Abstract
SARS-CoV membrane protein could be detected easily using Western blotting in non-denaturing condition but not regular denaturing treatment. Boiling treatment, causing the aggregation of SARS-CoV membrane protein in the stacking gels, results in the failure to detect the membrane protein in the separating gels. Aggregated membrane proteins could not be dissociated by 1% Triton-X 100, 6 M urea, or 2% SDS. The region with amino acid residues from 51 to 170 is responsible for thermal aggregation of SARS-CoV membrane protein. Hydrophobic regions with amino acid residues from 61 to 90, from 91 to 100, from 136 to 170, are essential for this protein aggregation. Thermal aggregation of SARS-CoV membrane protein is not unique among structural proteins of coronaviruses. However, SARS-CoV membrane protein seems to be more sensitive to heat treatment, since the membrane protein of MHV-JHM, another member of the Coronaviridae, would not aggregate after the same treatment. Therefore, if SARS-CoV membrane protein needs to be analyzed using SDS-PAGE, boiling should be avoided. Thermal aggregation of SARS-CoV membrane protein may be one of the reasons for the inactivation of this virus by heat. The unusual property of SARS-CoV membrane protein aggregation induced by heat also provides a model for the study of protein aggregation.
1. Introduction
The outbreak of severe acute respiratory syndrome (SARS) in 2003 (Vijayanand et al., 2004) is associated with a newly discovered coronavirus, SARS-associated coronavirus (SARS-CoV) (Rota et al., 2003). Coronaviruses are exceptionally large RNA viruses and employ complex regulatory mechanisms to express their genomes (Holmes and Lai, 1996). The complete genome sequences of several SARS-CoV isolates have been determined (Marra et al., 2003, Rota et al., 2003). The genome structure, gene expression pattern and protein profiles of SARS-CoV are similar to those of other coronaviruses (Holmes and Enjuanes, 2003, Thiel et al., 2003). Nine SARS-CoV specific RNAs were synthesized in virus-infected cells (Thiel et al., 2003). These RNAs were predicted to encode two large replicative polyprotein (pp1a and pp1ab), four structural proteins (spike, membrane, envelope, and nucleocapsid proteins), and other auxiliary proteins. The four structural proteins of coronaviruses (S, E, M, and N) play roles in virion morphogenesis (Holmes and Lai, 1996). N binds to viral RNA to form nucleocapsid. Co-expression of M and E proteins together can form virus-like particles (de Haan et al., 1998a). Interactions between the M and E proteins and nucleocapsids result in virus budding through cellular membrane. By interaction with M protein, the S protein is incorporated into the viral envelope and the mature virions are released from the cells. It has also been demonstrated that virus-like particles of SARS-CoV could be formed by expressing M and E proteins in insect cells (Ho et al., 2004).
The mechanisms of SARS pathogenesis are largely unknown and may involve both viral cytocidal effects on the cells and immune-mediated mechanisms (Lai, 2003). To understand the humoral immune response against SARS-CoV infection, the antibody profile against various SARS-CoV structural proteins in different time course during SARS-CoV infection needs to be examined. It has been demonstrated that different treatments (e.g. heating, 2-ME) would promote the unfolding of structural proteins in other coronaviruses (Callebaut and Pensaert, 1980, Deregt et al., 1987, Hogue and Brian, 1986, Sturman et al., 1980, Wege et al., 1979). Therefore, both denaturing (Ma et al., 2002, Makowski and Ramsby, 1997) and non-denaturing treatments (Lin et al., 1998) should be used for antigen preparations. Nucleocapsid and spike proteins can be detected using Western blotting analysis by either denaturing condition (sample buffer containing 67.5 mM Tris–HCl (pH 6.8), 5% 2-mercaptoethanol, 3% SDS, 0.1% bromophenol blue, 10% glycerol, treatment at 100 °C for 10 min) or non-denaturing conditions (sample buffer containing 50 mM Tris–HCl (pH 6.8), 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol; no boiling treatment) while membrane protein could only be detected in non-denaturing but not denaturing condition (Lo et al., 2005). In the present study, factors affecting the detection of SARS-CoV membrane protein in SDS-PAGE were investigated.
2. Materials and methods
2.1. Plasmid construction
SARS-CoV membrane gene fragment was derived from the serum of one SARS patient (Lo et al., 2005) by RT-PCR (reverse transcriptase-polymerase chain reaction). PCR primers used in this study are listed in Table 1 . PCR primers (M-S2 and M-A2) were used to amplify the membrane gene fragment. After PCR reaction, DNA fragment was digested by restriction enzymes (EcoRI and XbaI) and cloned into pcDNA3 (Invitrogen, USA) expression vector (linearized by EcoRI/XbaI). The sequences of this membrane gene are identical to the corresponding gene sequences of CUHK-W1 isolate (GI: 30027610) except one nucleotide variation (C in the nucleotide 26585 of CUHK-W1 isolate while T in this isolate, causing amino acid change from Ala to Val in position 68). This expression plasmid encodes full-length membrane protein (221 a.a., labeled as “M”).
Table 1.
PCR primers used in this study
| Name | Sequence |
|---|---|
| M-S2 | (5′-CCGGAATTCATGGCAGACAACGGTACTA-3′) |
| M-S3 | |
| M-A2 | |
| M-A3 | |
| M-A4 | |
| M-C63A-S | |
| M-C63A-AS | |
| M-C85A-S | |
| M-C85A-AS | |
| M-C158A-S | |
| M-C158A-AS | |
| M51-S | (5′-CGGAATTCATGGTTTTCCTCTGGCTCTT-3′) |
| M170-AS | |
| M170-AS3 | |
| L50/171-S | (5′-TACATAATAAAGCTTACATCACGAACGCTT-3′) |
| L50/171-AS | (5′-AAGCGTTCGTGATGTAAGCTTTATTATGTA-3′) |
| L46/61-S | (5′-AACAGGTTTTTGTACCTTGCTTGTTTTGTG-3′) |
| L46/61-AS | (5′-CACAAAACAAGCAAGGTACAAAAACCTGTT-3′) |
| L90/101-S | (5′-ATTGTAGGCTTGATGCTGTTTGCTCGTACC-3′) |
| L90/101-AS | (5′-GGTACGAGCAAACAGCATCAAGCCTACAAT-3′) |
| L60/91-S | (5′-TTGTGGCCAGTAACATGGCTTAGCTACTTC-3′) |
| L60/91-AS | (5′-GAAGTAGCTAAGCCATGTTACTGGCCACAA-3′) |
| L100/136-S | (5′-GTTGCTTCCTTCAGGGAACTTGTCATTGGT-3′) |
| L100/136-AS | (5′-ACCAATGACAAGTTCCCTGAAGGAAGCAAC-3′) |
| L135/171-S | (5′-CCGCTCATGGAAAGTACATCACGAACGCTT-3′) |
| L135/171-AS | (5′-AAGCGTTCGTGATGTACTTTCCATGAGCGG-3′) |
| CC1-S | |
| CC115-AS | |
| HCV-1 | (5′-CGGAATTCAGGTCTCGTAGACCG-3′) |
| CC115-AS2 | |
| JHMM-S2 | (5′-CGGAATTCATGAGTAGTACCACTCAGG-3′) |
| JHMM-A2 |
Note: GAATTC, 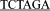 , and
, and 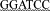 are the recognition sequences for EcoRI, XbaI, and BamHI, respectively.
are the recognition sequences for EcoRI, XbaI, and BamHI, respectively.  means point mutation while bolded nucleotides represent stop codon.
means point mutation while bolded nucleotides represent stop codon.
To mutate amino acid 63 of membrane protein from Cys to Ala, PCR primers (M-S2 and M-C63A-AS) were used to amplify the 5′-end of the membrane gene fragment while PCR primers (M-C63A-S and M-A2) were used to amplify the 3′-end fragment. These two DNA fragments were linked together by PCR using primers (M-S2 and M-A2). After PCR, the DNA fragment was digested by restriction enzymes (EcoRI/XbaI) and cloned into pcDNA3 (linearized by EcoRI/XbaI) expression vector. This expression plasmid encodes full-length membrane protein with mutation in amino acid 63 from Cys to Ala. To mutate amino acids 85, 158 of membrane protein from Cys to Ala, the same approach was used except that M-C85A-AS (or M-C158A-AS) primer instead of M-C63A-AS and M-C85A-S (or M-C158A-S) instead of M-C63A-S were used.
PCR primers (M-S2 and M-A3) were used to amplify the membrane gene and the PCR product was cloned into pcDNA3.1/V5-His A (Invitrogen, USA) expression vector (linearized by EcoRI/XbaI). This expression plasmid encodes the full-length membrane protein plus V5 and His tag encoded from vector sequence (M-V5-His, labeled as “M*”). Similarly, PCR primer pairs (M51-S with M-A3, M-S2 with M170-AS3, and M51-S with M170-AS3) were used to amplify the corresponding DNA fragments and cloned into pcDNA3.1/V5-His A expression vector (linearized by EcoRI/XbaI) to get the expression plasmids encode membrane protein with deletion of first 50 amino acids (M51-221*), with deletion of last 51 amino acids (M1-170*), or membrane protein from amino acid 51 to amino acid 170 (M51-170*) respectively, plus V5 and His tag.
To clone the DNA fragment of the membrane protein with deletion from amino acid 51 to 170 into pcDNA3.1/V5-His A (MΔ51-170*), primers (M-S2 and L50/171-AS) were used to amplify the 5′-end of the membrane gene fragment while primers (L50/171-S and M-A3) were used to amplify the 3′-end fragment. These two DNA fragments were linked together by PCR using primers (M-S2 and M-A3). After PCR, the DNA fragment was digested by restriction enzymes (EcoRI/XbaI) and cloned into pcDNA3.1/V5-His A expression vector (linearized by EcoRI/XbaI). A similar approach was used to clone the DNA fragments of the membrane protein with deletion in different regions (amino acids from 47 to 60, from 61 to 90, from 91 to 100, from 101 to 135, and from 136 to 170) into pcDNA3.1/V5-His A (linearized by EcoRI/XbaI) using antisense primer (L46/61-AS, L60/91-AS, L90/101-AS, L100/136-AS, or L135/171-AS) to replace L50/171-AS and sense primer (L46/61-S, L60/91-S, L90/101-S, L100/136-S, or L135/171-S) to replace L50/171-S.
To clone the expression plasmid encoding a fusion protein with the order of the first 115 amino acids of HCV core protein (Ma et al., 2002), full-length membrane protein, plus V5 and His tag (C-M-V5-His), PCR primers (HCV-1 and CC115-AS2) were used to amplify the gene fragment of the first 115 amino acids of HCV core protein while PCR primers (M-S3 and M-A3) were used to amplify the entire membrane gene fragment. After PCR reaction, two DNA fragments were digested by restriction enzymes (EcoRI/BamHI or BamHI/XbaI) separately and cloned into pcDNA3.1/V5-His A (linearized by EcoRI/XbaI) expression vector together. Similarly, to clone the expression plasmid encoding a fusion protein with the order of full-length membrane protein, the first 115 amino acids of HCV core protein, plus V5 and His tag (M-C-V5-His), PCR primers (M-S2 and M-A4) were used to amplify the entire membrane gene fragment while PCR primers (CC1-S and CC115-AS) were used to amplify the gene fragment of the first 115 amino acids of HCV core protein.
To clone the DNA fragment of the membrane protein of mouse hepatitis virus (GI: 58968), virus RNA isolated from JHM strain (Weiner, 1973) was converted to cDNA using random hexamers. PCR primers (JHMM-S2 and JHMM-A2) were used to amplify the membrane gene fragment. After PCR reaction, DNA fragment was digested by restriction enzymes (EcoRI and XbaI) and cloned into pcDNA3 (Invitrogen, USA) expression vector (linearized by EcoRI/XbaI).
All the expression plasmids were verified by sequencing and in vitro transcription/translation (Promega, USA).
2.2. Protein expression in Vero E6 cells
Vero E6 cells were maintained in RPMI 1640 medium containing 10% fetal calf serum, 1% Glutamine (200 mM, Biological Industries, USA), and 100 μg/ml penicillin/streptomycin (Gibco BRL, USA). 2.5 × 105 to 2.7 × 105 cells were plated in a 35 mm dish. After an overnight incubation, cells were infected with a recombinant vaccinia virus carrying the T7 phage RNA polymerase gene (Fuerst et al., 1986). Two hours after infection, cells were transfected with 0.4 μg plasmid DNA by using Effectene transfection reagent (Qiagen, Germany). Twenty-one hour after transfection, recombinant proteins in the cells were analyzed.
2.3. Western blotting analysis
For Western blotting analysis, cells were dissolved in sample preparation buffers after washing with PBS twice. Two sample preparation buffers were used: denaturing buffer containing β-mercaptoethanol (Ma et al., 2002, Makowski and Ramsby, 1997) and non-denaturing buffer without β-mercaptoethanol (Lin et al., 1998). The samples were treated at room temperature or 100 °C in the sample buffer for 10 min before electrophoresis. Usually, 4.5% (acrylamide percentage) gel was used as the stacking gel and 12% gel as the separating gel in this study. When proteins with smaller size were analyzed (e.g. deletion mutants of membrane protein), a 15% gel was used as the separating gel. SDS-PAGE gel after electrophoresis was transferred to PVDF paper (Pall Corporation, USA). All procedures were carried out at room temperature. The PVDF paper was blocked in PBST (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4, 0.1% Triton-X 100) with 5% milk for 3 h. After blocking, the PVDF paper was incubated with one SARS patient's serum diluted 1000-fold in PBST with 5% milk, for 3 h. The PVDF paper was then washed three times in PBST for 10 min. Afterwards, goat anti-human IgG conjugated with horseradish peroxidase (Amersham Biosciences Ltd., USA), which had been diluted 2500-fold in PBST with 5% milk, was added for another 1 h of incubation. After three more 10 min washes with PBST, the signal was developed by the “Western Lightning™ Chemiluminescence Reagent Plus” kit (Perkin-Elmer Life Sciences, USA). If rabbit anti-HCV core polyclonal antibody (Ma et al., 2002) or anti-V5/AP antibody (Invitrogen, USA) were used as the primary antibodies to carry out the assay, the previous published procedures (Ma et al., 2002) were followed.
2.4. Immunofluorescence analysis
Cells with recombinant protein expression were treated at room temperature or at 100 °C for 10 min. After that, samples were washed with PBS and then fixed with 4 °C acetone:methanol (1:1) for 10 min. Fixed cells were washed with incubation buffer (0.05% NaN3, 0.02% saponin, 1% skim milk in PBS) twice for 2 min each time, then incubated with anti-His monoclonal antibody (or anti-HCV core polyclonal antibody) at 37 °C for 30 min. Samples were washed with PBS three times (5 min each time at room temperature), then incubated with FITC-conjugated goat anti-mouse (or anti-rabbit) IgG antibody in 20X dilution at 37 °C for 30 min. Again, samples were washed with PBS three times (5–10 min each time at room temperature). Then, DAPI (Merck, Germany) was used to stain DNA as the localization of nucleus. Samples were observed under a confocal microscope.
2.5. In vitro transcription and translation
An amount of 0.5 μg of expression DNA (in the case of JHMM, plasmid was linearized by XbaI first) was in vitro translated and labeled with [S35] Methionine by TNT T7 Quick Couples Transcription/Translation system (Promega, USA) in a total reaction volume of 15 μl following manufacturer's instructions. After 1 h incubation at 30 °C, PBS were added in the reaction mixtures to a total volume of 60 μl. Four aliquots (10 μl per aliquot) were treated at different temperatures (20, 60, 80, or 100 °C) for 10 min. After treatment, 10 μl of sample preparation buffer was added in the aliquots before electrophoresis. Samples were then analyzed by SDS-PAGE and visualized by Phosphoimage (Fujitsu, Japan).
3. Results
3.1. SARS-CoV membrane protein could not be detected easily using Western blotting with boiling treatment
SARS-CoV spike and nucleocapsid proteins could be detected using Western blotting (WB) analysis under either regular denaturing or non-denaturing condition while membrane protein could be detected only under non-denaturing but not regular denaturing condition (Lo et al., 2005) (Fig. 1A). The absence of membrane protein in WB could be due to conformational change through the breaking of intramolecular disulfide bond within this protein under denaturing condition. To verify this possibility, three Cys residues within the membrane protein (a.a. 63, 85, 158) were mutated into Ala residues individually. However, mutation of Cys residues within the membrane protein individually does not affect the detection of this protein (Fig. 1A). Thus, intramolecular disulfide bond formation is not responsible for the failure to detect the membrane protein in WB analysis. Heating at 100 °C for 10 min (boiling) in denaturing condition was suspected to be responsible for the failure to detect the membrane protein by WB analysis. To verify this possibility, membrane protein was treated under different conditions (without 2-ME but boiling, with 2-ME but no boiling) and then analyzed by WB (Fig. 1B). The results shown in Fig. 1B confirm that boiling but not 2-ME treatment results in failure to detect the membrane protein in WB analysis. The effect of heating at different temperatures on the membrane protein was also studied (Fig. 1C). It is failure to detect SARS-CoV membrane protein as monomers after treated with temperature over 80 °C.
Fig. 1.
SARS-CoV membrane protein could not be easily detected using Western blotting with boiling treatment. (A) Membrane proteins in either wild type or mutation in individual Cys residue could easily be detected in non-denaturing (nd) but not denaturing (d) treatment. Protein transiently expressed in Vero E6 cells were equally aliquoted and treated in different conditions. Serum of one SARS patient was used as first antibody in WB analysis. Proteins marked by the thick arrow are the glycosylated SARS-CoV membrane protein, the same size as the membrane protein from purified SARS-CoV particles (virions). (B) Boiling but not 2-ME treatment is responsible for the inefficient detection of membrane protein in WB analysis. Sample preparation and detection are the same as that of (A). (C) Thermal effect on the SARS-CoV membrane protein. Sample preparation and detection are the same as that of (A).
The failure to detect the membrane protein in WB with boiling could be due to conformational change (resulting in the failure of antibody recognition), protein degradation, or aggregation of this protein. The possibility of conformational change causing recognition by the antibody is ruled out by the observation that in vitro translated, S35-labeled membrane protein would also lose its signal at the expected size after boiling (data not shown). This conclusion is strengthened further when different tag peptides added to the N- or C-terminus of membrane protein (M-V5-His; C-M-V5-His; M-C-V5-His) do not affect the detection of membrane protein in WB using antibodies against these tag peptides (data not shown).
3.2. Thermal aggregation of SARS-CoV membrane protein
To study whether boiling causes directly the failure to detect the membrane protein in WB analysis but not through activating other protease(s), the membrane protein transiently expressed in mammalian cells was treated with urea (1, 3, or 6M) or SDS (0.5, 1, or 2%) to inactivate possible proteases before boiling. Similar to the un-treated sample, the membrane proteins in pre-treated samples were detected at a lower level as monomers in WB analysis after boiling (data not shown, Fig. 2A) though 2% SDS pre-treatment did reduce the effect. This result suggests that boiling treatment directly causes the failure to detect the membrane protein in WB analysis. Boiling will not breakdown the primary structure of proteins in the regular denaturing buffer (Schultz and Liebman, 1997). Furthermore, no suspected degraded products from the membrane protein (i.e. proteins smaller than the membrane protein) were detected after boiling (data not shown and Fig. 2B). Therefore, protein aggregation is probably the cause that leads the failure to detect the membrane protein in WB analysis. To test this possibility, the recombinant membrane protein fused with V5-His tag were expressed transiently in mammalian cells and analyzed by WB using monoclonal antibody against V5 tag in both stacking and separating gels. In Fig. 2B, the recombinant fusion membrane protein could be detected in stacking gels but not separating gels after boiling treatment. The same results were obtained when different recombinant fusion proteins (M-C-V5-His or C-M-V5-His) were used for the assays (data not shown). The existence of recombinant fusion protein after boiling could also be demonstrated by immunofluorescence of M-C-V5-His or C-M-V5-His (data not shown) using antibody against core protein. Therefore, the failure to detect the membrane protein in WB analysis is due to the thermal aggregation of this protein. The protein band marked by the thin arrow (Fig. 2A) is unglycosylated M* protein while the other one marked by the thick arrow is glycosylated M* protein since the latter protein will reduce its size to that of the former one after deglycosylation treatment (Ma et al., 2005).
Fig. 2.
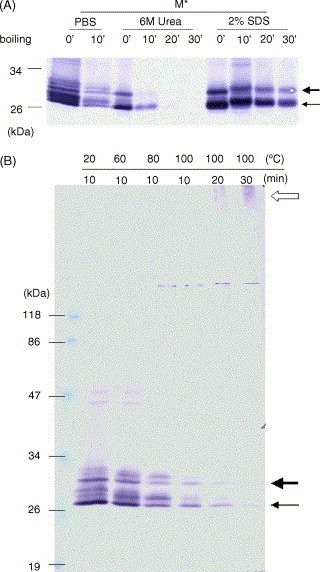
Thermal aggregation of SARS-CoV membrane protein. (A) Effect of boiling treatment on SARS-CoV membrane protein in the presence of 6M urea or 2% SDS. Sample preparation and detection are the same as those of Fig. 1 except samples were treated with either 6 M urea or 2% SDS before boiling treatment. There are many distinct protein bands other than unglycosylated and glycosylated M proteins. These protein bands may represent the cellular proteins interacting with M protein during non-denaturing condition. The interaction between cellular proteins and M protein is disrupted in the presence of denaturants (6 M urea or 2% SDS). (B) Recombinant membrane protein (M*) was detected in stacking gel after boiling treatment. Sample preparation and detection are the same as those of Fig. 1 except whole SDS-PAGE including stacking gel was transferred to perform the WB assay. The empty arrow indicates the membrane protein aggregates.
3.3. Domains of SARS-CoV membrane protein important for the thermal aggregation
To determine which domain(s) of the membrane protein is responsible for the thermal aggregation, different regions of the membrane protein were removed and the recombinant membrane proteins with deletion were analyzed using WB with or without heat treatment. Like full-length membrane protein, the recombinant truncated membrane proteins with amino acid residues from 51 to 170 (e.g. M51-221*, M1-170*, and M51-170*) do not exist as monomers after heat treatment while the monomer amount of recombinant membrane protein with deletion in this region (MΔ51-170*) does not seem to be affected by heat treatment (Fig. 3A). Therefore, the region with amino acid residues from 51 to 170 is responsible for the thermal aggregation of SARS-CoV membrane protein. To narrow down further the amino acid residues essential for this aggregation, different recombinant membrane proteins with deletion in this region were expressed and analyzed. Recombinant membrane proteins with deletion from amino acid 47 to 60 or from 101 to 135 (MΔ47-60* or MΔ101-135*) were detected as monomers with less amount after heat treatment while recombinant proteins with deletion from 61 to 90, from 91 to 100, and from 136 to 170 (MΔ61-90*, MΔ91-100*, and MΔ136-170*) were detected as monomers with the same amount before or after heat treatment (Fig. 3B). Therefore, regions with amino acid residues from 61 to 90, from 91 to 100, and from 136 to 170, are essential for the thermal aggregation of SARS-CoV membrane protein.
Fig. 3.
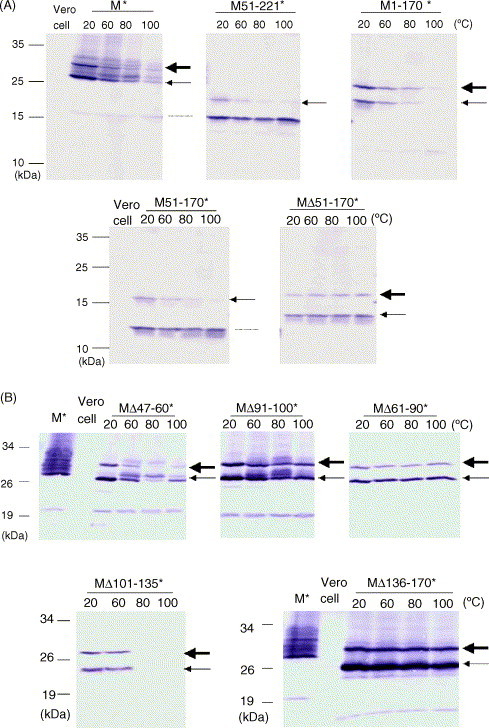
Domains important for the thermal aggregation of SARS-CoV membrane protein. (A) Various recombinant fusion proteins (M*, M51-221*, M1-170*, M51-170*, and MΔ51-170*) were transiently expressed in the cells. Proteins were collected and heated at different temperatures (20, 60, 80, and 100 °C) for 10 min. After that, the samples were analyzed by WB using anti-V5 monoclonal antibody. The proteins marked by the thin arrow are unglycosylated recombinant membrane proteins while the other ones marked by the thick arrow are glycosylated recombinant proteins. The protein smaller than unglycosylated membrane protein (marked by dotted line) containing the C-terminus of recombinant membrane protein may be derived by cleavage from precursor recombinant membrane proteins or translation by internal initiation. In any case, it was not produced by heat treatment since its concentration did not increase after heat treatment. (B) Same as (A), different recombinant fusion proteins (MΔ47-60*, MΔ61-90*, MΔ91-100*, MΔ101-135*, and MΔ136-170*) were used.
3.4. The mechanism of thermal aggregation of SARS-CoV membrane protein
To study the mechanism of the thermal aggregation of SARS-CoV membrane protein, various recombinant membrane proteins with deletion in different regions were co-expressed in mammalian cells and analyzed by WB. MΔ51-170* could not be aggregated with M51-170* after boiling treatment when these two proteins were co-expressed in the cells (left panel of Fig. 4A) as the amount of MΔ51-170* protein monomer remains unchanged. Similar results were observed when MΔ51-170*, MΔ61-90*, MΔ136-170* were co-expressed with full-length membrane protein (M*) (data not shown, right panel of Fig. 4A). Co-expression of MΔ91-100* with MΔ61-90*, MΔ91-100* with MΔ136-170*, MΔ61-90* with MΔ136-170* would not induce the aggregation of these proteins after boiling (data not shown).
Fig. 4.
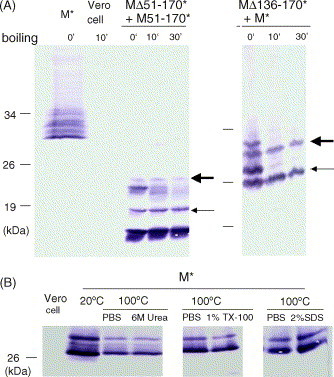
Characteristics of thermal aggregation of SARS-CoV membrane protein. (A) Two proteins (M51-170* and MΔ51-170* in the left panel while M* and MΔ136-170* in the right panel) were transiently expressed in the cells and protein samples heated at 100 °C for 0, 10, or 30 min were detected by anti-V5 monoclonal antibody. MΔ51-170* (left panel) and MΔ136-170* (right panel) were marked by the arrows. (B) Aggregation of M* protein was induced by treatment at 100 °C for 10 min. After cooling to the room temperature, different denaturants (6 M urea, 1% TX-100, or 2% SDS in final concentration) were used to dissociate the aggregated membrane proteins into monomers.
To study whether the aggregated membrane protein could be dissociated by denaturants, the transiently expressed membrane proteins (M*) in mammalian cells, were treated with 1% Triton-X 100, 6 M urea, or 2% SDS, after boiling, and analyzed by WB. The membrane proteins could not be detected more in the expected size as monomers after denaturant treatment (Fig. 4B). Thus, the membrane proteins could not dissociate from aggregates to form monomers by these denaturants.
The structural properties of the aggregated membrane protein may be different from those of the authentic membrane protein. For example, recombinant M-V5-His fusion protein could be detected before but not after boiling treatment by immunoflurescence using anti-His antibody though M-C-V5-His or C-M-V5-His could be detected by anti-core antibody (data not shown).
3.5. Mouse hepatitis virus JHM strain (MHV-JHM) membrane protein is not as sensitive as SARS-CoV membrane protein to boiling treatment
To study whether the membrane protein of mouse hepatitis virus (MHV) (Weiner, 1973) would also aggregate after the same heat treatment, full-length membrane gene derived from JHM strain RNA by RT-PCR was in vitro translated. The in vitro translated, S35-labeled membrane protein was analyzed in SDS-PAGE after 10 min heating treatment at different temperatures (20, 60, 80, and 100 °C). MHV-JHM membrane protein monomer amount remains at the similar level after different treatments (data not shown). Thus, MHV-JHM membrane protein is not as sensitive as SARS-CoV membrane protein to boiling.
4. Discussion
In this study, thermal aggregation of SARS-CoV membrane protein was demonstrated. These results suggest that detection of the membrane protein with SDS-PAGE should avoid boiling. In other coronaviruses, membrane protein is an abundant structural protein in the virions (de Haan et al., 1998a). In a previous study (Rota et al., 2003), SARS-CoV membrane protein, unlike nucleocapsid or spike proteins, is hardly detected when virus particles were analyzed by SDS-PAGE. It is possible that SARS-CoV membrane protein is still abundant in the virions and could not enter the separating gel due to aggregation induced by boiling. It has also been demonstrated that heat can inactivate the SARS-CoV (Rabenau et al., 2004). The thermal aggregation of the membrane protein may be one of the reasons for this inactivation since protein aggregation often results in loss of their functions (Hoffner and Djian, 2002, Ishimaru et al., 2003, Lin et al., 2004, Murphy, 2002).
The mouse hepatitis coronavirus membrane protein is O-glycosylated (de Haan et al., 1998b). The N-terminal sequences of SARS-CoV membrane protein containing Asn-Gly-Thr (a.a. 4–6) is a defined concensus sequence (Asn-X-(Ser/Thr)) for N-linked glycosylation (Ma et al., 2005). Glycosylation is not required for the thermal aggregation of SARS-CoV membrane protein since in vitro translated, unglycosylated membrane protein (data not shown) and recombinant membrane protein deleting the first 50 a.a. (M51-221*) expressed transiently in the cells could still aggregate after heat treatment (Fig. 3A).
SARS-CoV membrane protein could be detected by WB analysis in non-denaturing condition but not regular denaturing condition using antibodies derived from one SARS patient's serum (Fig. 1A). Mutation of Cys residues within the membrane protein individually does not affect the protein detection by the same serum (Fig. 1A). This result indicates that intramolecular disulfide bond formation of membrane protein is not the only epitopic determinant for the recognition of antibodies in this SARS patient. On the other hand, glycosylated but not unglycosylated membrane protein was recognized by antibodies derived from this SARS patient's serum in either virus particles or transient expression in mammalian cells (Fig. 1A–C) even unglycosylated membrane protein (M*) is more abundant than glycosylated one analyzed by anti-V5 antibody (data not shown). Therefore, glycosylation does play a major role in the formation of epitopic determinant for the recognition of antibodies in this SARS patient.
SARS-CoV membrane protein pretreated with 6 M urea or 2% SDS would still aggregate after boiling treatment though 2% SDS treatment did reduce this aggregation since more protein monomers were detected (Fig. 2A). Moreover, the thermally aggregated membrane proteins were not dissociated by 1% Triton-X 100, 6 M urea, or 2% SDS (Fig. 4B). These results indicate that extensive interaction between membrane proteins were involved during the aggregation. This possibility is strengthened when various recombinant membrane proteins with deletion in different regions were co-expressed in mammalian cells and analyzed by WB to study the interaction of different protein molecules (Fig. 4A). Similarly, membrane proteins with smaller size (marked by dotted line in Fig. 3A and B) possibly derived from full-length protein also could not be aggregated in the presence of full-length protein. These results suggest that thermal aggregation of membrane protein requires extensive interaction between hydrophobic regions of membrane protein. This interaction may cause the conformational change of the aggregated membrane proteins. This possibility was supported by the observation that recombinant M-V5-His fusion protein could be detected before but not after boiling treatment by immunoflurescence using anti-His antibody (though M-C-V5-His or C-M-V5-His could be detected by anti-core) (data not shown). The size of the aggregates seems to be heterogeneous in the stacking gels (Fig. 2B). However, the stoichiometry and constitutes of these aggregates remain uncharacterized.
Thermal aggregation of SARS-CoV membrane protein could occur using purified virus particles, recombinant proteins transiently expressed in mammalian cells (Fig. 1A), and even in vitro translated product (data not shown). This indicates that the concentration of membrane protein required for this aggregation is very low and there is no need for the protein to be located in a particular location, e.g. endoplasmic reticulum (no microsome was added in the in vitro translation reaction). There are many β-sheet structures within membrane protein (Hu et al., 2003). These structural characteristics are similar to those of proteins linked to neurodegenerative diseases: β-amyloid, prion, and huntingtin (Murphy, 2002). However, it is still unknown whether SARS-CoV membrane protein could aggregate without boiling (it may occur with a slower rate). Further studies are needed to address this issue. Myoglobin (Yan et al., 2003), hemoglobin (Yan et al., 2004), and oat globulin (Zhao et al., 2004) were demonstrated to aggregate by heat treatment at different temperatures. At present, it is not known whether a similar mechanism is involved in the aggregation process of these proteins. To study the assembly mechanism or the atomic structures of the aggregated membrane protein, purified recombinant SARS-CoV membrane proteins are needed. On the other hand, unlike the aggregation of light neurofilament (Lin et al., 2004), the SARS-CoV membrane proteins could not dissociate from aggregates to monomers by 1% Triton-100 (Fig. 4B). Study on the aggregation of SARS-CoV membrane protein should help us understand more about the protein aggregation.
The monomer amount of recombinant SARS-CoV membrane proteins with deletion from amino acid 47 to 60 (MΔ47-60*) was less abundant after treatment at 60 or 80 °C but not at 100 °C (Fig. 3B). In this case, the thermal aggregation of MΔ47-60* may be similar to that of oat globulin whose hexamers and trimers were dissociated into monomers upon heating at 100 °C (Zhao et al., 2004).
A temperature-dependent conformational change in E1 protein of coronavirus A59 resulting in the aggregation of this protein was reported previously (Sturman et al., 1980). Therefore, thermal aggregation of SARS-CoV membrane protein is not unique among structural proteins of coronaviruses. However, SARS-CoV membrane protein seems to be more sensitive to the heat treatment since the membrane protein of MHV-JHM, another member of the Coronaviridae, would not aggregate after the same condition treatment (data not shown). It is interesting that SARS-CoV, but not MHV-JHM, membrane protein aggregates after boiling since these two proteins share high degree of homology with each other.
In summary, thermal aggregation of SARS-CoV membrane protein was demonstrated in this study, suggesting that the analysis of this protein using SDS-PAGE should avoid regular boiling. Furthermore, regions with amino acid residues from 61 to 90, from 91 to 100, from 136 to 170, are essential for the thermal aggregation of membrane protein.
Acknowledgements
We thank Dr. Yue-Li Juang and Ms. Pei-Chi Yeh for making photos of confocal microscopy. We also thank Dr. Yi-Cheng Chen and Dr. Huichun Li for helpful discussion and critical review of this manuscript. This work has been supported by grants from National Science Council of Taiwan (NSC 92-2314-B-320-006) and from Tzu Chi University (TCMRC 9232-01) to Dr. Shih-Yen Lo.
References
- Callebaut P.E., Pensaert M.B. Characterization and isolation of structural polypeptides in haemagglutinating encephalomyelitis virus. J. Gen. Virol. 1980;48(1):193–204. doi: 10.1099/0022-1317-48-1-193. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72(8):6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Roestenberg P., de Wit M., de Vries A.A., Nilsson T., Vennema H., Rottier P.J. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J. Biol. Chem. 1998;273(45):29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- Deregt D., Sabara M., Babiuk L.A. Structural proteins of bovine coronavirus and their intracellular processing. J. Gen. Virol. 1987;68(Pt 11):2863–2877. doi: 10.1099/0022-1317-68-11-2863. [DOI] [PubMed] [Google Scholar]
- Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 1986;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318(4):833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffner G., Djian P. Protein aggregation in Huntington's disease. Biochimie. 2002;84(4):273–278. doi: 10.1016/s0300-9084(02)01398-6. [DOI] [PubMed] [Google Scholar]
- Hogue B.G., Brian D.A. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5(2–3):131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V., Enjuanes L. Virology. The SARS coronavirus: a postgenomic era. Science. 2003;300(5624):1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- Holmes K.V., Lai M.M. Coronaviridae: the viruses and their replication. In: Fields B.N., Knipe D.M., Howley P.M., editors. 3rd ed. vol. 1. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 1075–1093. (Fields Virology). [Google Scholar]
- Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J., Tian X., Jiang F., Zhao X., Liu S., Zeng C., Yang H. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinf. 2003;1(2):118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D., Andrade L.R., Teixeira L.S., Quesado P.A., Maiolino L.M., Lopez P.M., Cordeiro Y., Costa L.T., Heckl W.M., Weissmuller G., Foguel D., Silva J.L. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42(30):9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- Lai M.M. SARS virus: the beginning of the unraveling of a new coronavirus. J. Biomed. Sci. 2003;10(6 Pt 2):664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Zhai J., Canete-Soler R., Schlaepfer W.W. 3′-untranslated region in a light neurofilament (NF-L) mRNA triggers aggregation of NF-L and mutant superoxide dismutase-1 proteins in neuronal cells. J. Neurosci. 2004;24(11):2716–2726. doi: 10.1523/JNEUROSCI.5689-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.L., Liao C.L., Chen L.K., Yeh C.T., Liu C.I., Ma S.H., Huang Y.Y., Huang Y.L., Kao C.L., King C.C. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 1998;72(12):9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S.Y., Yang, H.H., Ma, H.C., Chen, L.K., 2005. The profile of specific antibodies to individual SARS-CoV structural proteins in SARS sero-converted patients, submitted for publication.
- Ma H.C., Ke C.H., Hsieh T.Y., Lo S.Y. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J. Gen. Virol. 2002;83(Pt 12):3085–3092. doi: 10.1099/0022-1317-83-12-3085. [DOI] [PubMed] [Google Scholar]
- Ma, H.C., Lee, Y.N., Yang, H.H., Chen, L.K., Lo, S.Y., 2005. Expression and antibody response of SARS-CoV M protein, submitted for publication.
- Makowski G.S., Ramsby M.L. Protein molecular weight determination by sodium dodecyl sulfate polyacrylamide gel electrophoresis. In: Creighton T.E., editor. 2nd ed. vol. 1. Oxford University Press; New York: 1997. pp. 1–27. (Protein Structure. The Practical Approach Series). [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Murphy R.M. Peptide aggregation in neurodegenerative disease. Annu. Rev. Biomed. Eng. 2002;4:155–174. doi: 10.1146/annurev.bioeng.4.092801.094202. [DOI] [PubMed] [Google Scholar]
- Rabenau, H.F., Cinatl, J., Morgenstern, B., Bauer, G., Preiser, W. and Doerr, H.W., 2004. Stability and inactivation of SARS coronavirus. Med. Microbiol. Immunol. (Berl). [DOI] [PMC free article] [PubMed]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Schultz R.M., Liebman M.N. Protein I: compostion and structure. In: Devlin T.M., editor. 4th ed. vol.1. Wiley-Liss Inc.; New York: 1997. pp. 23–85. (Textbook of Biochemistry with Clinical Correlations). [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33(1):449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Vijayanand P., Wilkins E., Woodhead M. Severe acute respiratory syndrome (SARS): a review. Clin. Med. 2004;4(2):152–160. doi: 10.7861/clinmedicine.4-2-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Nagashima K., ter Meulen V. Structural polypeptides of the murine coronavirus JHM. J. Gen. Virol. 1979;42(1):37–47. doi: 10.1099/0022-1317-42-1-37. [DOI] [PubMed] [Google Scholar]
- Weiner L.P. Pathogenesis of demyelination induced by a mouse hepatitis. Arch. Neurol. 1973;28(5):298–303. doi: 10.1001/archneur.1973.00490230034003. [DOI] [PubMed] [Google Scholar]
- Yan Y.B., Wang Q., He H.W., Hu X.Y., Zhang R.Q., Zhou H.M. Two-dimensional infrared correlation spectroscopy study of sequential events in the heat-induced unfolding and aggregation process of myoglobin. Biophys. J. 2003;85(3):1959–1967. doi: 10.1016/S0006-3495(03)74623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.B., Wang Q., He H.W., Zhou H.M. Protein thermal aggregation involves distinct regions: sequential events in the heat-induced unfolding and aggregation of hemoglobin. Biophys. J. 2004;86(3):1682–1690. doi: 10.1016/S0006-3495(04)74237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Mine Y., Ma C.Y. Study of thermal aggregation of oat globulin by laser light scattering. J. Agric. Food Chem. 2004;52(10):3089–3096. doi: 10.1021/jf030735y. [DOI] [PubMed] [Google Scholar]


