Highlights
► A set of primers specific to the BVDV genome was designed and used in the RT-LAMP. ► Both genotypes of BVDV can be detected by the RT-LAMP method. ► The RT-LAMP is a rapid, sensitive and specific diagnostic assay.
Keywords: Bovine viral diarrhea virus (BVDV), 5′ untranslated region (5′UTR), Reverse transcription loop-mediated isothermal amplification (RT-LAMP)
Abstract
A reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed and optimized to detect bovine viral diarrhea viral (BVDV) RNA. The RT-LAMP assay is highly sensitive and able to detect 4.67 × 100 copies of BVDV RNA. Additionally, the RT-LAMP method is capable of detecting both genotypes of BVDV. No cross-reaction with other bovine viruses was observed. The ability of RT-LAMP to detect BVDV RNA from bovine fecal swabs was also evaluated. Of the 88 fecal swabs, 38 were found to be positive by RT-LAMP assay, whereas 39 were positive by real-time RT-PCR. Taken together, the BVDV specific RT-LAMP method is highly specific and sensitive and can be used as a rapid and direct diagnostic assay for testing clinical samples.
1. Introduction
Bovine viral diarrhea virus (BVDV) is a positive-sense, single-stranded RNA virus with a genome size of approximately 12.5 kb. BVDV is a member of the genus Pestivirus in the family Flaviviridae. BVDV is classified into two biotypes, cytopathogenic and noncytopathogenic, based on the presence or absence of the cytopathogenic effects in cell cultures. In addition, there are two major genotypes of BVDV (BVDV1 and BVDV2) based on the genetic relatedness (Ridpath et al., 1994).
BVDV has a high prevalence rate and low mortality, leading to significant economic losses (Houe, 1995). BVDV infected animals may develop fever, mild diarrhea, and leukopenia. Infection of pregnant animals with noncytopathogenic BVDV during the first trimester may cause abortion, stillborn, or persistently infected claves (Mahony et al., 2005). Noncytopathogenic BVDV may spontaneously mutate to the cytopathogenic biotype, resulting in the onset of fatal mucosal disease (Meyers et al., 1991). Calves infected persistently are a major source of virus shedding since they usually do not exhibit any apparent clinical signs.
Identification of persistently infected calves, in combination with a proper vaccination program, is essential to the successful control of BVDV. The currently available diagnostic methods for BVDV include virus isolation, immunoassay, electron microscopy (EM), nuclei acid hybridization, and reverse transcription-polymerase chain reaction (RT-PCR) (Deregt et al., 2002, Ridpath et al., 2002, Fulton et al., 2006, Youssef, 2006). Although RT-PCR is a highly sensitive and specific test for detecting BVDV RNA, it has the intrinsic disadvantage of requiring a high-precision instrument such as a thermocycler for amplification and a time-consuming and complicated detection method (gel electrophoresis unit). In recent years, many studies have demonstrated the potential application of loop-mediated isothermal amplification (LAMP) or reverse transcription (RT)-LAMP assay for rapid detection of viral DNA or RNA (Parida et al., 2004, Enomoto et al., 2005, Cho et al., 2006, Chen et al., 2008, Komiyama et al., 2009, Rovira et al., 2009, Fan et al., 2010, Yin et al., 2010). Like RT-PCR, the RT-LAMP technique amplifies the target viral RNA sequence (Notomi et al., 2000). In this study, a RT-LAMP assay for detection of BVDV RNA is developed. Since the 5′ untranslated region (5′UTR) of BVDV is among the most conserved regions and has been chosen as a preferred target region for detection of BVDV RNA by RT-PCR (Letellier et al., 1999, Fan et al., 2010), a set of six primers was designed to amplify 6 target sequences at the 5′UTR of the BVDV genome for the RT-LAMP assay.
2. Materials and methods
2.1. BVDV reference strains, field isolates, and other bovine pathogens
BVDV reference strains, field isolates, and other bovine pathogens used in the study are listed in Table 1 . Three BVDV genotype 1 (BVDV1) reference strains, 13 BVDV1 field isolates, and 1 BVDV genotype 2 (BVDV2) reference strain were used to develop and optimize the RT-LAMP conditions. The BVDV reference virus strains were propagated in Madin-Darby bovine kidney (MDBK) cells in Dulbecco's modification of Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Shijiqing, China, free of BVDV and antibody to BVDV). Thirteen field isolates were isolated from calves with typical BVDV clinical signs such as diarrhea, miscarriages and stillborn. For virus isolation, a total of 1 g of fresh aseptically collected liver tissues was homogenized in 4 ml of phosphate-buffered saline (PBS, pH 7.2). A 1:10 dilution of the supernatants was then inoculated onto MDBK monolayer cultures and incubated for 6 days. The presence or absence of cytopathic effect was recorded. After 3 times of freezing and thawing, culture supernatants were collected after centrifugation and stored at −70 °C for further characterization with indirect immunoperoxidase (IPX) test and electron microscopy (EM). All 13 field isolates were BVDV positive as revealed by both positive IPX staining and the typical BVDV morphology (40–60 nm in diameter) under an electron microscope.
Table 1.
Virus and samples used in the RT-LAMP assay.
| Name | Source | RT-LAMP result |
|
|---|---|---|---|
| Agarose gel electrophoresis | Color change after adding dye | ||
| BVDV1 | |||
| Oregon CV24 | CVCC, China | + | + |
| NADL | CVCC, China | + | + |
| AV68 | CVCC, China | + | + |
| GX-BVDV1 | GVRI | + | + |
| GX-BVDV2 | GVRI | + | + |
| GX-BVDV3 | GVRI | + | + |
| GX-BVDV4 | GVRI | + | + |
| GX-BVDV5 | GVRI | + | + |
| GX-BVDV6 | GVRI | + | + |
| GX-BVDV7 | GVRI | + | + |
| GX-BVDV8 | GVRI | + | + |
| GX-BVDV9 | GVRI | + | + |
| GX-BVDV10 | GVRI | + | + |
| GX-BVDV11 | GVRI | + | + |
| GX-BVDV12 | GVRI | + | + |
| GX-BVDV13 | GVRI | + | + |
| BVDV2 | |||
| GX-041 | GVRI | + | + |
| Other bovine pathogens | |||
| Bovine rotavirus (NCDV strain, G6P10 genotype) | CVCC, China | − | − |
| Mycobacterium bovis (MB332, Guangxi field isolate) | CVCC, China | − | − |
| Classical swine fever virus (Guangming stain, AV64, North American genotype) | CVCC, China | − | − |
| Classical swine fever virus (Shimen stain, AV1411, North American genotype) | |||
| Classical swine fever virus (79105 strain, AV63, North American genotype) | |||
| Infective bovine rhinotracheitis virus (Bartha Nu/67 strain, AV20) | CVCC, China | − | − |
| Bovine Coronavirus (GX-BC-125, Guangxi field isolate) | GVRI | − | − |
| Negative tissue samples | |||
| Healthy bovine nasal swab | GVRI | − | − |
| Healthy bovine blood sample | GVRI | − | − |
| Positive tissue sample | |||
| BVDV bovine nasal swab | GVRI | + | − |
| BVDV bovine blood sample | GVRI | + | − |
| Fecal negative samples | |||
| A/Holstein cow/Guangxi/NN1732/2009 | NN | − | − |
| A/Holstein cow/Guangxi/NN3363/2009 | NN | − | − |
| A/Holstein cow/Guangxi/NN4523/2009 | NN | − | − |
| A/Holstein cow/Guangxi/NN4462/2009 | NN | − | − |
| A/Holstein cow/Guangxi/NN21/2009 | NN | − | − |
| A/Holstein cow/Guangxi/NN12/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN789/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN35/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN4520/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN7/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN3620/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN49/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN332/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN28/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN0137/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN703/2009 | NN | − | − |
| A/Water baffalo/Guangxi/NN46/2009 | NN | − | − |
| A/Yellow cowGuangxi/LZ789/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ45/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ733/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ719/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ776/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ782/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ713/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ708/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ730/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ744/2010 | LZ | − | − |
| A/Yellow cowGuangxi/LZ760/2010 | LZ | − | − |
| Cell cultures | |||
| MDBK 1 | GVRI | − | − |
| MDBK 2 | GVRI | − | − |
| MDBK 3 | GVRI | − | − |
| MDBK 4 | GVRI | − | − |
| MDBK 5 | GVRI | − | − |
CVCC: Chinese Veterinary Culture Collection Center; GVRI: Guangxi Veterinary Research Institute; NN: Jinguang Diary Farm, Nanning, Guanxi Province; LZ: Huangshi Cattle Farm, Liuzhou, Guanxi Province; +: positive; −: negative.
Five other bovine pathogens, 2 negative tissue samples (1 healthy bovine nasal swab and 1 healthy bovine blood sample), 28 fecal negative samples, and 5 individual MDBK cell culture samples were included as negative controls to test the specificity of the assay (Table 1). All the BVDV positive and negative samples were validated by both the BVDV antibody test kit (IDEXX, USA) and an RT-PCR assay.
2.2. RNA/DNA extraction
The genomic viral RNA was extracted from 250 μl of BVDV-infected culture supernatant by using the TRIZOL RNA extract reagent (Invitrogen, America) in accordance with the manufacture's protocol. DNA was extracted using phenol:chloroform:isoamyl alcohol (1:1:24, v/v/v) according to the Gibco BRL manufacturer's protocol (Gibco BRL, Grand Island, NY, USA). The extracted RNA and DNA were eluted in distilled water and stored at −70 °C until use.
2.3. Primer design
A multiple sequence alignment was performed for 40 randomly selected BVDV1 isolates and 15 BVDV2 isolates from GenBank database. The 5′UTR of BVDV, which is sufficiently conserved among the randomly selected BVDV isolates, was chosen for primer design. Primer Explored V4 software was used to design the RT-LAMP primers (http://primerexplorer.jp/e/). The two outer primers are known as the forward outer primer (F3) and the backward outer primer (B3), which helps in strand displacement. The inner primers are known as the forward inner primer (FIP) and the backward inner primer (BIP), respectively. Each inner primer has two distinct sequences corresponding to the sense and antisense sequences of the target, one for priming during the early stage and the other for self-priming during late stage of LAMP. FIP contains F1C (complementary to F1) and F2 sequence. BIP contains the B1C sequence (complementary to B1) and B2 sequence. Both FIP and BIP were high-performance liquid chromatography purified. FIP and BIP contained 3 R(C+G) and 1 Y(A+T) to facilitate the amplification of both BVDV1 and BVDV2 (Table 2 and Fig. 1 ). All primers were synthesized by Invitrogen (GuangZhou, China). The nucleotide sequences and locations of the primers for both Oregon CV24 (GenBank accession no. 0911605.1) and New York 93 (GenBank accession no. AF502399.1) are shown in Table 2 and Fig. 1.
Table 2.
Primers used in RT-LAMP.
The positions of the primers are based on the nucleotide sequence of Oregon CV24 (GenBank accession no. AF0911605.1).
a Each inner primer of RT-LAMP contains two connected primers. The R and Y are shown in red color.
Fig. 1.

Locations of the primers used in RT-LAMP. The GenBank accession numbers for Oregon CV24 and New York 93 are 0911605.1 and 502399.1, respectively. The nucleotide sequences of primers are underlined. The mismatched nucleotides between the two strains are marked in red.
2.4. RT-PCR
RT-PCR was performed as described previously (Hamel et al., 1995). Briefly, RT-PCR was performed with the Quant One Step RT-PCR Kit (Qiagen Inc., Beijing, China) by using 1 μl (20 ng) of RNA template and 50 pmol of each primer in a 25 μl reaction volume by following the manufacturer's protocol with the following cycling times and temperatures: 94 °C for 5 min and 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. RT-PCR product was analyzed by agarose gel electrophoresis.
2.5. Real-time RT-PCR
Real-time RT-PCR was performed using the following primers and probe, which detects a 102 bp product of the 5′UTR of BVDV. The primers are 5′-TAGCCATGCCCTTAGTAGGACT-3′ and 5′-GAACCACTGACGACTACCCTGT-3′. The probe is FAM-CAGTGGTGAGTTCGTTGGATGGCT-BHQ1. Real-time RT-PCR amplification was carried out with the Real-time One Step RT-PCR Kit (Takara, Dalian, China) as described previously (Fan et al., 2010). Briefly, 10 μl of 2× One Step RT buffer III, 0.5 μl of TaKaRa Ex Taq HS (5 U/μl), 0.5 μl of PrimeScript RT Enzyme Mix II (5 U/μl), 0.5 μl of each primer (4 μM), 0.5 μl of probe (4 μM), 1 μl of RNA template, and 7 μl of DNase/RNase-free water were added into a 0.2 ml tube. Real-time PCR was performed in a Light Cycler 2.0 (Roche Diagnostic, Indianapolis, IN) using the following conditions: reverse transcription reaction at 42 °C for 5 min, 95 °C for 10 s, and 40 cycles of 95 °C for 5 s, 60 °C for 30 s. Cycle threshold (CT) was manually set up to reflect the best kinetic parameter.
2.6. RT-LAMP
The RT-LAMP reaction was performed in a 25 μl reaction mixture containing 1.4 μmol of each deoxyribonucleotide triphosphate, 0.8 mmol of betaine (Sigma Chemical Co., Beijing, China), 2.5 μl of 10× Thermo buffer, 8 mmol MgSO4, 8 U of Bst DNA polymerase (large fragment; New England Biolabs), 0.125 U of enhanced Avian myeloblastosis virus reverse transcriptase (Takara, Dalian, China), and 1 μl of the extracted target RNA. Various concentrations of the FIP, BIP, F3, B3, loop F, and loop B primers were used to optimize the ratio of primers. To determine the optimal temperature and incubation time for the RT-LAMP assay, the reaction mixtures were incubated in a water bath at 60 °C, 61.5 °C, 63 °C, 64.5 °C, and 66 °C for 20, 40, 60 and 80 min, respectively. The reaction was terminated by heating at 80 °C for 2 min.
2.7. Analysis of RT-LAMP product
The RT-LAMP products were analyzed by three methods. The first and the most direct method was to visually inspect the turbidity of the samples formed due to the accumulation of magnesium pyrophosphate, a byproduct of the DNA amplification reaction (Cho et al., 2006, Thekisoe et al., 2009). The second method was to add a fluorescent dye such as SYBR Green I (Solarbio, Beijing, China) or Genefinder™ (Boiv, Xiamen, China) to the samples and to observe the color change under an ultraviolet (UV) hand lamp at a 365 nm wavelength. Samples which were yellow-green were considered positive, while samples which were orange were considered negative. Samples were compared with a negative control to allow for background fluorescence (Rovira et al., 2009, Fan et al., 2010). Finally, the RT-LAMP products were detected by agarose gel electrophoresis. The RT-LAMP reaction generates a combination of DNA fragments of different sizes. The presence of a smear or a pattern of multiple bands of different molecular weights was considered a positive result. A molecular marker was used to estimate the sizes of amplified products.
2.8. Specificity and sensitivity of RT-LAMP
To evaluate the specificity of the RT-LAMP assay, experiments were performed initially on a panel of reference BVDV virus strains including both genotype 1 and 2 (Table 1). In addition, bovine rotavirus (BRV), Mycobacterium bovis (Golemba et al., 2008), classical swine fever virus (CSFV), infectious bovine rhinotracheitis virus (BoHV-1), bovine coronavirus (BC), bovine negative fecal samples, healthy bovine nasal swab and blood sample, and MDBK cells were included as negative controls.
The detection limit of RT-LAMP was tested and compared with conventional RT-PCR and real-time RT-PCR by using the same templates at identical RNA concentrations (4.67 × 108 to 4.67 × 10−1 copies of RNA/μl). Additionally, the healthy cattle fecal swab samples spiked with known amount of viral RNA of BVDV strain Oregon CV24 were quantitated by real-time RT-PCR and RT-LAMP. Briefly, BVDV RNA derived from Oregon CV24 was serially diluted from 4.67 × 108 to 4.67 × 10−1 copies of RNA/μl and added to the healthy fecal samples. RNAs extracted from these artificial positive samples were subjected to RT-LAMP and real-time PCR.
2.9. Fecal specimens
A total of 88 fecal swab samples were collected from calves with diarrhea, which came from different dairy farms in the Guangxi province (Table 3 ). A written informed consent was obtained from each participating farm owner. The veterinarians of the participating farms collected fecal swab samples from calves of 6 to 48 months old. No official review and approval of the animal protocol by Guangxi Veterinary Research Institute was needed. The samples were diluted in 1 ml of sterilized water, followed by RNA extraction as described in Section 2.2. BVDV-specific RT-LAMP assay was performed as describe in Section 2.6. The results of BVDV specific RT-LAMP were compared with the results of conventional RT-PCR and real-time RT-PCR. The real-time RT-PCR products were cloned into a PMD-18T (Takara, Dalian, China) vector and sequenced. The phylogenetic analysis of the 102 bp real-time RT-PCR products of these BVDV positive samples was performed using MegAlign (DNAStar 5.0).
Table 3.
Comparison of RT-PCR, real-time RT-PCR, and RT-LAMP methods for detection of BVDV from clinical fecal samples.
| Location of samples | Number of samples | Number of positive samples |
||
|---|---|---|---|---|
| RT-PCR | Real-time RT-PCR | RT-LAMP | ||
| Nanning | 23 | 9 | 13 | 12 |
| Liuzhou | 15 | 5 | 7 | 7 |
| Fangcheng | 10 | 1 | 2 | 2 |
| Shangsi | 9 | 2 | 3 | 3 |
| Guilin | 20 | 9 | 9 | 9 |
| Hengxian | 11 | 4 | 5 | 5 |
| Total | 88 | 30 | 39 | 38 |
3. Results
3.1. Optimization of RT-LAMP
Following standardization and optimization, the optimal ratio of primer concentrations for RT-LAMP reaction was found to be 8:1:4 (1.6, 0.2 and 0.8 mmol) for inner:outer:loop primers. The RT-LAMP assay amplified a 228 bp target sequence of the 5′UTR of BVDV after incubation at 63 °C in 60 min. The RT-LAMP products were observed as a ladder-like pattern on the agarose gel. This is due to the formation of a mixture of stem-loop DNAs with various stem lengths and cauliflower-like structures by annealing between alternately inverted repeats of the target sequence in the same strand.
3.2. Sensitivity and specificity of RT-LAMP
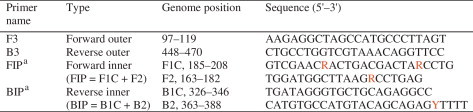
The RT-LAMP specifically detected BVDV Oregon CV24, NADL, AV68, 13 Guangxi field isolates and GX-041. No cross-reactivity with other bovine viruses was observed (Table 1). This specificity was further confirmed by agarose gel electrophoresis and the color change after adding a fluorescent dye (data not shown). The sensitivity of this method was determined by using a 10-fold serial dilution of BVDV RNA. The detection limit of RT-LAMP and real-time RT-PCR were 4.67 × 100 copies, whereas the detection limit of RT-PCR was 4.67 × 103 copies (Fig. 2 ). The detection limit of RT-LAMP for spiking negative samples was 4.67 × 101 copies, whereas the detection limit of real-time RT-PCR for spiking negative samples was 4.67 × 103 copies (data not shown).
Fig. 2.
The sensitivities of RT-LAMP, RT-PCR and real-time RT-PCR assays. A serial 10-fold dilution of BVDV RNA derived from CV24 reference strain was used in the RT-LAMP, RT-PCR and real-time RT-PCR. (A) Results of the RT-LAMP products were viewed under ultraviolet (UV) light. (B) Results of the real-time RT-PCR analysis. (C) Results of the RT-PCR analysis. Lane M: 2000-bp DNA marker; Lane N: negative control; Lane 1: 4.67 × 108 copies/tube; Lane 2: 4.67 × 107 copies/tube; Lane 3: 4.67 × 106 copies/tube; Lane 4: 4.67 × 105 copies/tube; Lane 5: 4.67 × 104 copies/tube; Lane 6: 4.67 × 103 copies/tube; Lane 7: 4.67 × 102 copies/tube; Lane 8: 4.67 × 101 copies/tube; Lane 9: 4.67 × 100 copies/tube; Lane 10: 4.67 × 10−1 copies/tube. All experiments were repeated three times and similar results were obtained.
3.3. Detection of BVDV from clinical samples
The sensitivity of the RT-LAMP method in detecting BVDV RNA from 88 clinical fecal samples was compared with those of RT-PCR and real-time RT-PCR methods. Results are shown in Table 3. Thirty of 88 fecal swab samples (34.1%) were positive by RT-PCR analysis, whereas 39 of 88 fecal swab samples (44.3%) were positive by real-time RT-PCR, and 38 of 88 (43.2%) were positive by RT-LAMP (Table 3). Thirty samples (34.1%) were positive by three methods. Eight samples (9.1%) were positive by both real-time RT-PCR and RT-LAMP, but negative by RT-PCR analysis. No sample (0%) was positive by RT-PCR and negative by RT-LAMP. All RT-PCR positive samples are also RT-LAMP positive. However, a sample was positive by real-time PCR, but negative by RT-LAMP. All the real-time RT-PCR positive samples were true positive as evidenced by DNA sequence analysis, suggesting that no nonspecific amplification in RT-LAMP reaction occurred. A phylogenetic analysis of the 102 bp real-time RT-PCR products of 39 samples in comparison with those of Oregon CV24 and New York 93 is shown in Fig. 3 .
Fig. 3.
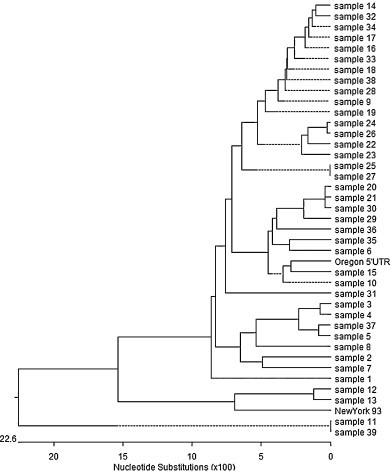
The phylogenetic analysis of the 102 bp real-time RT-PCR products of the 39 BVDV positive field samples in comparison with Oregon CV24 and New York 93 reference strains by using MegAlign (DNAStar 5.0).
4. Discussion
RT-LAMP has been used in detecting viral RNA molecules due to its simplicity and high sensitivity for a number of viruses including avian influenza virus, classical swine fever virus, West Nile virus, and porcine reproductive and respiratory syndrome virus (Parida et al., 2004, Chen et al., 2008, Chen et al., 2010, Rovira et al., 2009). In this study, an RT-LAMP method was developed for detection of both BVDV1 and BVDV2. The RT-LAMP method exhibited the same sensitivity as real-time RT-PCR and did not detect classical swine fever virus and other bovine viruses. Similarly, an RT-RAMP method for classical swine fever virus did not detect BVDV (Chen et al., 2010), suggesting the specificity of each RT-LAMP assay. Twenty-one BVDV1 isolates and 17 BVDV2 isolates were selected at random from the GenBank database and the sequence homology in the 5′UTR region upon which the RT-LAMP primers were designed was analyzed. It is estimated that the RT-LAMP would detect 95% of BVDV1 and 70.6% of BVDV2. The lower detection rate for BVDV2 is primarily due to the higher genetic variations observed in the 5′UTR of BVDV2 compared to BVDV1 isolates. All 39 BVDV positive samples confirmed by real-time RT-PCR were clustered within the BVDV1 genotype as revealed by DNA sequence analysis of the 102 bp PCR product (Fig. 3). The RT-LAMP method will be particularly useful in detecting BVDV from cattle herds in China since BVDV1 is the major type of BVDV circulating in the cattle herds (Li et al., 1983, Xue et al., 2010). BVDV2 was first isolated from cattle in China in 2010 (Xie et al., 2011). Due to the inherent genetic heterogeneity of BVDV, it remains to be a challenge to develop a universal molecular test that is capable of detecting all BVDV isolates.
The BVDV specific RT-LAMP method is 1000 times more sensitive than RT-PCR in detecting BVDV RNA. In contrast, real-time RT-PCR has the same sensitivity as RT-LAMP (4.67 copies of viral RNA). The RT-LAMP failed to detect a true BVDV positive clinical fecal sample, which was confirmed by real-time RT-PCR and DNA sequencing. There were 8 fecal samples positive by RT-LAMP and real-time RT-PCR but negative by RT-PCR. It is possible that the 8 fecal samples may have had few viral RNA molecules, which were below the detection limit of RT-PCR analysis. It is interesting to note that the primers for RT-PCR (100–342 nt), real-time RT-PCR (104–205 nt), and RT-RAMP (97–470 nt) described in the study detected the same 5′UTR region of BVDV Oregon CV24 (GenBank accession # AF0911605.1).
SYBR Green I and GeneFinder™ were used for the detection of the amplified products and their detection efficiency was compared. Both fluorescent dyes give consistent results. Some previous studies indicated that false positive results could occur due to the high amplification efficiency, short target sequences, or contaminations associated with adding the fluorescent dye (Thekisoe et al., 2009). The false positive results associated with contamination can be avoided by adding SYBR green I directly to the reaction mixture.
In conclusion, the BVDV-specific RT-LAMP is a simple, rapid, highly sensitive and specific assay. This technique has potential application in both clinical diagnosis and field surveillance of BVDV since it does not require the use of sophisticated equipment.
Acknowledgements
This work was partly supported by Guangxi Natural Science Foundation (2011GXNSFA018096) and by Guangxi Government Senior Scientist Foundation (2011B020).
Contributor Information
Zhixun Xie, Email: xiezhixun@126.com.
Xiuqing Wang, Email: xiuqing.wang@sdstate.edu.
References
- Chen H.T., Zhang J., Sun D.H., Ma L.N., Liu X.T., Cai X.P., Liu Y.S. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. Journal of Virological Methods. 2008;151:200–203. doi: 10.1016/j.jviromet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Chen L., Fan X.Z., Wang Q., Xu L., Zhao Q.Z., Zhou Y.C., Liu J., Tang B., Zou X.Q. A novel RT-LAMP assay for rapid and simple detection of classical swine fever virus. Virologica Sinica. 2010;25:59–64. doi: 10.1007/s12250-010-3043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.S., Kang J.I., Park N.Y. Detection of canine parvovirus in fecal samples using loop-mediated isothermal amplification. Journal of Veterinary Diagnostic Investigation. 2006;18:81–84. doi: 10.1177/104063870601800111. [DOI] [PubMed] [Google Scholar]
- Deregt D., Carman P.S., Clark R.M., Burton K.M., Olson W.O., Gilbert S.A. A comparison of polymerase chain reaction with and without RNA extraction and virus isolation for detection of bovine viral diarrhea virus in young calves. Journal of Veterinary Diagnostic Investigation. 2002;14:433–437. doi: 10.1177/104063870201400516. [DOI] [PubMed] [Google Scholar]
- Enomoto Y., Yoshikawa T., Ihira M., Akimoto S., Miyake F., Usui C., Suga S., Suzuki K., Kawana T., Nishiyama Y., Asano Y. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. Journal of Clinical Microbiology. 2005;43:951–955. doi: 10.1128/JCM.43.2.951-955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Xie Z., Liu J., Pang Y., Deng X., Xie Z., Peng Y., Xie L., Khan M.I. Establishment of real-time fluorescent quantitative PCR for detection of bovine viral diarrhea virus. Progress in Veterinary Medicine. 2010;31:10–14. [Google Scholar]
- Fulton R.W., Hessman B., Johnson B.J., Ridpath J.F., Saliki J.T., Burge L.J., Sjeklocha D., Confer A.W., Funk R.A., Payton M.E. Evaluation of diagnostic tests used for detection of bovine viral diarrhea virus and prevalence of subtypes 1a, 1b, and 2a in persistently infected cattle entering a feedlot. Journal of the American Veterinary Medical Association. 2006;228:578–584. doi: 10.2460/javma.228.4.578. [DOI] [PubMed] [Google Scholar]
- Golemba M.D., Parreno V., Jones L.R. Simple procedures to obtain exogenous internal controls for use in RT-PCR detection of bovine pestiviruses. Molecular and Cellular Probes. 2008;22:212–214. doi: 10.1016/j.mcp.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Hamel A.L., Wasylyshen M.D., Nayar G.P. Rapid detection of bovine viral diarrhea virus by using RNA extracted directly from assorted specimens and a one-tube reverse transcription PCR assay. Journal of Clinical Microbiology. 1995;33:287–291. doi: 10.1128/jcm.33.2.287-291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houe H. Epidemiology of bovine viral diarrhea virus. Veterinary Clinics of North America. Food Animal Practice. 1995;11:521–547. doi: 10.1016/s0749-0720(15)30465-5. [DOI] [PubMed] [Google Scholar]
- Komiyama C., Suzuki K., Miura Y., Sentsui H. Development of loop-mediated isothermal amplification method for diagnosis of bovine leukemia virus infection. Journal of Virological Methods. 2009;157:175–179. doi: 10.1016/j.jviromet.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Letellier C., Kerkhofs P., Wellemans G., Vanopdenbosch E. Detection and genotyping of bovine diarrhea virus by reverse transcription-polymerase chain amplification of the 5′ untranslated region. Veterinary Microbiology. 1999;64:155–167. doi: 10.1016/S0378-1135(98)00267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu Z., Wu Y. Isolation and identification of bovine viral diarrhea virus-mucosal disease virus strain Changchun 184. Chinese Journal of Veterinary Science. 1983;3:546–553. [Google Scholar]
- Mahony T.J., McCarthy F.M., Gravel J.L., Corney B., Young P.L., Vilcek S. Genetic analysis of bovine viral diarrhoea viruses from Australia. Veterinary Microbiology. 2005;106:1–6. doi: 10.1016/j.vetmic.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Meyers G., Rumenapf T., Tautz N., Dubovi E.J., Thiel H.J. Insertion of cellular sequences in the genome of bovine viral diarrhea virus. Archives of Virology, Supplement. 1991;3:133–142. doi: 10.1007/978-3-7091-9153-8_15. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. Journal of Clinical Microbiology. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath J.F., Bolin S.R., Dubovi E.J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- Ridpath J.F., Hietala S.K., Sorden S., Neill J.D. Evaluation of the reverse transcription-polymerase chain reaction/probe test of serum samples and immunohistochemistry of skin sections for detection of acute bovine viral diarrhea infections. Journal of Veterinary Diagnostic Investigation. 2002;14:303–307. doi: 10.1177/104063870201400405. [DOI] [PubMed] [Google Scholar]
- Rovira A., Abrahante J., Murtaugh M., Munoz-Zanzi C. Reverse transcription loop-mediated isothermal amplification for the detection of Porcine reproductive and respiratory syndrome virus. Journal of Veterinary Diagnostic Investigation. 2009;21:350–354. doi: 10.1177/104063870902100308. [DOI] [PubMed] [Google Scholar]
- Thekisoe O.M., Bazie R.S., Coronel-Servian A.M., Sugimoto C., Kawazu S., Inoue N. Stability of Loop-Mediated Isothermal Amplification (LAMP) reagents and its amplification efficiency on crude trypanosome DNA templates. Journal of Veterinary Medical Science. 2009;71:471–475. doi: 10.1292/jvms.71.471. [DOI] [PubMed] [Google Scholar]
- Xie Z., Tang Y., Fan Q., Liu J., Pang Y., Deng X., Xie Z., Peng Y., Xie L., Khan M.I. Rapid detection of Group I avian adenoviruses by a loop-mediated isothermal amplification. Avian Diseases. 2011;55:575–579. doi: 10.1637/9719-031611-Reg.1. [DOI] [PubMed] [Google Scholar]
- Xue F., Zhu Y.M., Li J., Zhu L.C., Ren X.G., Feng J.K., Shi H.F., Gao Y.R. Genotyping of bovine viral diarrhea viruses from cattle in China between 2005 and 2008. Veterinary Microbiology. 2010;143:379–383. doi: 10.1016/j.vetmic.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Yin S., Shang Y., Zhou G., Tian H., Liu Y., Cai X., Liu X. Development and evaluation of rapid detection of classical swine fever virus by reverse transcription loop-mediated isothermal amplification (RT-LAMP) Journal of Biotechnology. 2010;146:147–150. doi: 10.1016/j.jbiotec.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Youssef B.Z. Comparative study between ELISA, immuno-diffusion and cell bound immuno assay techniques for detection of anti-bovine viral diarrhea antibodies in calves of some farms in Alexandria and Behira governorates. Journal of the Egyptian Public Health Association. 2006;81:29–41. [PubMed] [Google Scholar]


