Abstract
Turkey coronavirus (TCoV) infection causes acute atrophic enteritis in the turkey poults, leading to significant economic loss in the U.S. turkey industry. Rapid detection, differentiation, and quantitation of TCoV are critical to the diagnosis and control of the disease. A specific one-step real-time reverse transcription-polymerase chain reaction (RRT-PCR) assay for detection and quantitation of TCoV in the turkey tissues was developed using a dual-labeled fluorescent probe. The fluorogenic probe labeled with a reporter dye (FAM, 6-carboxytetramethylrhodamin) and a quencher dye (AbsoluteQuencher™) was designed to bind to a 186 base-pair fragment flanked by the two PCR primers targeting the 3′ end of spike gene of TCoV. The assay was performed on different avian viruses and bacteria to determine the specificity as well as serial dilutions of TCoV for the sensitivity. Three animal trials were conducted to further validate the assay. Ten-day-old turkey poults were inoculated orally with 100 EID50 of TCoV. Intestinal tissues (duodenum, jejunum, ileum, cecum), feces from the cloacal swabs, or feces from the floor were collected at 12 h, 1, 2, 3, 5, 7, and/or 14 days post-inoculation (DPI). RNA was extracted from each sample and subjected to the RRT-PCR. The designed primers and probe were specific for TCoV. Other non-TCoV avian viruses and bacteria were not amplified by RRT-PCR. The assay was highly sensitive and could quantitate between 102 and 1010 copies/μl of viral genome. The viral RNA in the intestine segments reached the highest level, 6 × 1015 copies/μl, in the jejunum at 5 DPI. Eighty-four intestine segments assayed by the developed RRT-PCR and immunofluorescence antibody assay (IFA) revealed that there were 6 segments negative for TCoV by both assays, 45 positive for TCoV by IFA, and 77 positive for TCoV by RRT-PCR. Turkey coronavirus was detected in the feces from the cloacal swabs or floor 1–14 DPI; however, the viral RNA load varied among different turkey poults at different intervals from different trials. The highest amount of viral RNA, 2.8 × 1010 copies/μl, in the feces was the one from the cloacal swab collected at 1 DPI. The average amount of TCoV RNA in the cloacal fecal samples was 10 times higher than that in the fecal droppings on the floor. Taken together, the results indicated that the developed RRT-PCR assay is rapid, sensitive, and specific for detection, differentiation, and quantitation of TCoV in the turkey tissues and should be helpful in monitoring the progression of TCoV induced acute enteritis in the turkey flocks.
Keywords: Turkey coronavirus, Real-time RT-PCR, TaqMan probe, Spike gene
1. Introduction
Turkey coronaviral enteritis, due to turkey coronavirus (TCoV) infection, causes atrophic enteritis, decreased weight gain, increased mortality and uneven flock growth in the turkeys leading to significant economic loss in the turkey industry. The occurrence of turkey coronaviral enteritis is still reported in many states of the U.S. There are no effective vaccine and treatment available for prevention and treatment of the disease. Therefore, rapid detection, differentiation, and quantitation of TCoV are very critical to monitor and control TCoV infection in the turkey flocks.
Turkey coronavirus belongs to the antigenic group 3 in the family Coronaviridae. It is a pleomorphic enveloped virus with the size about 80–160 nm in diameter. There are 20 nm long club-shaped projections around the virions. The genomic nucleic acid is a linear, positive sense, single stranded RNA. The major structural proteins include spike (S) glycoprotein, membrane (M) protein and nucleocapsid (N) protein. The S protein is highly variable among different coronaviruses while M and N protein are more conserved between different antigenic groups. The S proteins of coronaviruses have been reported to form peplomer structures on the surface of coronavirus, which bind to cell receptors and induce cell fusion and neutralizing antibodies (Lu et al., 2004). The similarity of S gene nucleotide and amino acid sequence among coronaviruses varies. Although TCoV and infectious bronchitis virus (IBV) are in the same antigenic group, the S gene nucleotide sequence of TCoV only share 30–40% homology to that of IBV (Lin et al., 2004). The amino terminal S1 region of several coronaviruses has been reported to contain the receptor binding domain. It can bind to cellular receptors, induce neutralizing antibodies, and determine host specificity and tropism (Hwang et al., 2006, Sheahan et al., 2008). The carboxyl terminal S2 region consisting of transmembrane domain can induce cell fusion and viral assembly (Broer et al., 2006, Howard et al., 2008). The S2 gene is more conserved than S1 gene between different coronaviruses and between different strains or isolates of the same coronavirus (Cavanagh, 2005, Lin et al., 2004, Loa et al., 2006a, Loa et al., 2006b). Therefore, S2 gene is a better target to detect TCoV isolates and differentiate TCoV from other coronaviruses.
Conventional diagnostic methods for detecting TCoV include virus isolation (VI), immunofluorescent antibody assay (IFA), immunoperoxidase assay (IP), and reverse transcriptase polymerase chain reaction (RT-PCR) (Breslin et al., 2000, Sekiguchi et al., 2004, Spackman et al., 2005). Because there is no cell culture system for TCoV, the virus isolation has to use turkey eggs for propagation of the virus, which is time consuming and labor intensive. Immunofluorescent antibody assay and IP are antibody-based methods. They are simpler and faster than virus isolation but the sensitivity is very low. Since the sequence information of TCoV was available, RT-PCR has been developed and it has high specificity and sensitivity. It has been applied to detect TCoV RNA in fecal samples and intestinal contents of infected turkeys (Breslin et al., 2000, Loa et al., 2006a). Nevertheless, none of the above-mentioned methods can rapidly differentiate and quantitate TCoV.
Real-time RT-PCR (RRT-PCR) uses a dual-labeled probe combined with the 5′ → 3′ exonuclease activity of Taq polymerase to increase the release of reporter dye fluorescence in the course of the PCR amplification (Holland et al., 1991). Quantitative data can be accessed by the standard curve established with serial dilutions of standard RNA. This method has been applied to quantitative detection of many coronaviruses, including canine coronavirus (CCov), feline infectious peritonitis virus (FIPV) and severe acute respiratory syndromes coronavirus (SARS CoV) (Decaro et al., 2004, Gut et al., 1999, Hui et al., 2004). The procedure dose not need post-PCR electrophoresis so the processing time can be saved and the risks for carry-over and cross-contamination between samples can be lessen. The purpose of the present study was to develop a sensitive and specific one-step RRT-PCR to detect, differentiate, and quantitate TCoV RNA in the feces and tissues.
2. Material and methods
2.1. Turkey eggs and poults
Turkey eggs and 1-day-old turkey poults (British United Turkey of America, BUTA) of both sexes were obtained from Perdue Farm (Washington, IN, USA). They were free of recognized pathogens for turkeys, including TCoV. Turkey poults were housed in the isolated floor pens. Feed and water were provided ad libitum. The protocol for care and use of turkey eggs and turkey poults in the present study was approved by Purdue University Animal Care and Use Committee.
2.2. Viruses
Turkey coronavirus (TCoV isolate 540) was isolated from the intestines of 28-day-old turkey poults with outbreaks of acute enteritis in Indiana. Affected intestines were homogenized with 5-fold volume of phosphate-buffered saline (PBS), clarified by centrifugation at 3000 × g for 10 min at 4 °C, and filtered through 0.45 and 0.22 μm membrane filters (Millipore Products Division, Bedford, MA), respectively. Twenty-two days old embryonated turkey eggs were inoculated with 200 μl of the filtrate via amniotic route and embryo intestines were harvested after 3 days of incubation. Harvested embryo intestines were processed and propagated as described above for 5 passages. Intestines of TCoV-infected turkey embryos in the fifth passage were prepared as 20% suspension in PBS. The suspension was homogenized, clarified, filtered as described above, and used as inoculum. The titer of virus in the inoculum was determined by inoculation of the suspension at 10-fold dilutions into groups of five 22-day-old embryonated turkey eggs. The 50% embryo infectious dose (EID50) was calculated by the method of Reed and Muench (Loa et al., 2001, Reed and Muench, 1938, Senne, 2008). An inoculum containing 100 EID50/200 μl was prepared and used to infect experimentally turkey poults.
2.3. Primers and probe
A forward primer QS1F (5′-TCGCAATCTATGCGATATG-3′), a reverse primer QS1R (5′-CAGTCTTGGGCATTACA C-3′), and a TaqMan dual-labeled probe QS1P (5′-AbsoluteQuencher-TCTGTGGCAATGGTAGCCATGTTC-FAM-3′) were designed to amplify and detect 186 base-pair fragment of the S2 gene of the TCoV isolate 540. The primers and probe were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
2.4. RNA extraction
The starting material consisted of 100 mg of feces from the cloacal swab samples or 100 mg of feces from the floor was mixed with 500 μl of PBS solution. The mixture was centrifugated at 3000 rpm for 2 min and 140 μl of supernatant was subjected to RNA extraction by using QIAamp® viral RNA mini kit (Qiagen, Valencia, CA, USA). For intestine tissue samples, 40 mg of tissue was weighted and mixed with lysis solution for further homogenization. RNA was extracted from the homogenate with the Vergene tissue RNA purification kit (Gentra, Minneapolis, MN, USA) following the manufacturer's instruction.
2.5. Real-time RT-PCR for TCoV
A total 25 μl of reaction mixture contained 900 nM of forward primer QS1F, 300 nM of reverse primer QS1R, 200 nM of probe QS1P, 12.5 μl of Platinum® Quantitative RT-PCR ThermoScript™ One-Step System 2x reaction mix, 0.5 μl of SuperScript™ III reverse transcriptase/Platinum Taq polymerase mix (Invitrogen, Carlsbad, CA, USA), and 5 μl of RNA template. The reaction was carried out in Rotor-Gene 3000™ real-time thermocycler (Corbett Research, Sydney, Australia) at 50 °C for 30 min, 94 °C for 5 min, and 45 cycles of 94 °C for 20 s and 61 °C for 1 min to acquire the fluorescence FAM. Signals were regarded as positive if the fluorescence intensity exceeded 10 times the standard deviation of the baseline fluorescence.
2.6. Specificity of RRT-PCR for TCoV
RNA of Salmonella, Escherichia coli, avian reovirus, turkey herpesvirus, avian influenza virus, Newcastle disease virus, enterovirus, rotavirus, transmissible gastroenteritis virus (TGEV), infectious feline peritonitis virus (FIPV), canine coronavirus (CCoV), bovine coronavirus (BCoV), infectious bronchitis virus (IBV) strain Ark99, IBV strain Connecticut, IBV strain M41, TCoV isolate 517, TCoV isolate 540, TCoV isolate ATCC, TCoV isolate 310, TCoV isolate 1440, and TCoV isolate 428 were extracted by the Vergene tissue RNA purification kit (Gentra). The extracted RNA was used in RRT-PCR with primers QS1F and QS1R and the probe QS1P. In addition, the amplicons of RRT-PCR were electrophoresed on 1% agarose gel.
2.7. Sensitivity and standard curve of RRT-PCR for TCoV
The carboxyl S gene segment of TCoV isolate 540 encompassing the fragment targeted by the primers QS1F and QS1R was amplified by forward primer 6F (5′-GACCATGGGATT TGTTGAA-3′) and reverse primer 6R (5′-TTTTTAATGGCATCTTTTGA-3′) and constructed into pTriEx3 vector (Novagen, San Diego, CA, USA) designed as pTriEx3-6F/6R. To obtain a standard for quantitative RRT-PCR for TCoV, the in vitro transcripts of pTriEx3-6F/6R were generated by using RiboMAXTM large scale RNA production system-T7 (Promega, Madison, WI, USA). The pTriEx-6F/6R transcripts were treated with DNase, purified by Zymo DNA clean and concentrate kit (Zymo, Orange, CA, USA), and quantified by spectrophotometer at 260 nm wavelength. Ten-fold serial dilutions of the RNA transcripts were made in RNase-free water and analyzed by RRT-PCR with the procedures described above to establish standard curve for quantification and determine the limit for detection. The standard curves were created by plotting the Ct value against each dilution of known concentration. The formula used to calculate the copy numbers per μl is as followed: copies/μl = (ng/μl × 10−9)/(MW of transcript) × (6.023 × 1023).
2.8. Experimental infections of turkeys with TCoV
In trial 1, there were 14 nine-day-old turkeys in the negative control group and 21 turkeys in the experimentally infected group. Turkeys were inoculated orally with 100 EID50 of TCoV isolate 540. Duodenum, jejunum, ileum, and cecum from 2 control and 3 infected turkeys were collected at 12 h post-inoculation (HPI), 1 day post-inoculation (DPI), 2 DPI, 3 DPI, 5 DPI, 7 DPI, and 14 DPI. Tissue samples were frozen for IFA and RNA was extracted from each sample for quantitative RRT-PCR for TCoV. In trial 2, 3 nine-day-old turkeys were inoculated orally with 100 EID50 of TCoV isolate 540. Cloacal fecal sample was collected in the swab from each turkey everyday from 1 to 14 DPI. In trial 3, 3 nine-day-old turkeys were in the negative control group and 3 turkeys inoculated with 100 EID50 of TCoV isolate 540 were in the experimentally infected group. The fecal droppings on the floor were collected everyday from 1 to 14 DPI and stored frozen until analyzed. RNA was extracted from each fecal swab or dropping sample for quantitative RRT-PCR for TCoV.
2.9. Immunofluorescence antibody assay
Frozen intestinal samples were embedded in the embedding medium and sectioned at −20 °C. Frozen intestinal sections were fixed in acetone for 10 min and incubated subsequentially with turkey anti-TCoV antiserum (1:40 dilution) at room temperature for 1 h. After washing with PBS for 10 min, the sections were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-turkey IgG (H + L) (KPL, Gaithersburg, MA, USA) at room temperature in dark for 1 h. Slides were washed in PBS for 10 min, air dried, and mounted by the cover glasses in Vectashield® mounting medium for fluorescence (Vector Laboratories, Burlingame, CA, USA). The slides were examined by a fluorescent microscope (Optiphot-2, Nikon, Melville, NY, USA).
2.10. Statistical analysis
SPSS 15.0 (SPSS Inc., Chicago, IL) was used to analyze the data. One-way ANOVA was employed to evaluate the concentration of TCoV in the intestinal samples. Tukey test was further used to investigate the relationships between TCoV level measured in different intestinal fragments at different time points. Statistical significance was set at p < 0.05.
3. Results
3.1. Specificity of RRT-PCR for TCoV
Primers QS1F and QS1R were highly specific to TCoV and conserved among known TCoV isolates (Fig. 1 ). Expected 186-bp band was amplified from TCoV RNA of TCoV isolates used in the present study, including Indiana TCoV isolates 517 and 540, Minnesota TCoV isolates ATCC and 310, North Carolina TCoV isolate 1440, and Arkansas TCoV isolate 428. No bands of the expected size were amplified from the other coronaviruses (CCoV, BCoV, TGEV, FIPV, IBV Ark99, IBV Connecticut, IBV M41), enteric bacteria (Salmonella and E. coli), and avian viruses (avian influenza virus, Newcastle disease virus, avian reovirus, turkey herpesvirus, enterovirus, and rotavirus).
Fig. 1.
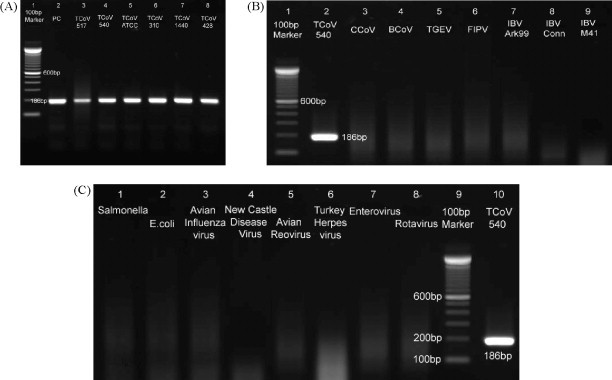
Specificity of real-time RT-PCR (RRT-PCR) for turkey coronavirus (TCoV). (A) The products of RRT-PCR with the primers QS1F and QS1R and the probe QS1P performed on RNA extracted from TCoV isolate 517, 540, ATCC, 310, 1440, and 428 are shown in 1% agarose gel from lanes 3 to 8, respectively. Lane 1 is DNA marker 100-base pair (bp) (Invitrogen, Carlsbad, CA, USA) and lane 2 is RNA in vitro transcribed from plasmid pTriEx3-6F/6R as the positive control. (B) The products of RRT-PCR performed on RNA extracted from TCoV isolate 540, canine coronavirus (CCoV), bovine coronavirus (BCoV), transmissible gastroenteritis virus (TGEV), feline infectious peritonitis virus (FIPV), infectious bronchitis virus (IBV) Ark99, IBV Conn, and IBV M41 are shown on 1% agarose gel from lanes 2 to 9, respectively. Lane 1 is 100-bp DNA marker. (C) The products of RRT-PCR performed on RNA extracted from Salmonella, Escherichia coli, avian influenza virus, Newcastle disease virus, avian reovirus, turkey herpesvirus, enterovirus, and rotavirus are shown on 1% agarose gel from lanes 1 to 8, respectively. Lane 9 is 100-bp DNA marker and lane 10 is TCoV 540 as the positive control.
3.2. Sensitivity and detection limit
A linear standard curve of template copy number against threshold cycle time (Ct) value was established using serial 10-fold diluted pTriEx3-6F/6R RNA transcripts. The coefficient of linear regression (R 2) for the standard curve was 0.995 and the linear range was between 102 and 1010 copies/μl (Fig. 2 ). The detection limit was as low as 102 copies/μl.
Fig. 2.
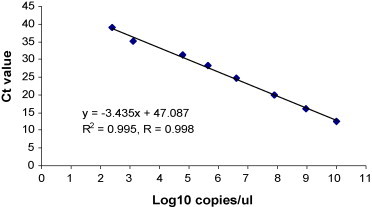
Standard curve of real-time RT-PCR (RRT-PCR) assay for turkey coronavirus (TCoV). The X-axis shows the concentration of serially diluted RNA in vitro transcribed from standard pTriEx3-6F/6R in log 10 value and the Y-axis indicates the corresponding threshold cycle (Ct) value. The linear quantitative correlation was established from 102 to 1010 copies/μl with 99.8% coefficient. The detection limit is as low as 102 copies/μl.
3.3. TCoV in intestinal samples from experimentally infected turkeys
For a total 84 intestinal samples from turkeys infected experimentally with TCoV collected at 7 time points. Seventy-seven segments were detected positive for TCoV by RRT-PCR. The results of 51 samples were in agreement by both RRT-PCR and IFA assays: 45 positive and 6 negative for TCoV (Fig. 3 ). The 6 samples tested negative for TCoV by both assays were collected at 12 h and 1 day post-inoculation.
Fig. 3.
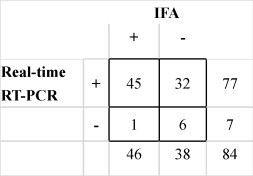
Detection of turkey coronavirus (TCoV) in the intestinal tissue samples from experimentally infected turkeys in the trial 1. The results by immunofluorescent antibody assay (IFA) are compared to those by real-time RT-PCR (RRT-PCR). The numbers indicate the numbers of samples positive (+) or negative (−) for TCoV.
Regarding the TCoV level measured by RRT-PCR, TCoV can be detected as early as 12 HPI (Fig. 4 ). The highest concentration, 5 × 1015 copies/μl, was detected in jejunum at 5 DPI. Except in duodenum showing the highest TCoV level at 1 DPI, the highest concentration of TCoV in jejunum, ileum, or cecum was measured at 5 DPI. The TCoV concentration was significantly lower in duodenum than that in the other 3 intestinal segments at 3, 5, and 7 DPI (p < 0.05). The TCoV level was significantly higher in ileum than that in the other 3 intestinal segments at 3 and 14 DPI (p < 0.05). Greater variation in the results of RRT-PCR and IFA was seen in the tissue samples collected at early time points when there was lower TCoV concentration in the sample.
Fig. 4.
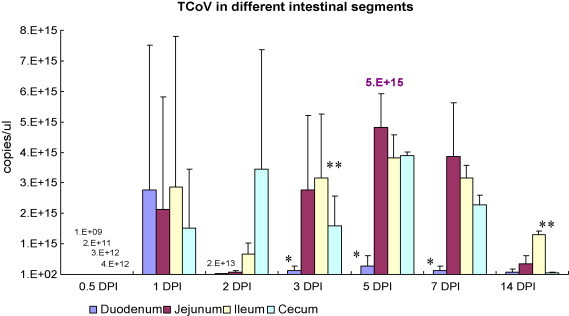
The turkey coronavirus (TCoV) levels in the intestinal tissue samples from experimentally infected turkeys determined by real-time RT-PCR (RRT-PCR) in the trial 1. Different intestinal segments are indicated by different columns. The TCoV level is calculated and shown as copies/μl. The bar in each column is standard deviation. Asterisks indicate significantly differences (p < 0.05) found among different intestinal segments collected at the same time point.
3.4. TCoV in cloacal fecal swabs from experimentally infected turkeys
TCoV was detected in cloacal fecal swabs collected daily 1–14 DPI from the same 3 turkeys inoculated with TCoV. Higher levels of TCoV in the fecal cloacal swabs were detected in 3 turkeys at 1 and 7 DPI, respectively. The highest concentration of TCoV, 2.88 × 1010 copies/μl, was seen on day 1. The kinetics of the viral load from each turkey based on the TCoV level in the fecal sample from 1 to 14 DPI is shown in Fig. 5 .
Fig. 5.
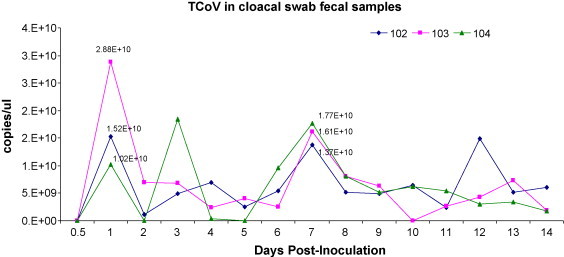
The concentration of turkey coronavirus (TCoV) in the cloacal fecal swab samples collected sequentially after inoculation from the same three turkeys for 14 days in the trial 2. Three lines with different shapes (♦, ■, ▴) represent three different turkeys (individual number 102, 103, and 104). The TCoV level is calculated and shown as copies/μl.
3.5. TCoV in fecal droppings from experimentally infected turkeys
The TCoV levels of the fecal droppings on the floor assayed by RRT-PCR ranged from 104 to 2.8 × 106 copies/μl from 1 to 14 DPI (Fig. 6 ). The viral level in the feces on the floor was 2 log less than that from the cloacal fecal swabs. Higher levels of TCoV in the fecal droppings on the floor were detected at 3, 8, and 13 DPI, respectively.
Fig. 6.
The concentration of turkey coronavirus (TCoV) in the fecal droppings on the floor assayed by real-time RT-PCR (RRT-PCR) from 1 to 14 days after inoculation in the trial 3. The blue line with shape (♦) represents samples of TCoV-infected turkeys and the pink line with shape (■) represents samples of the negative control group. The TCoV level is calculated and shown as copies/μl. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
The development and evaluation of a TCoV-specific RRT-PCR assay using dual-labeled fluorescent probes for the detection and quantification of TCoV RNA from the intestinal tissues, cloacal fecal swabs, and fecal droppings on the floor was reported in the present study. The designed primers and probe can distinguish TCoV from other pathogens, including E. coli, Salmonella, rotavirus, reovirus, enterovirus, avian influenza virus, Newcastle disease virus, turkey herpesvirus, and other coronaviruses (CCoV, BCoV, TGEV, FIPV, and IBV). Based on the studies by IFA, immunoperoxidase (IP) procedures, enzyme-linked immunosorbent assay (ELISA), and sequence analysis, there is a close antigenic and genomic relationship between TCoV and IBV (Akin et al., 2001, Guy et al., 1997, Lin et al., 2002, Loa et al., 2000, Stephensen et al., 1999). In addition, experimental infection studies using the animals have showed that TCoV could replicate in the enteric tissues of chickens without causing clinical signs (Gelb et al., 1987, Guy, 2000), BCoV could replicate in the intestines of turkeys with no or mild clinical diseases (Dea et al., 1991, Ismail et al., 2001), and IBV could maintain in the blood of infected turkeys up to 48 h via intravenous inoculation (Gelb et al., 1987). Thus, rapid, accurate, and specific detection of TCoV is critical to the control and eradication of turkey enteritis in the turkey flocks. Two RRT-PCR assays for the detection of avian enteritis viruses have been reported and could be applied to the detection of TCoV. One assay was designed to target M gene of TCoV and the other was targeted 5′ un-translated region of IBV genome. However, both gene sequences are conserved between TCoV and IBV (Callison et al., 2006, Spackman et al., 2005). Therefore, both assays could not detect TCoV differentially. The present study targeted the unique region of TCoV S gene that is specific to TCoV and conserved among various TCoV isolates and could detect, differentiate, and quantitate TCoV specifically.
The sensitivity and reproducibility of the RRT-PCR assay in the present study was evaluated by generating an external standard curve with in vitro T7-transcribed sense RNA of plasmid inserted with gene fragment encoding carboxyl terminal spike protein of TCoV. The standard curve was generated for each independent run of samples for the accuracy of absolute quantification. The limit of detection of TCoV-specific RRT-PCR assay was as low as 102 copies/μl, more sensitive than that by using RRT-PCR assay targeting M gene of TCoV in which the detection limit for a single target test in TCoV was 1100 copies and for multiplex with the genes of other enteric viruses was 2200 copies (Spackman et al., 2005). In addition, one advantage of the TCoV-specific RRT-PCR assay in the present study was the wide range of linearity of standard curve from 102 to 1010 copies, rendering the application of such assay to different kinds of specimens with various levels of virus load.
It is critical to validate a newly developed molecular assay by testing with a number of known negative and positive samples. In the present study, experimentally infected intestinal tissues, cloacal fecal samples, and/or fecal droppings were collected and subjected to the analysis by IFA and RRT-PCR, respectively. Six out of the total 84 samples collected at 12 h and 1 day post-inoculation in the trial 1 were tested negative for TCoV by both assays. The viral load in those samples could be expected too low to be detected by these two assays. Thirty-two samples that had low concentration of TCoV were only detected positive by RRT-PCR but not by IFA, indicating RRT-PCR is more sensitive than IFA on the detection of TCoV. There was one sample positive for TCoV by IFA, but negative by RRT-PCR. It may be caused by the loss of TCoV RNA during processing of intestine or during RNA extraction. Furthermore, in addition to specific and sensitive detection of TCoV, RRT-PCR can provide absolute quantitative data of TCoV in tissue specimens.
In the trial 1, TCoV can be detected as early as 12 h post-inoculation by the developed RRT-PCR. In duodenum, the highest concentration of TCoV was measured on day 1. The TCoV concentration was significantly lower in the duodenum than that in the jejunum, ileum and cecum. At 3 and 14 dpi, the TCoV level was significantly higher in the ileum than that in the other 3 segments. Therefore, the viral load varied through 14 days after inoculation, but TCoV were concentrated in the jejunum and ileum after 3 dpi. The results were consistent with previous studies showing that TCoV could be detected in the intestinal contents of experimentally inoculated turkeys for up to 6 weeks post-inoculation by virus isolation and 7 weeks post-inoculation by a RT-PCR procedure (Breslin et al., 2000).
In the trial 2, TCoV was detected from the cloacal swab samples sequentially for 14 days from the same three turkeys inoculated with 100 EID50 of TCoV. The dynamic changes of viral load from each turkey from 1 to 14 dpi were most likely related to the replication of the virus, reintroduction of the virus from the environment, the damage of intestinal epithelial cells, or the regeneration of epithelial cells. In the trial 3, TCoV in the fecal droppings on the floor could be detected and quantified by RRT-PCR, but TCoV levels in the fecal droppings were 2 logs less than those from the cloacal fecal swabs. Such findings indicated the possibilities of TCoV RNA degradation in the fecal droppings exposed in the environment. The feasibility of using the cloacal swabs and fecal droppings as the sample sources for specific detection and quantitation of TCoV by RRT-PCR increases the practical application of RRT-PCR in the diagnosis of TCoV in the field situation greatly because necropsy of animals to collect intestinal segments for RRT-PCR can be avoided and the time to carry out RRT-PCR can be shortened.
In conclusion, the TCoV S gene-targeted RRT-PCR developed in the present study can detect, differentiate, and quantify TCoV specifically in the intestines, cloacal fecal swabs, and fecal droppings on the floor.
Acknowledgement
The authors thank the support provided by the United States Department of Agriculture.
References
- Akin A., Lin T.L., Wu C.C., Bryan T.A., Hooper T., Schrader D. Nucleocapsid protein gene sequence analysis reveals close genomic relationship between turkey coronavirus and avian infectious bronchitis virus. Acta Virol. 2001;45:31–38. [PubMed] [Google Scholar]
- Breslin J.J., Smith L.G., Barnes H.J., Guy J.S. Comparison of virus isolation, immunohistochemistry, and reverse transcriptase-polymerase chain reaction procedures for detection of turkey coronavirus. Avian Dis. 2000;44:624–631. [PubMed] [Google Scholar]
- Broer R., Boson B., Spaan W., Cosset F.L., Corver J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 2006;80:1302–1310. doi: 10.1128/JVI.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E., Jackwood M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138:60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Dea S., Verbeek A., Tijssen P. Transmissible enteritis of turkeys: experimental inoculation studies with tissue-culture-adapted turkey and bovine coronaviruses. Avian Dis. 1991;35:767–777. [PubMed] [Google Scholar]
- Decaro N., Pratelli A., Campolo M., Elia G., Martella V., Tempesta M., Buonavoglia C. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J. Virol. Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb J., Fries P.A., Jr., Crary C.K., Donahoe J.P., Jr., Roessler D.E. Sentinel bird approach to isolating infectious bronchitis virus. J. Am. Vet. Med. Assoc. 1987;190:1628. [Google Scholar]
- Gut M., Leutenegger C.M., Huder J.B., Pedersen N.C., Lutz H. One-tube fluorogenic reverse transcription-polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods. 1999;77:37–46. doi: 10.1016/S0166-0934(98)00129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S. Turkey coronavirus is more closely related to avian brochitis virus than to mammalian coronaviruses. Avian Pathol. 2000;29:206–212. doi: 10.1080/03079450050045459. [DOI] [PubMed] [Google Scholar]
- Guy J.S., Barnes H.J., Smith L.G., Breslin J. Antigenic characterization of a turkey coronavirus identified in poult enteritis- and mortality syndrome-affected turkeys. Avian Dis. 1997;41:583–590. [PubMed] [Google Scholar]
- Holland P.M., Abramson R.D., Watson R., Gelfand D.H. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.W., Travanty E.A., Jeffers S.A., Smith M.K., Wennier S.T., Thackray L.B., Holmes K.V. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell–cell fusion. J. Virol. 2008;82:2883–2894. doi: 10.1128/JVI.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui R.K., Zeng F., Chan C.M., Yuen K.Y., Peiris J.S., Leung F.C. Reverse transcriptase PCR diagnostic assay for the coronavirus associated with severe acute respiratory syndrome. J. Clin. Microbiol. 2004;42:1994–1999. doi: 10.1128/JCM.42.5.1994-1999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W.A., Liddington R.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail M.M., Cho K.O., Ward L.A., Saif L.J., Saif Y.M. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. 2001;45:157–163. [PubMed] [Google Scholar]
- Lin T.L., Loa C.C., Wu C.C. Existence of gene 5 indicates close genomic relationship of Turkey coronavirus to infectious bronchitis virus. Acta Virol. 2002;46:107–116. [PubMed] [Google Scholar]
- Lin T.L., Loa C.C., Wu C.C. Complete sequences of 3′ end coding region for structural protein genes of turkey coronavirus. Virus Res. 2004;106:61–70. doi: 10.1016/j.virusres.2004.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T., Thacker H.L., Hooper T., Schrader D. Humoral and cellular immune responses in turkey poults infected with turkey coronavirus. Poult. Sci. 2001;80:1416–1424. doi: 10.1093/ps/80.10.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Hooper T.A., Schrader D.L. Differential detection of turkey coronavirus, infectious bronchitis virus, and bovine coronavirus by a multiplex polymerase chain reaction. J. Virol. Methods. 2006;131:86–91. doi: 10.1016/j.jviromet.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loa C.C., Lin T.L., Wu C.C., Bryan T.A., Thacker H.L., Hooper T., Schrader D. Detection of antibody to turkey coronavirus by antibody-capture enzyme-linked immunosorbent assay utilizing infectious bronchitis virus antigen. Avian Dis. 2000;44:498–506. [PubMed] [Google Scholar]
- Loa C.C., Wu C.C., Lin T.L. Comparison of 3′-end encoding regions of turkey coronavirus isolates from Indiana, North Carolina, and Minnesota with chicken infectious bronchitis coronavirus strains. Intervirology. 2006;49:230–238. doi: 10.1159/000091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Manopo I., Leung B.P., Chng H.H., Ling A.E., Chee L.L., Ooi E.E., Chan S.W., Kwang J. Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Sekiguchi Y., Shirai J., Taniguchi T., Honda E. Development of reverse transcriptase PCR and nested PCR to detect porcine hemagglutinating encephalomyelitis virus. J. Vet. Med. Sci. 2004;66:367–372. doi: 10.1292/jvms.66.367. [DOI] [PubMed] [Google Scholar]
- Senne D.A. Virus propagation in embryonating eggs. In: Swayne D.E., Senne D.A., Beard C.W., editors. A Laboratory Manual for the Isolation and Identification of Avian Pathogens. 5th ed. America Association of Avian Pathologists, Inc.; Athens, Georgia: 2008. pp. 204–208. [Google Scholar]
- Sheahan T., Rockx B., Donaldson E., Corti D., Baric R. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J. Virol. 2008;82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E., Kapczynski D., Sellers H. Multiplex real-time reverse transcription-polymerase chain reaction for the detection of three viruses associated with poult enteritis complex: turkey astrovirus, turkey coronavirus, and turkey reovirus. Avian Dis. 2005;49:86–91. doi: 10.1637/7265-082304R. [DOI] [PubMed] [Google Scholar]
- Stephensen C.B., Casebolt D.B., Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


