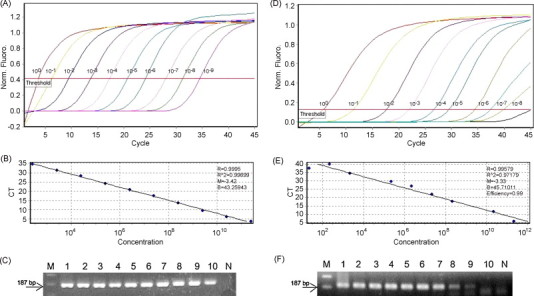
Standards for the SYBR Green real-time RT-PCR assay for the quantitation of BToV and PToV cRNA. (A) Amplification of 100, 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, 10−7, 10−8 and 10−9 dilutions of the cRNA standard used in parallel with each SYBR Green-based real-time RT-PCR assay. (B) Standard curves of the real-time RT-PCR based on serial dilutions of BToV cRNA standards. In the standard curve of these dilutions, each dot represents the result of duplicate amplification of each dilution. The coefficient of determination (R2) and the slope(s) of the regression curve are indicated. (C) SYBR Green real-time RT-PCR products using serially diluted in vitro transcripts. M, molecular marker; lanes 1–10: 2.54 × 1011, 2.54 × 1010, 2.54 × 109, 2.54 × 108, 2.54 × 107, 2.54 × 106, 2.54 × 105, 2.54 × 104, 2.54 × 103 and 2.54 × 102 viral copies/reaction; N, negative control. (D) Amplification of 100, 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, 10−7, 10−8 and 10−9 dilutions of cRNA standard used in parallel with each SYBR Green-based real-time RT-PCR assay. (E) Standard curves of the real-time RT-PCR based on serial dilutions of PToV cRNA standards. In the standard curve of these dilutions each dot represents the result of duplicate amplification of each dilution. The coefficient of determination (R2) and the slope (s) of the regression curve are indicated. (F) SYBR Green real-time RT-PCR products using serially diluted in vitro transcripts. M, molecular marker; lanes 1–10: 2.17 × 1011, 2.17 × 1010, 2.17 × 109, 2.17 × 108, 2.17 × 107, 2.17 × 106, 2.17 × 105, 2.17 × 104 and 2.17 × 103 viral copies/reaction; N, negative control.
3.2. Comparison of the detection limits of in vitro transcripts of BToV and PToV RNAs by conventional RT-PCR, nested PCR, and SYBR Green real-time RT-PCR
For the comparison and assessment of SYBR Green real-time RT-PCR sensitivity and its end point with two conventional assays, 10-fold serial dilutions of the in vitro transcripts described above were tested. The lowest detection limits of conventional RT-PCR and nested PCR using the serial dilutions were, respectively, 2.54 × 105 and 2.54 × 104 viral copies/reaction for BToV and 2.17 × 107 and 2.17 × 105 viral copies/reaction for PToV. Analysis of the dilution series showed that the SYBR Green real-time RT-PCR assay could detect dilutions 100 times lower than nested RT-PCR for both ToVs, however, it could detect 1000 and 10,000 times lower than RT-PCR for BToV and PToV, respectively. Non-specific reactions were not evident with any of the three PCR approaches (data not shown).
3.3. Comparison of BToV and PToV detection rates by SYBR Green real-time RT-PCR, conventional RT-PCR, and nested PCR from archived stool samples
The prevalence of BToVs and PToVs and their genetic properties in the diarrhea specimens of calves (n = 121) and piglets (n = 86) was reported previously (Park et al., 2008, Shin et al., 2010). Using the fecal samples, the detection rates of BToV and PToV by real-time RT-PCR were compared with those of conventional RT-PCR and nested PCR (Table 2, Table 3 ). One (0.8%) bovine and 7 (8.1%) porcine samples were positive by both conventional RT-PCR and real-time RT-PCR, 21 (17.4%) bovine and 24 (27.9%) porcine samples were positive by real-time RT-PCR and negative by the conventional RT-PCR. No samples that were negative by real-time RT-PCR were positive by conventional RT-PCR. Ninety-nine bovine and 55 porcine samples were negative by both tests (Table 2, Table 3). The percentage agreements between these two assays were 82.6% for BToV and 72.1% for PToV detection. The sensitivity and specificity of real-time RT-PCR compared with conventional PCR for BToV were 100% and 82.5%, respectively, and for PToV were 100% and 69.6%, respectively. The agreement beyond chance was calculated with a Kappa statistic of 0.072 for BToV and 0.272 for PToV, meaning a statistically fair agreement. The detection rate of real-time PCR was markedly higher than that of conventional RT-PCR (P < 0.05).
Table 2.
Comparison of the detection rates of BToV by real-time RT-PCR, conventional RT-PCR, and nested PCR assay.
| RT-PCRa |
Nested PCRb |
|||||||
|---|---|---|---|---|---|---|---|---|
| + | − | Total | + | − | Total | |||
| Real-time RT-PCR | + | 1 | 21 | 22 | 13 | 9 | 22 | |
| − | 0 | 99 | 99 | 0 | 99 | 99 | ||
| Total | 1 | 120 | 121 | 13 | 108 | 121 | ||
Percent observed agreement (Po) = (1+99)/121 = 82.6%. Sensitivity = 1/1+0 = 100%. Specificity = 99/21+99 = 82.5%. Kappa = 0.072.
Po = (13+99)/121 = 92.6%. Sensitivity = 13/13+0 = 100%. Specificity = 99/9+99 = 91.7%. Kappa = 0.702.
Table 3.
Comparison of the detection rates of PToV by real -time RT-PCR, conventional RT-PCR, and nested PCR assay.
| RT-PCRa |
Nested PCRb |
|||||||
|---|---|---|---|---|---|---|---|---|
| + | − | Total | + | − | Total | |||
| Real-time RT-PCR | + | 7 | 24 | 31 | 17 | 14 | 31 | |
| − | 0 | 55 | 55 | 0 | 55 | 55 | ||
| Total | 7 | 79 | 86 | 17 | 69 | 86 | ||
Percent observed agreement (Po) = (7+55)/86 = 72.1%. Sensitivity = 7/(7+0) = 100%. Specificity = 55/(24+55) = 69.6%. Kappa = 0.272.
Po = (17+55)/86 = 83.7%. Sensitivity = 17/(17+0) = 100%. Specificity = 55/(14+55) = 79.7%. Kappa = 0.608.
Comparing nested PCR and real-time RT-PCR, 13 (10.7%) bovine and 17 (19.8%) porcine samples were positive in both methods, nine (7.4%) bovine and 14 (16.3%) porcine samples were positive by real-time RT-PCR and negative by nested PCR. No samples were negative by real-time RT-PCR and positive by nested PCR, and 99 bovine and 55 porcine samples were negative by both tests (Table 2, Table 3). The percentage agreements between the two assays were 92.6% for BToV and 83.7% for PToV detection. The sensitivity and specificity of real-time RT-PCR compared with nested PCR were, respectively, 100% and 91.7% for BToV, and 100% and 79.7% for PToV. The agreement beyond chance was calculated with a Kappa statistic of 0.702 in BToV and 0.608 in PToV, meaning that it was statistically significant. The detection rate of real-time PCR was significantly higher in BToV and moderately higher in PToV than that of nested PCR (P < 0.05).
To confirm whether the samples positive by the SYBR Green real-time RT-PCR assay but negative by both RT-PCR and nested PCR assays were true positive, 21 bovine and 24 porcine amplicons positive by the real-time RT-PCR assay were sequenced directly. The nucleotide sequence of all amplicons matched that of the bovine and porcine M gene (data not shown), indicating that the positive reaction by the real-time RT-PCR assay was true positive.
3.4. Viral copy numbers in the stool samples as evaluated by SYBR Green real-time RT-PCR
The efficacy and reliability of the SYBR Green real-time RT-PCR assay for the detection and quantitation of BToV and PToV in the 121 bovine and 86 porcine stool samples were evaluated. All positive samples subjected to SYBR Green real-time RT-PCR presented a fluorescence signal and Ct values between 19.0 and 35.0 for BToV and 20.0 and 36.0 for PToV (data not shown).
BToV and PToV amplicons displayed melting temperatures (T m) between 74 and 80 °C (data not shown). The range of T m values for BToV and PToV using real-time RT-PCR SYBR Green chemistry of in vitro transcripts showed 78.2–79.7 °C for BToV and 76.5–77.8 °C for PToV-positive samples. However, the non-template controls were 72 °C. The viral copy numbers of positive samples per assay as measured by SYBR Green real-time RT-PCR ranged from 3.4 × 102 to 5.4 × 107 for BToV, and 3.1 × 103 to 1.3 × 105 for PToV. One BToV and seven PToV samples that tested positive in both real-time RT-PCR and conventional RT-PCR contained 5.4 × 107 and 9.9 × 104–1.3 × 105 viral copies per reaction, respectively. Thirteen bovine and 17 porcine samples that tested positive by real-time RT-PCR and nested PCR had 5.4 × 102–4.0 × 104 and 3.6 × 103–6.4 × 104 viral copies per reaction, respectively. However, the nine bovine and 14 porcine fecal samples that tested positive only by real-time RT-PCR possessed comparatively lower viral copies, ranging from 3.4 × 102 to 1.2 × 104 and 3.1 × 103 to 3.9 × 104 viral copies per reaction, respectively.
The specificity of BToV by real-time RT-PCR was assessed using bovine fecal samples that had tested positive for other enteric pathogens including groups A, B and C bovine rotaviruses (BRV A–C), bovine coronavirus (BCoV), bovine norovirus, bovine viral diarrhea virus (BVDV), Salmonella spp., Clostridium spp., Campylobacter spp., shiga-toxin-producing Escherichia coli, Coccidium spp. and Cryptosporidium spp. (Asakura et al., 1998, Park et al., 2008). The specificity to PToV by real-time RT-PCR was assessed using porcine diarrhea specimens that had tested positive for porcine rotaviruses A, B, C (PRV A–C), and porcine sapovirus. There was no positive signal recorded for any of these pathogens.
4. Discussion
In this study, an optimized real-time RT-PCR was developed using SYBR Green chemistry to detect BToV or PToV in bovine or porcine fecal samples for the efficient and rapid screening of samples on a large scale. The primer pair for the SYBR Green-based real-time RT-PCR assay was designed to target sequences centered on the conserved M gene. The conserved stretches in a portion of the M gene have shown high homology among the published BToV and PToV isolates and, with the nucleocapsid gene, was thought to be the most sensitive and specific detection genes for both viruses in fecal samples (Ito et al., 2007, Pignatelli et al., 2009). Using both target genes, the detection of BToV or PToV by real-time RT-PCR was demonstrated to be specific and sensitive in this study and in another study (Pignatelli et al., 2010). The findings of the present study proved the application and efficiency of using one primer pair for the detection of two viruses. However, the results showed a higher analytical sensitivity of BToVs over PToVs. The detection of a larger amplicon size that was >70 bp (187 bp), and the positive weaker amplicons in the case of PToVs could have possibly affected the melt rate and phenomenon of dye translocation, which consequently affected SYBR Green binding and detection (Varga and James, 2006).
To refine further the method developed, it was first intended to find ways of distinguishing the two viruses according to the melt curve analysis. The M gene region has different GC/AT ratios in BToV and PToV. Accordingly, the functions of the GC/AT ratio, length and sequence can be demonstrated by melting curve analysis and can be used to differentiate amplification products with <2 °C intervals in melting temperature (Ririe et al., 1997). Melting curve analysis of the in vitro transcripts showed a difference of only 1 °C between BToV and PToV, and, using the field samples, melting curve analysis showed overlapping temperature ranges for BToV (74.7–79.7 °C) and PToV (73.9–78.1 °C). From this, it can be inferred that the differentiation between BToV and PToV by the current real-time RT-PCR assay becomes more difficult to determine and insignificant. Nonetheless, the desired products can be distinguished from undesirable products, and in many cases the need for gel electrophoresis is eliminated (Ririe et al., 1997). The samples that showed close melting temperature with the no-template control were reconsidered positive as they showed a good level of viral copies and a clear band after electrophoresis. In this study, all of the real-time RT-PCR samples tested were verified by electrophoresis and sequencing.
The ideal method for the detection of BToV or PToV in stool samples should have a high degree of sensitivity and specificity, low risk of contamination (Koopmans and Horzinek, 1994), and consistency of performance in the laboratory. At present, the SYBR Green RT-PCR assay is 1000 and 10,000 times more sensitive than BToV and PToV detection by conventional RT-PCR, respectively, and 100 times more sensitive than nested PCR for both viruses when using cRNA. Therefore, the one-step SYBR Green quantitative real-time RT-PCR assay is significantly superior (P < 0.05) to the other PCR assays in terms of sensitivity, specificity and quantitative linearity. The short amplicons in the real-time PCR assays used in this study likely resulted in more efficient amplification and higher analytical sensitivity. Despite the shorter amplicon size in PToV nested PCR, the real-time PCR reproduced amplicons with greater analytical sensitivity. Taken together, these observations indicate that the SYBR Green-based RT-PCR assay is a useful and reliable method for the detection of BToV and PToV.
The reported fecal prevalence of BToVs in bovine diarrhea specimens by conventional RT-PCR or nested PCR was 36.4% in Canada (Duckmanton et al., 1998), 5.2% in Austria (Haschek et al., 2006), 9.7% in the United States (Hoet et al., 2003), 3.6% in Hungary (Matiz et al., 2002), 15.2–23.7% in Japan (Kiriwasa et al., 2007) and 2.9% in South Korea (Park et al., 2008). Porcine toroviruses were detected in 5% of the diarrhea specimens of piglets in Hungary (Matiz et al., 2002), 23.7% in Japan (Kiriwasa et al., 2007), and 6.4% in South Korea (Shin et al., 2010). Using the same South Korean bovine and porcine fecal samples (Park et al., 2008, Shin et al., 2010), SYBR Green real-time RT-PCR assay detected BToVs in 18.2% and PToVs in 36.0% of the samples. These findings support the above suggestion that SYBR Green real-time RT-PCR is more sensitive than conventional RT-PCR and nested PCR. In addition, these observations indicate that the fecal prevalence of BToV and PToV in calves and piglets might be higher than previously reported in South Korea (Park et al., 2008, Shin et al., 2010).
Toroviruses in calves and piglets have been identified in a number of outbreaks in bovine and porcine herds with severe and recurring diarrhea negative for routine virologic, bacteriologic and parasitologic agents (Hoet et al., 2003, Kiriwasa et al., 2007, Park et al., 2009, Shin et al., 2010). Interestingly, those fecal samples that tested positive only by SYBR Green real-time RT-PCR contained lower numbers of BToV and PToV RNA (3.4 × 102–1.2 × 104 and 3.1 × 103–3.9 × 104 viral copies per reaction, respectively). These diarrhea specimens with lower number of ToVs were co-infected with other enteric pathogens, including BCoV, BRVA and/or BVDV in the bovine fecal samples, and PRV A–C in the porcine fecal samples (data not shown). Therefore, it can be presumed that BToV or PToV can either induce diarrhea alone, or can play a role in accelerating the clinical and pathological presentation of diarrhea with the other pathogens (Hoet et al., 2003, Kiriwasa et al., 2007, Park et al., 2009, Shin et al., 2010).
In conclusion, a rapid, sensitive, specific and reproducible one-step SYBR Green real-time RT-PCR was developed for the detection and quantitation of BToV and PToV in bovine and porcine stool samples. This high-throughput assay may be useful for the establishment of a surveillance system and diagnostic purposes for BToV and PToV infections worldwide.
Acknowledgements
This study was supported by a grant (2009-0081752) from the Korea Science and Engineering Foundation (KOSEF) and the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE), Republic of Korea. The authors would like to acknowledge a graduate fellowship from the Korean Ministry of Education and Human Resources Development through the Brain Korea 21 project.
Contributor Information
Sang-Ik Park, Email: animal000@hanmail.net.
Kyoung-Oh Cho, Email: choko@chonnam.ac.kr.
References
- Asakura H., Makino S., Shirahata T., Tsukamoto T., Kurazono H., Ikeda T., Takeshi K. Detection and genetical characterization of shiga toxin-producing Escherichia coli from wild deer. Microbiol. Immunol. 1998;42:815–822. doi: 10.1111/j.1348-0421.1998.tb02356.x. [DOI] [PubMed] [Google Scholar]
- Beards G.M., Green J., Hall C., Flewett T.H., Lamouliatte F., Du Pasquier P. An enveloped virus in stools of children and adults with gastroenteritis resembles the Breda virus of calves. Lancet. 1984;2:1050–1052. doi: 10.1016/S0140-6736(84)91454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.O., Hasoksuz M., Nielsen P.R., Chang K.O., Lathrop S., Saif L.J. Cross protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001;146:2401–2419. doi: 10.1007/s007050170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun Y.H., Jeong Y.J., Park S.I., Hosmillo M., Shin D.J., Kwon H.J., Kang S.Y., Woo S.K., Kang M.I., Park S.J., Cho K.O. Development of one-step real-time reverse transcription polymerase chain reaction assays for rapid detection of porcine group C rotaviruses. J. Vet. Diagn. Invest. 2010;22:74–77. doi: 10.1177/104063871002200113. [DOI] [PubMed] [Google Scholar]
- Duckmanton L., Carman S., Nagy E., Petric M. Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 1998;36:1266–1270. doi: 10.1128/jcm.36.5.1266-1270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham P.J.K., Hassard L.E., Norman G.R., Yemen R.L. Viruses and virus-like particles detected during examination of faeces from calves and piglets with diarrhea. Can. Vet. J. 1989;30:876–881. [PMC free article] [PubMed] [Google Scholar]
- Glass R.I., Noel J., Ando T., Fankhauser R., Belliot G., Mounts A., Parashar U.D., Bresee J.S., Monroe S.S. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 2000;181(Suppl. 2):S254–S261. doi: 10.1086/315588. [DOI] [PubMed] [Google Scholar]
- Haschek B., Klein D., Benetka V., Herrera C., Sommerfeld-Stur I., Vilcek S., Moestl K., Baumgartner W. Detection of bovine torovirus in neonatal calf diarrhea in Lower Austria and Styria (Austria) J. Vet. Med. B: Infect. Dis. Vet. Public Health. 2006;53:160–165. doi: 10.1111/j.1439-0450.2006.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet A.E., Nielsen P.R., Hasoksuz M., Thomas C., Wittum T.E., Saif L.J. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Invest. 2003;15:205–212. doi: 10.1177/104063870301500301. [DOI] [PubMed] [Google Scholar]
- Hoet A.E., Saif L.J. Bovine torovirus (Breda virus) revisited. Anim. Health Res. Rev. 2004;5:157–171. doi: 10.1079/ahr200498. [DOI] [PubMed] [Google Scholar]
- Ito T., Okada N., Fukuyama S.I. Epidemiological analysis of bovine torovirus in Japan. Virus Res. 2007;126:32–37. doi: 10.1016/j.virusres.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriwasa R., Takeyama A., Koiwa M., Hiroshi I. Detection of bovine torovirus in fecal specimens of calves with diarrhea in Japan. J. Vet. Med. Sci. 2007;69:471–476. doi: 10.1292/jvms.69.471. [DOI] [PubMed] [Google Scholar]
- Koopmans M., Horzinek M.C. Toroviruses of animals and human: a review. Adv. Virus Res. 1994;43:233–272. doi: 10.1016/S0065-3527(08)60050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroneman A., Cornelissen L.A., Horzinek M.C., de Groot R.J., Egberink H.F. Identification and characterization of a porcine torovirus. J. Virol. 1998;72:3507–3511. doi: 10.1128/jvi.72.5.3507-3511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiz K., Kecskemeti S., Kiss I., Adam Z., Tanyi J., Nagy B. Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 2002;50:293–296. doi: 10.1556/AVet.50.2002.3.5. [DOI] [PubMed] [Google Scholar]
- Park S.J., Oh E.H., Park S.I., Kim H.H., Jeong Y.J., Lim G.K., Hyun B.H., Cho K.O. Molecular epidemiology of bovine toroviruses circulating in South Korea. Vet. Microbiol. 2008;126:364–371. doi: 10.1016/j.vetmic.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.I., Park D.H., Saif L.J., Jeong Y.J., Shin D.J., Chun H.Y., Park S.J., Kim H.Y., Hosmillo M., Kwon H.J., Kang M.I., Cho K.O. Development of SYBR Green real-time RT-PCR for rapid detection, quantitation and diagnosis of unclassified bovine enteric calicivirus. J. Virol. Methods. 2009;159:64–68. doi: 10.1016/j.jviromet.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli J., Jiménez M., Grau-Roma L., Rodríguez D. Detection of porcine torovirus by real time RT-PCR in piglets from a Spanish farm. J. Virol. Methods. 2010;163:398–404. doi: 10.1016/j.jviromet.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Pignatelli J., Jimenez M., Luque J., Rejas M.T., Lavazza A., Rodriguez D. Molecular characterization of a new PToV strain. Evolutionary implications. Virus Res. 2009;143:33–43. doi: 10.1016/j.virusres.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ririe K.M., Rasmussen R.P., Wittwer C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- Shin D.J., Park S.I., Jeong Y.J., Hosmillo M., Kim H.H., Kim H.J., Kwon H.J., Kang M.I., Park S.J., Cho K.O. Detection and molecular characterization of porcine toroviruses in Korea. Arch. Virol. 2010;155:417–422. doi: 10.1007/s00705-010-0595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., Lavazza A., Matiz K., Horzinek M.C., Koopmans M.P., de Groot R.J. Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 2003;77:9567–9577. doi: 10.1128/JVI.77.17.9567-9577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga A., James D. J. Real-time RT-PCR and SYBR Green I melting curve analysis for the identification of Plum pox virus strains C, EA, and W: effect of amplicon size, melt rate, and dye translocation. J. Virol. Methods. 2006;132:146–153. doi: 10.1016/j.jviromet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Weiss M., Steck F., Horzinek M.C. Purification and partial characterization of a new enveloped RNA virus (Berne virus) J. Gen. Virol. 1983;64:1849–1858. doi: 10.1099/0022-1317-64-9-1849. [DOI] [PubMed] [Google Scholar]
- Woode G.N., Reed D.E., Runnels P.L., Herrig M.A., Hill H.T. Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 1982;7:221–240. doi: 10.1016/0378-1135(82)90036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


