Highlights
► Non-proliferating CV-A16 pseudoviruses were generated by reverse genetics. ► A novel pseudovirus-based CV-A16 neutralization assay (pCNA) was developed. ► The kinetics of CVA16 pseudovirus infectivity was determined in vitro. ► Screening with the pCNA correlated well with a conventional neutralization assay. ► The pCNA may be a rapid screening test for CV-A16 infection.
Keywords: Coxsackievirus A16, Pseudovirus, Luciferase, Neutralizing antibody assay
Abstract
Serum neutralizing antibody titers are indicative of protective immunity against Coxsackievirus A16 (CV-A16) and Enterovirus 71 (EV71), the two main etiological agents of hand, foot and mouth disease (HFMD), and provide the basis for evaluating vaccine efficacy. The current CV-A16 neutralization assay based on inhibition of cytopathic effects requires manual microscopic examination, which is time-consuming and labor-intensive. In this study, a high-throughput neutralization assay was developed by employing CV-A16 pseudoviruses expressing luciferase for detecting infectivity in rhabdomyosarcoma (RD) cells and measuring serum viral neutralizing antibodies. Without the need to use infectious CV-A16 strains, the neutralizing antibody titer against CV-A16 could be determined within 15 h by measuring luciferase signals by this assay. The pseudovirus CV-A16 neutralization assay (pCNA) was validated by comparison with a conventional CV-A16 neutralization assay (cCNA) in testing 174 human serum samples collected from children (age <5 years). The neutralizing antibody titers determined by these two assays were well correlated (R2 = 0.7689). These results suggest that the pCNA can serve as a rapid and objective procedure for the measurement of neutralizing antibodies against CV-A16.
1. Introduction
Hand, foot and mouth disease (HFMD) is a common illness in children, but no effective vaccine or antiviral drug is yet available. Coxsackievirus A16 (CV-A16) and Enterovirus 71 (EV71) are two major etiological agents of HFMD (Hosoya et al., 2007, Ang et al., 2009, Solomon et al., 2010). The seroprevalence for individuals aged ≥1 year was found to be nearly 60% for CV-A16 and 40% for EV71 (Rabenau et al., 2010). CV-A16 has been classified into three genogroups and several subgroups (A, B1a, B1b, B2a, B2b and C) based on phylogenetic analyses of the VP4 or VP1 gene (Li et al., 2005, Hosoya et al., 2007, Perera et al., 2007, Zhou et al., 2010, Chen et al., 2012).
The illness caused by CV-A16 infection is usually mild (Chang et al., 1999), whereas EV71 infection is often associated with severe complications, such as brainstem encephalitis, severe pulmonary edema and shock, and significant mortality (Lee et al., 2009, Wong et al., 2010). However, recent findings suggest that the recombination between co-circulating CV-A16 and EV71 (Yip et al., 2010, Zhang et al., 2010) may have contributed to the increase of HFMD cases in China over the past few years (Zhang et al., 2009). Furthermore, CV-A16 infection is not always benign, as fatal cases associated with the virus have been reported (Cooper et al., 1989, Wang et al., 2004). Thus, further understanding of the virology and epidemiology of CV-A16 is important for developing improved diagnostic tests and vaccines.
A serum neutralizing antibody response is the major indicator of EV71 or CV-A16 infection and protective immunity and, thus, is used to evaluate vaccine efficacy for HFMD. The neutralization assay based on inhibition of cytopathic effects (CPE) is a standard method recommended by the World Health Organization (WHO) for measuring neutralizing antibodies against polioviruses and also has been applied widely to determine neutralizing antibody titers against CV-A16 and EV71 (Chang et al., 2002, Kung et al., 2007). However, the conventional CV-A16 neutralization assay (cCNA) is labor-intensive, subjective and time-consuming (at least 4 days). Therefore, it is not suitable for mass screening of protective antibodies in seroepidemiological studies. Recently, simplified neutralization assays employing pseudoviruses have been applied in detection of neutralizing antibodies against multiple enveloped viruses, including influenza, severe acute respiratory syndrome coronovirus (SARS-coV) (Temperton et al., 2005, Kobinger et al., 2007, Wright et al., 2008), hepatitis C virus (Bartosch et al., 2003, Keck et al., 2005, Netski et al., 2005), human immunodeficiency virus (HIV) (Montefiori, 2005) and rabies (Wright et al., 2008). However, packaging signals are poorly understood for non-enveloped viruses such as CV-A16, making it thus far difficult to package pseudoviral particles for detection of neutralizing antibodies in human serum.
The generation is described of non-proliferating CV-A16 pseudoviruses (luciferase-encoding CV-A16 replicons encapsidated by viral P1 capsid proteins) through triple transfection of 293T cells with a T7 RNA polymerase expression plasmid (pcDNA3.1-T7 RNA Pol), a plasmid containing the reverse-transcribed full-length CV-A16 genome encoding the firefly luciferase gene in place of the P1 gene (pBS-T7-CV-A16-luc) and a CV-A16 capsid expression plasmid (VR1012-5′ NTR-P1). Using these non-proliferating pseudoviruses, a novel CV-A16 neutralization assay (pCNA) was developed in which neutralizing antibody titers could be determined based on the luciferase signals in inoculated cells within 15 h. The results suggest that the pCNA may serve as a rapid and objective procedure for screening neutralizing antibody titers against CV-A16.
2. Materials and methods
2.1. Cells, viruses and human sera
RD cells (human embryonic rhabdomyosarcoma) and 293T cells (SV40 transfected human embryonic kidney 293 cells) were cultured as monolayers in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). The CV-A16 strain HN1129/CHN/2010 (B1b genotype, a gift kindly provided by the Henan Provincial Center for Disease Control and Prevention, Zhengzhou, China) was grown in RD cells and used for the cCNA in this study. Human sera were collected from children (age <5 years) at the Henan Provincial Center for Disease Control and Prevention and used for the detection of CV-A16 neutralizing antibodies by both the cCNA and pCNA.
2.2. Construction of plasmids
RNA was extracted from virus-infected RD cells using Trizol (Invitrogen, Carlsbad, CA, USA) and reverse transcribed using oligo(dT) primers and M-MLV reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The resulting cDNA was used for amplification of the CV-A16 5′ NTR-P1 fragment with forward primer 5′ATGCATGCGCGGCCGCTTAAAACAGCCTGTGGGTTG3′ and reverse primer 5′CTGGTCTAGATTACAGTGTTGTTATCTTGTATCTAC3′. The PCR product was subcloned into the NotI/XbaI sites of the VR1012 vector to produce VR1012-5′ NTR-P1.
The T7 RNA polymerase gene (GenBank ID: M38308) was commercially synthesized (Generay Bio Co. Ltd., Shanghai, China) and inserted into the NotI/XbaI sites of the pcDNA3.1(−) vector (Invitrogen). The resulting plasmid, pcDNA3.1-T7 RNA Pol, expresses T7 RNA polymerase under control of the strong human CMV immediate early promoter.
The commercially synthesized pBS-T7-CV-A16-luc plasmid (GenBank ID: JQ180469) contains the reverse transcribed full-length CV-A16 genome with the firefly luciferase gene in place of the P1 region (Arita et al., 2008), including a complete 5′ non-translated region (NTR) and 3′ NTR. The hepatitis delta virus (HDV) ribozyme (Rib) was added downstream of the 3′ NTR, followed by the T7 terminator (TT7) (Troupin et al., 2010). After transfection of this plasmid into 293T cells, the T7 RNA pol expressed by pcDNA3.1-T7 RNA Pol drives the transcription of the CV-A16 mRNA from the first nucleotide (T) of the 5′ end to the last nucleotide (A) of the 3′ end after ribozyme cleavage. Although this plasmid carries only a T7 promoter and no additional eukaryotic promoter sequences, spontaneous formation of pseudoparticles was observed after trans-encapsidation by P1 capsid proteins resulting from co-transfection with the VR1012-5′ NTR-P1 and pcDNA3.1-T7 RNA Pol plasmids (Fig. 1 ).
Fig. 1.
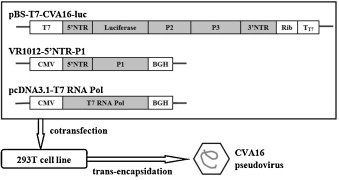
Generation of CV-A16 pseudoviruses. 293T cells were triple-transfected with the indicated plasmids to produce CV-A16 pseudoviruses containing luciferase. The pBS-T7-CV-A16-luc plasmid contains the reverse-transcribed full-length CV-A16 genome with the firefly luciferase gene in place of the P1 region. The VR1012-5′ NTR-P1 plasmid expresses the CV-A16 P1 gene. The pcDNA3.1-T7 RNA Pol plasmid expresses T7 RNA polymerase under control of the strong human CMV immediate early promoter. BGH: the bovine growth hormone polyadenylation (BGH-PolyA) signal, a specialized termination sequence for protein expression in eukaryotic cells.
2.3. Generation of CV-A16 pseudoviruses containing the firefly luciferase gene in 293T cells
A 70% confluent monolayer of 293T cells in a T75 flask (BD Biosciences, Franklin Lakes, NJ, USA) was triple-transfected with pcDNA3.1-T7 RNA Pol, pBS-T7-CV-A16-luc and pcDNA3.1-5′ NTR-P1 (3.3 μg each) using 30 μl Lipofectamine 2000 reagent (cat# 11668-019, Invitrogen) and then incubated at 37 °C in 15 ml of DMEM (10% FCS). The cells were washed, and the medium was replaced with DMEM (10% FCS) at 6 h post-transfection. After incubation for 48 h, the cells were harvested and stored at −80 °C.
2.4. Reverse transcription-PCR (RT-PCR)
Supernatants from the triple-transfected 293T cells were collected and extracted with an equal volume of chloroform, followed by RNase (15 mg/ml) and DNase (20 U/ml) treatment for 20 min at 37 °C. RT-PCR was performed using RevatraAce reverse transcriptase (TaKaRa, Dalian, China) and the primer pair: 5′ NTR-F (5′TTAAAACAGCCTGTGGGTTG3′) and Luc-R (5′CACGGCGATCTTTCCGCCCTTCT3′). The PCR products were purified by using a PCR purification kit (Tiangen, Beijing, China). Direct sequence analysis was carried out on the full-length genomic sequence of CV-A16 (GenBank ID: JQ180469) using DNA fragments amplified by RT-PCR as templates.
2.5. Transmission electron microscopy (TEM)
To perform electron microscopy, CV-A16 pseudoviruses were purified from the triple-transfected 293T cells. Briefly, after two rounds of freezing and thawing, the transfected 293T cell lysates were filtered through a 0.22-μm membrane. CV-A16 pseudoviruses were purified from the resulting suspension using DEAE-Sepharose (Amersham Pharmacia Biotech, Piscataway, NJ, USA) (Horodniceanu et al., 1979), followed by centrifugation at 100,000 × g for 2.5 h at 4 °C in a Beckman SW40 rotor through a 1-ml 30% sucrose cushion. The pellet was washed three times with distilled water and dissolved in 500 μl of phosphate-buffered saline (PBS). Samples were examined by TEM (JEM-1220; JEOL Datum, Tokyo, Japan) at an acceleration voltage of 80 kV, and the images were obtained at a magnification of 200,000×.
2.6. Western blotting
Purified virus stocks were mixed with SDS–PAGE sample buffer, boiled and separated on 13.5% polyacrylamide gels. Proteins were transferred onto PVDF membranes for Western blot analysis. Membranes were probed with a mouse anti-VP1 polyclonal antibody (a gift kindly provided by the National Institutes for Food and Drug Control, Zhengzhou, China) diluted 1:1000 in PBS with 0.05% Tween-20 and 1% skim milk, followed by a corresponding alkaline phosphatase (AP)-conjugated secondary antibody (Sigma, St. Louis, MO, USA) diluted 1:1000. The membranes were developed by chemiluminescence using nitro-blue tetrazolium (NBT, cat# DH2133, Dinguo, Beijing, China) and 5-bromo-4-chloro-3′-indolyphosphate (BCIP, cat# DH0326, Dinguo).
2.7. Titration of pseudovirus particles
The number of non-proliferating pseudovirus particles (pseudovirus titer) was determined by measuring the 50% cell culture infective dose (CCID50) using a microtitration assay performed with RD cells cultured in 96-well plates (BD Biosciences). Briefly, pseudoviruses from supernatants collected after freeze–thawing of the triple-transfected 293T cells were diluted serially 5-fold in DMEM (10% FCS) and used to inoculate RD cells (4 × 104 cells per well) in quadruplicate per dilution in 96-well plates (BD Biosciences). After incubation for 15 h, the cells were harvested for measuring luciferase activity using a luciferase assay kit (cat# E2650, Promega, Madison, WI, USA) according to the manufacturer's recommendations. Briefly, cells were lysed by addition of 100 μl of reporter lysis buffer (Promega), and the luciferase activity in cell lysates (100 μl per well) was measured using the Fluoroskan Ascent FL Microplate Fluorometer (Thermo Scientific, Waltham, MA, USA). CCID50 values were determined as the dilution that resulted in CV-A16 pseudovirus infectivity of 50% (Montefiori, 2005), and 2.5 times the control cell group value was the cut-off value to determine if a well was successfully infected. Here, “CCID50” reflects the titer of the non-proliferating pseudovirus, while “TCID50” reflects that of the replication-competent virus.
2.8. pCNA
Before carrying out the pCNA, the optimal incubation time with pseudoviruses was determined. Briefly, RD cells (4 × 104 per well) were seeded into 96-well microtiter plates using the medium described above and incubated with CV-A16 pseudoviruses at the multiplicity of infection (MOI) of 5 CCID50 per cell. The cells were washed at 2 h post-infection (hpi), and then luciferase activities in the cells were measured at designated time points (0–24 h).
For the pCNA, each human serum sample (30 μl) was initially diluted 8-fold with DMEM supplemented with 2% FCS, followed by 2-fold serial dilutions (1:8 to 1:1024 dilutions). Subsequently, 50 μl of diluted serum or DMEM supplemented with 2% FCS (mock treatment for the pseudovirus positive control) was added to a 96-well plate (2 wells per dilution in 1 plate). CV-A16 pseudoviruses (400 CCID50 in 50 μl) were added to each well of the plate and incubated at 37 °C for 1 h. After the incubation, RD cells (4 × 104 in 100 μl DMEM supplemented with 2% FCS) were added to each well, and then the plates were incubated at 37 °C for 15 h. Based on the results of the optimization experiment described above, luciferase activities of the infected cells were measured at 15 hpi. Infectivity was calculated as a percentage of the luciferase activity in the cells infected with CV-A16 pseudovirus pre-incubated with serum, relative to that of the cells infected with the pseudovirus positive control (set at 100%). A decrease of infectivity below a threshold value was considered positive neutralization. The reciprocal of the highest serum dilution showing an infectivity value equal to or less than the threshold value was determined to be the neutralizing antibody titer.
2.9. cCNA
A suspension of infectious CV-A16 was serially diluted 10-fold and incubated with RD cells in 96-well microtitration plates to determine viral infectivity titers. CPE was observed using an inverted microscope after an incubation period of 7 days. The 50% tissue culture infectious doses (TCID50) of CV-A16 were calculated by the Reed and Muench method (1938). Sera were heat-inactivated at 56 °C for 30 min, diluted serially 2-fold from 1:8 to 1:1024 and mixed with an equal volume of the CV-A16 inoculum (50 μl containing 100 TCID50/well of virus) at 37 °C for 1 h in 96-well microtiter plates. RD cells were added at 4 × 104/well and incubated for 7 days. Serum samples were tested in triplicate, and the neutralization titers were read as the highest dilution that completely inhibited CPE in over 50% of the wells. A positive control serum with a known titer was included in each run.
2.10. Statistical analysis
Results were obtained from at least three replicates and reported as the mean ± standard deviation (SD). All statistical analyses were carried out with the GraphPad Prism software package. Groups were compared by using Student's t-test. P values <0.05 were considered significant (Motulsky and Christopoulos, 2004).
3. Results
3.1. Generation of CV-A16 pseudoviruses containing firefly luciferase in 293T cells
To obtain CV-A16 pseudoviruses containing the firefly luciferase gene in place of the P1 gene (CV-A16-luc), 293T cells were triple-transfected with pT7-CV-A16-luc, pcDNA3.1-5′ NTR-P1 and pcDNA3.1-T7 RNA Pol. pcDNA3.1-5′ NTR-P1 provided the source of P1 proteins, while pcDNA3.1-T7 RNA Pol provided the source of T7 RNA polymerase for transcribing the CV-A16 replicon plasmid pT7-EV71-luc. Two days after the transfection, the 293T cells were lysed by freeze–thawing, and P1 protein encapsidated CV-A16 replicon particles expressing luciferase were collected in the supernatant after clarifying the lysate by centrifugation. This CV-A16 pseudovirus stock was titered at 1.0 × 105 CCID50/ml.
An RT-PCR with the primers 5′ NTR-F and Luc-R was performed to test for the production of CV-A16 pseudovirus particles in the supernatant of transfected 293T cells. A PCR product of 2.3 kb corresponding to the fragment expected with these primers was obtained (Fig. 2B, lane 2). The nucleotide sequence of the 2.3-kb segment was found to match completely with pBS-T7-CV-A16-luc by alignment (Sequencher 4.9 software). The formation of CV-A16 pseudoviruses was also confirmed by Western blotting (Fig. 2A), which revealed a specific band at 35 kDa (Fig. 2A, lane 3) corresponding to the expected size of viral VP1 proteins. Furthermore, TEM analyses revealed viral-like particles of 25–30 nm diameter, which were consistent in shape and size with wild-type CV-A16 virus (Fig. 2C).
Fig. 2.

Characterization of CV-A16 pseudoviruses. (A) Western blot analysis of purified CV-A16 pseudoviruses using mouse anti-VP1 mAb. 1, marker; 2, non-transfected 293T cell lysates; 3, CV-A16-luc. The band corresponding to viral VP1 proteins (VP1) is indicated with an arrow. (B) Electrophoretic profile of PCR products of CV-A16 pseudoviruses. Lane 1, supernatants collected from cells transfected with pBS-T7-CV-A16-luc and pcDNA3.1-T7 RNA Pol; lane 2, supernatants collected from cells transfected with pBS-T7-CV-A16-luc, pcDNA3.1-T7 RNA Pol and VR1012-5′ NTR-P1; M, 1 kb DNA ladder (New England Biolabs: N3232). (C) Transmission electron microscopic observation of purified CV-A16 pseudoviruses. Bar, 50 nm.
3.2. Determination of the optimal incubation time and virus input for pCNA
The kinetic pattern of CV-A16-luc infectivity was first determined by measuring luciferase activity of infected RD cells over a time course (Fig. 3A). At about 15 hpi, the luciferase activity peaked and then gradually decreased with time. Different doses of CV-A16 pseudovirus (200, 400 and 800 CCID50) were incubated with serial dilutions of a standard mouse anti-CV-A16 serum (containing 10 units of neutralizing antibodies against CV-A16 in 5 μl). Neutralization activity was evaluated by the cCNA. As shown in Fig. 3B, the standard anti-CV-A16 serum could neutralize infectivity of CV-A16 pseudoviruses over the range of titers tested, and there was a good dose–response relationship between luciferase signals and antiserum dilutions with the virus input of 400 CCID50/well. Therefore, this virus dose and the incubation time of 15 h were used in the subsequent assays.
Fig. 3.
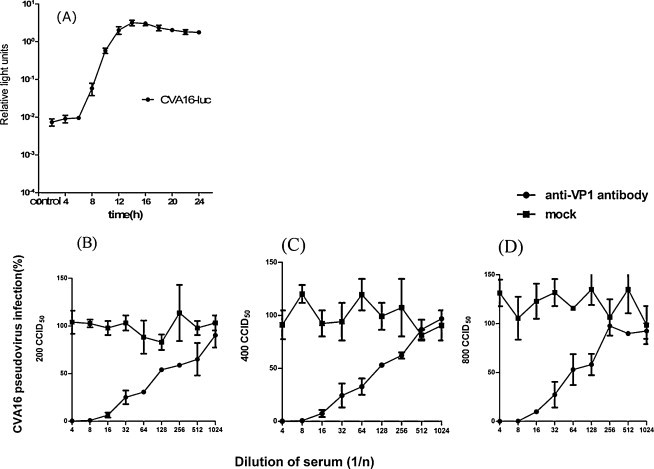
Determination of the optimal incubation time (A) and virus input (B–D) for pCNA. (A) Kinetics of CV-A16 pseudovirus infectivity in RD cells inoculated at MOI = 5 CCID50 per cell. The cells were washed at 2 hpi, and then the luciferase activities in the cells were measured at the indicated times. Data are shown from one out of three experiments with similar results, each performed in quadruplicate. (B–D) Optimization of the CV-A16 pseudovirus titer for the pCNA. Neutralization curves of CV-A16 pseudoviruses (200, 400 and 800 CCID50) with the standard anti-CV-A16 serum are shown. Error bars indicate standard deviations.
3.3. Comparison of cCNA and pCNA for measurement of neutralizing antibody titers against CV-A16 in human sera
The pCNA was carried out on 174 human serum samples, and the results were compared with those obtained by the cCNA. The data were analyzed using various threshold levels in order to determine the value which would give an anti-CV-A16 neutralizing antibody-positive rate in the samples (the percentage of samples that showed neutralizing antibody titers of ≥8) closest to that determined by the cCNA (Fig. 4 ). With the threshold value of 20% CV-A16-luc infectivity, a good correlation was observed between the neutralizing antibody titers determined by cCNA and pCNA (R 2 = 0.7689) (Fig. 5 ). No statistically significant difference was found between the neutralizing antibody titers by cCNA (4.471 ± 2.696) and pCNA (4.741 ± 2.942) (P < 0.0001, t = 4.679). Disagreement was observed in only three serum samples, which were cCNA negative and pCNA low positive (with titers of 8, 8 and 16) (Fig. 5). Thus, it seems likely that these three samples had extremely low titers to CV-A16 and that the pCNA was more sensitive than the cCNA. This assumption was also supported by the observation that the neutralizing antibody titers measured using the pCNA were higher than those measured by the cCNA (Fig. 6 ). However, 16 of the 174 sera had a >2-fold difference between the neutralizing antibody titers determined by these two methods (Fig. 5), and the neutralization titers of several serum samples exceeded the highest dilution (1:512 or 1:1024) used in this study. Therefore, the exact correlation coefficient could not be obtained.
Fig. 4.
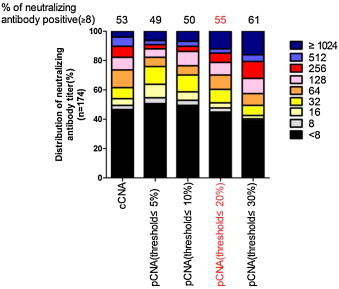
Determination of threshold values for CV-A16 psedoviruses in pCNA for human sera. Distribution of neutralizing antibody titers in human serum samples (n = 174) determined by the pCNA with a range of threshold values. The results were compared with those obtained by the cCNA. The neutralizing antibody-positive rates of the samples were determined for each threshold value and are shown above the graphs. The threshold value of 20% showed neutralizing-antibody positive rates closest to those obtained by the cCNA (highlighted in red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 5.
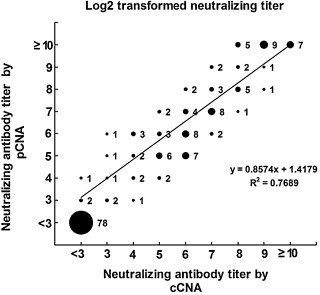
Correlation between CV-A16 neutralizing antibody titers from pCNA and cCNA. Neutralizing titers were measured by the cCNA and pCNA and log 2 transformed. The relative size of each spot corresponds to the number of the serum samples as indicated. R2 = 0.7689.
Fig. 6.
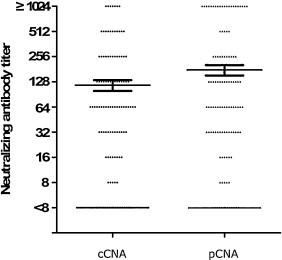
Distribution of neutralizing antibody titers measured by cCNA and pCNA in 174 serum samples.
4. Discussion
Pseudotype reporter viruses provide a safe, quantitative and high-throughput tool for assessing antibody neutralization for many enveloped viruses. Pseudoviruses undergo the steps of infection that include attachment, fusion and expression of the reporter gene, but they cannot form infectious progeny due to the lack of viral structural genes needed for several steps of the viral life cycle. Thus, pseudoviruses do not propagate beyond a single cycle of infection. In this study, non-proliferating CV-A16 pseudoviruses were generated by encapsidating luciferase-encoding CV-A16 replicons in viral P1 capsid proteins via triple transfection of 293T cells, and an assay based on these pseudoviruses for measurement of CV-A16 neutralizing antibody titers in human serum was developed.
A direct comparison of neutralizing antibody titers revealed a good correlation between the pCNA and cCNA (Fig. 5), but the pCNA appeared to be more sensitive (Fig. 6). The improved sensitivity is likely attributed to the inability of CV-A16 pseudoviruses to undergo multiple cycles of replication due to the lack of a P1 gene. Indeed, neutralization of the CV-A16 pseudoviruses can, therefore, be achieved with neutralizing antibodies which inhibit the initial binding of the viruses to the cells. When serum neutralization assays employ replication-competent viruses which can multiply after successfully entering the cell, typically greater levels of neutralizing antibodies are required to inhibit further viral spread, often resulting in an apparently lower neutralization titer. The cost of the cCNA is low, but requires at least 4 days to complete with manual microscopic examination and special expertise that may produce subjective variations among operators and laboratories. In this study, the processing time of the pCNA was shortened to 15 h (Fig. 3) by the use of CV-A16 pseudoviruses and detection of luciferase signals as the measure of infectivity. Therefore, the pseudovirus neutralization assay established in this study was demonstrated to be sensitive, safe, rapid, objective and therefore especially suited for rapid high-throughput screening of sera.
For other pseudovirus neutralization assays, threshold values arbitrarily set within a range of 10–50% generally give results that correlate well with those obtained by conventional assays, especially for determining the neutralization antibody-positive rates (Fukushi et al., 2006, Tamin et al., 2009, Wang et al., 2008). Fine-tuning of threshold values may be useful for linking the results obtained by different neutralization methods. In this study, the pCNA threshold value of 20% gave antibody-positive rates closest to those obtained by cCNA (Fig. 4).
CV-A16 is classified into three genogroups and several subgroups (A, B1a, B1b, B2a, B2b and C). In this study, only the P1 gene from the B1b genogroup was tested in the pCNA. However, it is possible to interchange the P1 gene with those of other genogroups, since the P1 capsid proteins from different CV-A16 genogroups and even from EV71 have been found to be capable of trans-encapsidating the CV-A16 replicon (data not shown). Therefore, the pCNA may be conveniently and quickly adapted to test antibody neutralization of all genogroups of CV-A16 and EV71 viruses, providing a solid platform for the development of a high-throughput surveillance system for HFMD that can be used in rural clinics in Asia. More importantly, this study has also paved the way for the development of a safe and inexpensive vaccine, which may be potentially used to protect children from infections with EV71 or CV-A16 of diverse genogroups.
5. Conclusions
Development of prophylactic vaccines against HFMD is a public health priority in many countries in Asia, and neutralizing antibody responses will be an important indicator for the evaluation of the immunogenicity of different CV-A16 vaccine candidates. In this study, a high-throughput CV-A16 neutralization assay using non-proliferating CV-A16 pseudovirus was developed, which may serve as a sensitive, safe, rapid and objective procedure for measurement of neutralizing antibody titers against CV-A16 in human serum. Thus, the pCNA can provide a robust method for coordinating global CV-A16 surveillance.
Competing interest
The authors declare no competing interests.
Ethical approval
This study was conducted according to the guidelines laid down in the Declaration of Helsinki and was approved by ethical review boards of Jilin University in Changchun, China. As all participants were below 5 years of age, written informed consent was obtained from their parents.
Acknowledgements
The authors would like to thank Xianghui Yu (College of Life Science, Jilin University) for editorial assistance. This work was supported in part by the National Natural Science Foundation of China (NSFC, project #: 20872048).
Contributor Information
Jun Jin, Email: jinjunjlu@126.com.
Hongxia Ma, Email: 64103168@qq.com.
Lin Xu, Email: xulinjlu@yahoo.cn.
Dong An, Email: andonginlab@sina.cn.
Shiyang Sun, Email: sunbaoshiyang@163.com.
Xueyong Huang, Email: huangxy@hncdc.com.cn.
Wei Kong, Email: weikong@jlu.edu.cn.
Chunlai Jiang, Email: jiangcl@jlu.edu.cn.
References
- Ang L.W., Koh B.K., Chan K.P., Chua L.T., James L., Goh K.T. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001–2007. Ann. Acad. Med. Singapore. 2009;38:106–112. [PubMed] [Google Scholar]
- Arita M., Ami Y., Wakita T., Shimizu H. Cooperative effect of the attenuation determinants derived from poliovirus sabin 1 strain is essential for attenuation of enterovirus 71 in the NOD/SCID mouse infection model. J. Virol. 2008;82:1787–1797. doi: 10.1128/JVI.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B., Bukh J., Meunier J.C., Granier C., Engle R.E., Blackwelder W.C., Emerson S.U., Cosset F.L., Purcell R.H. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.Y., King C.C., Hsu K.H., Ning H.C., Tsao K.C., Li C.C., Huang Y.C., Shih S.R., Chiou S.T., Chen P.Y., Chang H.J., Lin T.Y. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics. 2002;109:e88. doi: 10.1542/peds.109.6.e88. [DOI] [PubMed] [Google Scholar]
- Chang L.Y., Lin T.Y., Huang Y.C., Tsao K.C., Shih S.R., Kuo M.L., Ning H.C., Chung P.W., Kang C.M. Comparison of enterovirus 71 and coxsackie-virus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. Pediatr. Infect. Dis. J. 1999;18:1092–1096. doi: 10.1097/00006454-199912000-00013. [DOI] [PubMed] [Google Scholar]
- Chen L., Mou X., Zhang Q., Li Y., Lin J., Liu F., Yuan L., Tang Y., Xiang C. Detection of human enterovirus 71 and coxsackievirus A16 in children with hand, foot and mouth disease in China. Mol. Med. Rep. 2012;5:1001–1004. doi: 10.3892/mmr.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D.J., Shaw D.R., LaBrooy J.T., Blumbergs P., Gilbert J., Simmons A. Fatal rhabdomyolysis and renal failure associated with hand, foot and mouth disease. Med. J. Aust. 1989;151:232–234. doi: 10.5694/j.1326-5377.1989.tb115997.x. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Mizutani T., Saijo M., Kurane I., Taguchi F., Tashiro M., Morikawa S. Evaluation of a novel vesicular stomatitis virus pseudotype-based assay for detection of neutralizing antibody responses to SARS-CoV. J. Med. Virol. 2006;78:1509–1512. doi: 10.1002/jmv.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horodniceanu F., Panon G., Le Fur R., Barme M. Purification of poliovirus obtained from human diploid cells by means of DEAE-Sepharose. Dev. Biol. Stand. 1979;42:75–80. [PubMed] [Google Scholar]
- Hosoya M., Kawasaki Y., Sato M., Honzumi K., Hayashi A., Hiroshima T., Ishiko H., Kato K., Suzuki H. Genetic diversity of coxsackievirus A16 associated with hand, foot, and mouth disease epidemics in Japan from 1983 to 2003. J. Clin. Microbiol. 2007;45:112–120. doi: 10.1128/JCM.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck Z.Y., Li T.K., Xia J., Bartosch B., Cosset F.L., Dubuisson J., Foung S.K. Analysis of a highly flexible conformational immunogenic domain a in hepatitis C virus E2. J. Virol. 2005;79:13199–13208. doi: 10.1128/JVI.79.21.13199-13208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger G.P., Figueredo J.M., Rowe T., Zhi Y., Gao G., Sanmiguel J.C., Bell P., Wivel N.A., Zitzow L.A., Flieder D.B., Hogan R.J., Wilson J.M. Adenovirus-based vaccine prevents pneumonia in ferrets challenged with the SARS coronavirus and stimulates robust immune responses in macaques. Vaccine. 2007;25:5220–5231. doi: 10.1016/j.vaccine.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung S.H., Wang S.F., Huang C.W., Hsu C.C., Liu H.F., Yang J.Y. Genetic and antigenic analyses of enterovirus 71 isolates in Taiwan during 1998–2005. Clin. Microbiol. Infect. 2007;13:782–787. doi: 10.1111/j.1469-0691.2007.01745.x. [DOI] [PubMed] [Google Scholar]
- Lee T.C., Guo H.R., Su H.J., Yang Y.C., Chang H.L., Chen K.T. Diseases caused by enterovirus 71 infection. Pediatr. Infect. Dis. J. 2009;28:904–910. doi: 10.1097/INF.0b013e3181a41d63. [DOI] [PubMed] [Google Scholar]
- Li L., He Y., Yang H., Zhu J., Xu X., Dong J., Zhu Y., Jin Q. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 2005;43:3835–3839. doi: 10.1128/JCM.43.8.3835-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori D.C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005 doi: 10.1002/0471142735.im1211s64. 12.11.1–12.11.17. [DOI] [PubMed] [Google Scholar]
- Motulsky H., Christopoulos A. Oxford University Press; New York: 2004. Fitting Models to Biological Data Using Linear and Nonlinear Regression. A Practical Guide to Curve Fitting. [Google Scholar]
- Netski D.M., Mosbruger T., Depla E., Maertens G., Ray S.C., Hamilton R.G., Roundtree S., Thomas D.L., McKeating J., Cox A. Humoral immune response in acute hepatitis C virus infection. Clin. Infect. Dis. 2005;41:667–675. doi: 10.1086/432478. [DOI] [PubMed] [Google Scholar]
- Perera D., Yusof M.A., Podin Y., Ooi M.H., Thao N.T., Wong K.K., Zaki A., Chua K.B., Malik Y.A., Tu P.V., Tien N.T., Puthavathana P., McMinn P.C., Cardosa M.J. Molecular phylogeny of modern coxsackievirus A16. Arch. Virol. 2007;152:1201–1208. doi: 10.1007/s00705-006-0934-5. [DOI] [PubMed] [Google Scholar]
- Rabenau H.F., Richter M., Doerr H.W. Hand, foot and mouth disease: seroprevalence of Coxsackie A16 and Enterovirus 71 in Germany. Med. Microbiol. Immunol. 2010;199:45–51. doi: 10.1007/s00430-009-0133-6. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Solomon, Lewthwaite T., Perera P., Cardosa D., McMinn M.J., Ooi P., Virology M.H. Epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- Tamin A., Harcourt B.H., Lo M.K., Roth J.A., Wolf M.C., Lee B., Weingartl H., Audonnet J.C., Bellini W.J., Rota P.A. Development of a neutralization assay for Nipah virus using pseudotype particles. J. Virol. Methods. 2009;160:1–6. doi: 10.1016/j.jviromet.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperton N.J., Chan P.K., Simmons G., Zambon M.C., Tedder R.S., Takeuchi Y., Weiss R.A. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 2005;11:411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troupin C., Dehee A., Schnuriger A., Vende P., Poncet D., Garbarg-Chenon A. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J. Virol. 2010;84:6711–6719. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Y., Li Lu F., Wu M.H., Lee C.Y., Huang L.M. Fatal coxsackievirus A16 infection. Pediatr. Infect. Dis. J. 2004;23:275–276. doi: 10.1097/01.inf.0000115950.63906.78. [DOI] [PubMed] [Google Scholar]
- Wang W., Butler E.N., Veguilla V., Vassell R., Thomas J.T., Moos M., Jr., Ye Z., Hancock K., Weiss C.D. Establishment of retroviral pseudotypes with influenza hemagglutinins from H1, H3, and H5 subtypes for sensitive and specific detection of neutralizing antibodies. J. Virol. Methods. 2008;153:111–119. doi: 10.1016/j.jviromet.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Wong S.S., Yip C.C., Lau S.K., Yuen K.Y. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol. Infect. 2010;138:1071–1089. doi: 10.1017/S0950268809991555. [DOI] [PubMed] [Google Scholar]
- Wright E., Temperton N.J., Marston D.A., McElhinney L.M., Fooks A.R., Weiss R.A. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. J. Gen. Virol. 2008;89:2204–2213. doi: 10.1099/vir.0.2008/000349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C.C., Lau S.K., Zhou B., Zhang M.X., Tsoi H.W., Chan K.H., Chen X.C., Woo P.C., Yuen K.Y. Emergence of enterovirus 71 double-recombinant strains belonging to a novel genotype D originating from southern China: first evidence for combination of intratypic and intertypic recombination events in EV71. Arch. Virol. 2010;155:1413–1424. doi: 10.1007/s00705-010-0722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.M., Li C.R., Liu Y.J., Liu W.L., Fu D., Xu L.M., Xie J.J., Tan Y., Wang H., Chen X.C., Zhou B.P. To investigate pathogen of hand, foot and mouth disease in Shenzhen in 2008. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009;23:334–336. [PubMed] [Google Scholar]
- Zhang Y., Zhu Z., Yang W., Ren J., Tan X., Wang Y., Mao N., Xu S., Zhu S., Cui A., Yan D., Li Q., Dong X., Zhang J., Zhao Y., Wan J., Feng Z., Sun J., Wang S., Li D., Xu W. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol. J. 2010;7:94. doi: 10.1186/1743-422X-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Kong F., Wang B., McPhie K., Gilbert G.L., Dwyer D.E. Molecular characterization of enterovirus 71 and coxsackievirus A16 using the 5′ untranslated region and VP1 region. J. Med. Microbiol. 2010;60:349–358. doi: 10.1099/jmm.0.025056-0. [DOI] [PubMed] [Google Scholar]


