Abstract
A method of loop-mediated isothermal amplification (LAMP) was employed to develop a rapid and simple detection system for porcine circovirus type 2 (PCV2). The amplification could be finished in 60 min under isothermal condition at 64 °C by employing a set of four primers targeting the cap gene of PCV2. The LAMP assay showed higher sensitivity than the conventional PCR, with a detection limit of five copies per tube of purified PCV2 genomic DNA. No cross-reactivity was observed from the samples of other related viruses including porcine circovirus type 1 (PCV1), porcine parvovirus (PPV), porcine pseudorabies virus (PRV) and porcine reproductive and respiratory syndrome virus (PRRSV). The detection rate of PCV2 LAMP for 86 clinical samples was 96.5% and appeared greater than that of the PCR method. The LAMP assay reported can provide a rapid yet simple test of PCV2 suitable for laboratory diagnosis and pen-side detection due to ease of operation and the requirement of only a regular water bath or heat block for the reaction.
Keywords: Porcine circovirus type 2 (PCV2), Detection, Loop-mediated isothermal amplification (LAMP), Sensitivity, Specificity
1. Introduction
Porcine circovirus (PCV) is a small, non-enveloped, spherical single-stranded DNA virus, and can be classified as a member of the genus Circovirus of the family Circoviridae (Allan and Ellis, 2000). Two distinct genotypes of PCV, designated PCV type 1 (PCV1) and PCV type 2 (PCV2) have been identified. PCV1 shares an approximate 80% nucleotide sequence homology with PCV2 (Meehan et al., 1998). PCV1 was identified as a contaminant of porcine kidney cell cultures and considered non-pathogenic for swine (Allan et al., 1998). However, PCV2 is now generally accepted as the major infectious agent involved in postweaning multisystemic wasting syndrome (PMWS) (Bolin et al., 2001). Clinical signs of PCV2 infection in pigs include progressive weight loss, paleness, dyspnoea and, occasionally, diarrhoea and icterus. Histopathological findings include histocytic infiltration and lymphocyte depletion of lymphoid tissues, interstitial pneumonia and, less frequently, hepatitis and nephritis (Kim and Chae, 2003b, Segalés and Domingo, 2002).
Two major open reading frames (ORFs) have been recognized for the genome of PCV2; ORF1, called the rep gene, which encodes a protein of 35.7 kDa involved in virus replication (Mankertz et al., 1998), and ORF2, named the cap gene, which encodes the major immunogenic capsid protein of 27.8 kDa (Cheung, 2003, Nawagitgul et al., 2000). The capsid protein has the type-specific epitopes (Mahe et al., 2000), which suggests that ORF2 contributes to the development of PMWS and thus has potential for protective immunization in a vaccine (Liu et al., 2000), and for type-specific diagnosis (Blanchard et al., 2003). Epidemiologic data suggest that the virulence of PCV2 is strongly related to the presence of the capsid protein (Cho et al., 2006).
Accumulated evidence indicates that PCV2 replicates in the lymph nodes, lung, liver, spleen, heart and kidney of infected pigs. This results in impairment of the immune system through degradation of lymphoid tissues (Kennedy et al., 2000) and through changes in the proportions of lymphocyte subsets present in peripheral blood (Darwich et al., 2004). The presence of PCV2 in tissues of pigs with PMWS had been proven by virus isolation, polymerase chain reaction (PCR), in situ hybridization and immunohistochemistry (Allan et al., 1998, Kim and Chae, 2003a). Real-time PCR is a sensitive assay for the detection of PCV2 (Brunborg et al., 2004, Chung et al., 2005, Olvera et al., 2004). Although specialized equipment such as a thermal cycler is needed, PCR-based detection methods are commonly accepted because of their high sensitivity and specificity.
Loop-mediated isothermal amplification (LAMP) is a novel amplification method which was developed originally by Notomi et al. (2000). The most significant advantages of LAMP are the ability to amplify specific DNA sequences under isothermal conditions between 63 and 65 °C and a visible result within 30–60 min. The method has been applied successfully to the detection of human influenza A virus, severe acute respiratory syndrome coronavirus and Newcastle disease virus (Hong et al., 2004, Pham et al., 2005, Poon et al., 2005). However, the use of LAMP for detecting PCV2 has not been reported to date. In this study, we evaluated the potential of LAMP for the development of a simple and rapid detection system for PCV2.
2. Materials and methods
2.1. Viral strains and clinical samples
The PCV2-BJ vaccine strain was used to develop a LAMP method (Beijing Bio-pharmaceuticals Corporation). Monolayers of PK-15 cells (ATCC CCL-33) grown in 15-cm2 culture flasks were infected with an inoculum of PCV2 and cytopathic effects (CPE) were monitored daily. Upon observation of between 80 and 100% CPE, the supernatant from the infected culture was collected, centrifuged at 1000 × g for 10 min and stored in aliquots at −80 °C until use. Field isolates of PCV1, porcine parvovirus (PPV), pseudorabies virus (PRV), and porcine reproductive and respiratory syndrome virus (PRRSV) were identified by conventional PCR (or RT-PCR) and sequencing.
A total of 86 clinical samples that were diagnosed as PCV2-positive are shown in Table 1 , including peripheral blood and tissues of lymph nodes, lung, liver, kidney, heart and spleen. These clinical samples were taken from PCV2-antibody-positive pigs tested by ELISA. Among them, 78 samples were identified as positive by PCR and sequencing and the other eight samples were identified by virus isolation.
Table 1.
Sensitivity of LAMP and PCR assays for 86 clinical samples obtained from PCV2-infected pigs
| Type of tissue sample | No. of positive sample tested | % (no.) of positive samples for assay |
|
|---|---|---|---|
| LAMP | PCR | ||
| Blood | 16 | 100 (16) | 87.5 (14) |
| Lymph nodes | 12 | 100 (12) | 91.7 (11) |
| Lung | 15 | 100 (15) | 100 (15) |
| Liver | 10 | 80 (8) | 70 (7) |
| Kidney | 13 | 100 (13) | 100 (13) |
| Heart | 11 | 100 (11) | 100 (11) |
| Spleen | 9 | 88.9 (8) | 77.8 (7) |
| Total | 86 | 96.5 (83) | 90.7 (78) |
2.2. DNA and RNA extraction
DNA was extracted from blood, lymph nodes, lung, liver, kidney, heart and spleen samples taken from PCV2-infected and healthy pigs, using a DNeasy Tissue Kit (Qiagen) according to the manufacturer's instructions. After extraction, DNA was eluted in 60 μl of elution buffer and stored at −20 °C. RNA was extracted directly from PRRSV samples by using Trizol reagent (Invitrogen). Complementary DNA (cDNA) was synthesized using 15 μl of the eluted RNA with oligo(dT)18 primers and the reverse transcriptase kit (Takara Corp., Japan) according to the manufacturer's instructions.
2.3. Primer design and reaction protocol for LAMP and PCR
The highly conserved sequences in the capsid protein-coding region of PCV2 were selected as the target for LAMP and PCR. A set of four primers for the cap gene was designed for LAMP by alignment of six PCV2 genomic sequences (accession nos. AY874165, AY321998, DQ233257, DQ220737, EF493839 and EF524516). Primers F, B, FIP and BIP for LAMP are shown in Table 2 , and the F and B primers were also used in PCR.
Table 2.
Details of PCR and LAMP primers designed for detection of capsid protein coding sequences of PCV2
| Primer namea | Genome position | Squence |
|---|---|---|
| F | 596–617 | 5′-ATGGGCTGCTAATTTTGCAGAC-3′ |
| B | 961–982 | 5′-TCAATAGGAAATTCAGGGCATG-3′ |
| FIP | 762–783 | 5′-GTACAGTTCCACCTTTAGTCTC+TTTT+ |
| 659–680 | GGTTACCATGGTGAAGAAGTGG-3′ | |
| BIP | 840–861 | 5′-CAACTGCTGTCCCAGCTGTAGA+TTTT+ |
| 910–931 | TCCTCCGTGGATTGTTCTGTAG-3′ | |
The primers of F, B, FIP and BIP were for LAMP and each inner primer of LAMP has two binding regions connected by a TTTT spacer. Primers of F and B were also applied in PCR.
The LAMP reaction was carried out in a conventional water bath by mixing 2.0 μM each of FIP and BIP primer, 0.2 μM each of F and B primer, 1.0 mM each deoxynucleoside triphosphate, 8 U of Bst DNA polymerase (New England Biolabs) using the manufacturer's supplied 10× buffer (containing 2 mM of MgSO4, 0.8 M betaine) and 1 μl of extracted template DNA or cDNA in a 0.2 ml Eppendorf tube. The amplification reaction was performed at 64 °C for 60 min and then terminated by heating at 80 °C for 10 min. LAMP products were analyzed by 2.5% agarose gel electrophoresis. PCR was carried out in a 50 μl reaction volume containing 1.5 mM of each deoxynucleoside triphosphate, and 5 μl of 10× buffer, 5 U of Taq polymerase (Nippon Gene), 1 μM each of primers F and B, and 1.0 μl of extracted DNA or cDNA. The amplification regime was 5 min at 94 °C, followed by 30 cycles of 94 °C for 1 min, 55 °C for 30 s and 72 °C for 1 min, with a final elongation for 5 min at 72 °C. The PCR was carried out in the Gene Amp PCR system 9700 (Applied Biosystems). PCR products were subjected to electrophoresis on a 2.5% agarose gel.
2.4. Sensitivity and specificity of LAMP for PCV2
The detection limit of LAMP was tested and compared with PCR by using the same templates at identical concentrations. Serial dilutions of 1, 5, 25, 125, 625 and 3125 copies of DNA from PCV2-BJ strain were used in this assay. In addition, 86 clinical samples were analyzed with the LAMP reaction and the sensitivity of detection was compared between LAMP and PCR. To assess the specificity of LAMP, potential cross-reactions with DNA of PCV1, PPV, PRV and cDNA of PRRSV were examined. PCV2-BJ strain genomic DNA was used as the positive control and DNA extracted from healthy swine tissues was used as the negative control.
2.5. DNA sequencing
PCR products were sequenced with an automated ABI model 373A Stretch DNA sequencer. DNAStar software was applied to align the sequences and BLAST searching of GenBank was used to assess homology with the known capsid protein gene sequences of PCV2.
3. Results
3.1. Detection limit of the LAMP method
A successful LAMP reaction with PCV2-specific primers at 64 °C for 60 min produced many bands of different sizes upon agarose electrophoresis, since the LAMP products consisted of several inverted-repeat structures. The amplification by LAMP showed a ladder-like pattern, whereas the PCR product was a specific DNA band. The result indicated that the detection limit of the PCV2 LAMP was five copies per reaction whereas that of PCR was 25 copies (Fig. 1 ). The detection sensitivity of LAMP was therefore fivefold better than for conventional PCR.
Fig. 1.
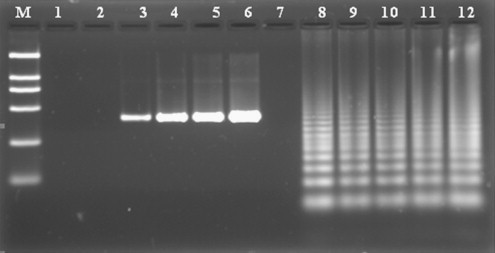
Comparative sensitivities of LAMP and PCR for the detection of PCV2 by agarose gel electrophoresis. From left to right: lane M, DNA Marker DL-2000 (Takara); lanes 1–6, different PCV2 copy numbers subjected to PCR (1, 5, 25, 125, 625 and 3125 copies/tube, respectively); lanes 7–12, different PCV2 copy numbers subjected to LAMP assay (1, 5, 25, 125, 625 and 3125 copies/tube, respectively). PCR products showed a specific amplification for the cap gene of PCV2-BJ with a detection limit of 25 copies, whereas detection limit for LAMP was five copies per reaction.
3.2. Comparative detection sensitivity of LAMP and PCR for PCV2 in clinical samples
In order to evaluate the optimal tissues for viral detection and to compare the sensitivity of PCV2 detection by LAMP and PCR, DNAs from tissue samples of blood, heart, lung, liver, kidney, lymph nodes and spleen from PCV2-infected pigs were extracted and subjected to LAMP and PCR. There was a 100% positive detection rate for both PCR and LAMP on extracts of lung, kidney and heart tissue. However, LAMP showed higher sensitivity than PCR for the detection of PVC2 DNA in blood, lymph nodes, liver and spleen tissue samples (Table 1). Overall, the detection rate of PCV2 LAMP for 86 clinical tissue samples was 96.5% and appeared better than that for the PCR method.
3.3. Analytical cross-reaction with PCV1, PPV, PRV and PRRSV of the PCV2 LAMP method
DNA extracted from tissues of healthy animals, pigs infected with PCV1, PPV and PRV, and cDNA from PRRSV were used as templates for PCV2 LAMP. Agarose gel electrophoresis analysis indicated that the PCV2 LAMP reaction did not detect PCV1, PPV, PRV, or PRRSV, and gave a negative reaction with tissues of healthy swine. Only with the PCV2-BJ DNA did the PCV2 primer set give a positive reaction (Fig. 2 ).
Fig. 2.
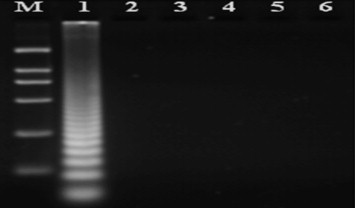
Electrophoretic analysis of cross-reaction in the PCV2 LAMP assay. PCV2-BJ, PCV1, PPV, PRV, and PRRSV were used as targets for the PCV2 LAMP assay. From left to right: lane M, DNA Marker DL-2000; lane 1, DNA of PCV2-BJ; lane 2, DNA of PCV1; lane 3, DNA of PPV; lane 4, DNA of PRV; lane 5, cDNA of PRRSV; lane 6, DNA from healthy swine tissue.
4. Discussion
PMWS was first observed in piglets of a high-health herd in Canada in 1991(Harding and Clark, 1997), and appeared to be an emerging disease that affected swine herds in many countries of North America, Europe and Asia (Allan et al., 1998, Choi et al., 2000). PCV2 (which differs markedly from PCV1) was commonly found in pigs with PMWS (Allan et al., 1998). Several researchers reported that PPV, PRV and PRRSV could also reproduce symptoms typical of PMWS (Ellis et al., 2000, Rodriguez et al., 1999, Rovira et al., 2002). Thus, the development of a simple and rapid diagnostic tool that could detect PCV2 and differentiate it from PCV1, PPV, PRV and PRRSV in the same samples would be of significance for epidemiological surveillance and prediction of the severity of PMWS outbreaks in swine herds.
LAMP is a new diagnostic method which is quite simple, requiring only a conventional water bath or heat block for incubation under isothermal conditions. Another useful feature of LAMP is that its products can be observed directly, by naked eye, because a white precipitate of magnesium pyrophosphate forms in the reaction tube (Mori et al., 2001). Adding SYBR Green I to LAMP reactions can increase the ease and sensitivity of detection by the naked eye (Iwamoto et al., 2003).
Some samples from blood, lymph nodes, liver and spleen that were positive by LAMP were not detected as positive by PCR. The greater sensitivity of LAMP (as compared to PCR) for detecting PCV2 detection accords with the sensitivity reported for LAMP methods used to detect Newcastle disease virus, Japanese encephalitis virus, mumps virus and West Nile virus (Okafuji et al., 2005, Parida et al., 2004, Parida et al., 2006, Pham et al., 2005). The lack of cross-reaction observed with PCV1, PPV, PRV and PRRSV suggest that the PCV2 LAMP system possesses reliable specificity in addition to high sensitivity. The presence of PCV2 in blood and many tissues following natural infection (Darwich et al., 2004, Kennedy et al., 2000) was confirmed by the PCV2 LAMP method using, in the present study, clinical samples of blood, lymph nodes, lung, liver, kidney, heart, and spleen. The optimal tissues for PCV2 LAMP are probably the blood, lymph nodes, lung, kidney and heart because these gave a 100% detection rate in the LAMP assay.
LAMP is a simple and timesaving procedure, allowing results to be obtained within 1 h, whereas the PCR method typically requires 2–4 h. Compared with PCR, the LAMP method appears to be a fast and sensitive tool for the clinical diagnosis of PCV2 infection. Nonetheless, the reliability of this assay should be further evaluated by large-scale investigation.
In conclusion, a PCV2 LAMP assay was developed successfully and shown to be a simple, highly sensitive, rapid and reliable method for the clinical diagnosis of PCV2 infection.
Acknowledgements
This work was supported in part by grants from the National Key Technologies R&D Program of China (no. 2006BAD06A03). This study was also supported by the National High-tech R&D Program (no. 2006AA10A208-1-1) and the National Natural Science Foundation of China (no.30671563 and no. 30700597).
References
- Allan G.M., Ellis J.A. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 2000;12:3–14. doi: 10.1177/104063870001200102. [DOI] [PubMed] [Google Scholar]
- Allan G.M., McNeilly F., Kennedy S., Daft B., Clarke E.G., Ellis J.A., Haines D.M., Meehan B.M., Adair B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Invest. 1998;10:3–10. doi: 10.1177/104063879801000102. [DOI] [PubMed] [Google Scholar]
- Blanchard P., Mahe D., Cariolet R., Truong C., Dimna M.L., Arnauld C., Rose N., Eveno E., Albina E., Madec F., Jestin A. An ORF2 protein-based ELISA for porcine circovirus type 2 antibodies in post-weaning multisystemic wasting syndrome. Vet. Microbiol. 2003;94:183–194. doi: 10.1016/S0378-1135(03)00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolin S.R., Stoffregen W.C., Nayar G.P., Hamel A.L. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J. Vet. Diagn. Invest. 2001;13:185–194. doi: 10.1177/104063870101300301. [DOI] [PubMed] [Google Scholar]
- Brunborg I.M., Moldal T., Jonassen C.M. Quantitation of porcine circovirus type 2 isolated from serum/plasma and tissue samples of healthy pigs and pigs with postweaning multisystemic wasting syndrome using a TaqMan-based real-time PCR. J. Virol. Methods. 2004;122:171–178. doi: 10.1016/j.jviromet.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Cheung A.K. Transcriptional analysis of porcine circovirus type 2. Virology. 2003;305:168–180. doi: 10.1006/viro.2002.1733. [DOI] [PubMed] [Google Scholar]
- Cho H.S., Kim T.J., Lee J.I., Park N.Y. Serodiagnostic comparison of enzyme-linked immunosorbent assay and surface plasmon resonance for the detection of antibody to porcine circovirus type 2. Can. J. Vet. Res. 2006;70:263–268. [PMC free article] [PubMed] [Google Scholar]
- Choi C., Chae C., Clark E.G. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J. Vet. Diagn. Invest. 2000;12:151–153. doi: 10.1177/104063870001200209. [DOI] [PubMed] [Google Scholar]
- Chung W.B., Chan W.H., Chaung H.C., Lien Y., Wu C.C., Huang Y.L. Real-time PCR for quantitation of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 in naturally infected and challenged pigs. J. Virol. Methods. 2005;124:11–19. doi: 10.1016/j.jviromet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Darwich L., Segalés J., Mateu E. Pathogenesis of postweaning multisystemic wasting syndrome caused by porcine circovirus 2: an immune riddle. Arch. Virol. 2004;149:857–874. doi: 10.1007/s00705-003-0280-9. [DOI] [PubMed] [Google Scholar]
- Ellis J.A., Bratanich A., Clark E.G., Allan G., Meehan B., Haines D.M., Harding J., West K.H., Krakowka S., Konoby C., Hassard L., Martin K., McNeilly F. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Invest. 2000;12:21–27. doi: 10.1177/104063870001200104. [DOI] [PubMed] [Google Scholar]
- Harding J.C., Clark E.G. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) Swine Health Prod. 1997;5:201–203. [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;43:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Sonobe T., Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Moffett D., McNeilly F., Meehan B., Ellis J., Krakowka S., Allan G.M. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 2000;122:9–24. doi: 10.1053/jcpa.1999.0337. [DOI] [PubMed] [Google Scholar]
- Kim J., Chae C. Multiplex nested PCR compared with in situ hybridization for the differentiation of porcine circoviruses and porcine parvovirus from pigs with postweaning multisystemic wasting syndrome. Can. J. Vet. Res. 2003;67:133–137. [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chae C. A comparison of the lymphocyte subpopulations of pigs experimentally infected with porcine circovirus 2 and/or parvovirus. Vet. J. 2003;165:325–329. doi: 10.1016/s1090-0233(02)00240-x. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang L., Willson P., Babiuk L.A. Quantitative, competitive PCR analysis of porcine circovirus DNA in serum from pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 2000;38:3474–3477. doi: 10.1128/jcm.38.9.3474-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe D., Blanchard P., Truong C. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 2000;81:1815–1824. doi: 10.1099/0022-1317-81-7-1815. [DOI] [PubMed] [Google Scholar]
- Mankertz A., Mankertz J., Wolf K., Buhk H.J. Identification of a protein essential for replication of porcine circovirus. J. Gen. Virol. 1998;79:381–383. doi: 10.1099/0022-1317-79-2-381. [DOI] [PubMed] [Google Scholar]
- Meehan B.M., McNeilly F., Todd D., Kennedy S., Jewhurst V.A., Ellis J.A., Hassard L.E., Clark E.G., Haines D.M., Allan G.A. Characterization of novel circovirus DNA associated with wasting syndromes in pigs. J. Gen. Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nawagitgul P., Morozov I., Bolin S.R., Harms P.A., Sorden S.D. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000;81:2281–2287. doi: 10.1099/0022-1317-81-9-2281. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafuji T., Yoshida N., Fujino M., Motegi Y., Ihara T., Ota Y., Notomi T., Nakayama T. Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:1625–1631. doi: 10.1128/JCM.43.4.1625-1631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera A., Sibila M., Calsamiglia M., Segales J., Domingo M. Comparison of porcine circovirus type 2 load in serum quantified by a real time PCR in postweaning multisystemic wasting syndrome and porcine dermatitis and nephropathy syndrome naturally affected pigs. J. Virol. Methods. 2004;117:75–80. doi: 10.1016/j.jviromet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of west Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M.M., Santhosh S.R., Dash P.K., Tripathi N.K., Saxena P., Ambuj S., Sahni A.K., Lakshmana R., Morita K. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 2006;44:4172–4178. doi: 10.1128/JCM.01487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H.M., Nakajima C., Ohashi K., Onuma M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J. Clin. Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Leung C.S.W., Chan K.H., Lee J.H.C., Yuen K.Y., Guan Y., Peiris J.S.M. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A.G.M., Segales J., Rosell C., Quintana J., Ayllon S., Camprodon A., Domingo M. Aujeszky's disease virus infection concurrent with postweaning multisystemic wasting syndrome in pigs. Vet. Rec. 1999;144:152–153. doi: 10.1136/vr.144.6.152. [DOI] [PubMed] [Google Scholar]
- Rovira A., Balasch M., Segales J., Garcia L., Plana-Duran J., Rosell C., Ellerbrok H., Mankertz A., Domingo M. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J. Virol. 2002;76:3232–3239. doi: 10.1128/JVI.76.7.3232-3239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J., Domingo M. Postweaning multisystemic wasting syndrome (PMWS) in pigs: a review. Vet. Q. 2002;24:109–124. doi: 10.1080/01652176.2002.9695132. [DOI] [PubMed] [Google Scholar]


