Abstract
Porcine hemagglutinating encephalomyelitis virus (PHEV), which causes porcine encephalomyelitis and is widespread among swine worldwide. RNA interference (RNAi) pathways have emerged as important regulators of virus–host cell interactions. In this study, two siRNA expression plasmids (shN1 and shN2) were generated to target two different coding regions of the nucleocapsid protein (N) of PHEV. The shRNAs were transiently transfected into a porcine kidney cell line, PK-15, to determine whether these constructs inhibited PHEV production. Our results revealed that both shRNAs were highly capable of inhibiting viral RNA genome replication, especially shN2. Next, stable transfection of shN2 was used to produce two siRNA stably expressing PK-15 cell clones (shN2-1 and shN2-2), and these two lines were infected with PHEV. The analysis of cytopathic effects (CPE) demonstrated that shN2-1 and shN2-2 were capable of protecting cells against PHEV infection with high specificity and efficiency. Furthermore, effective inhibition of viral replication persisted for up to 120 h by a TCID50 assay. These results indicated that RNAi targeting of the N gene could facilitate studies of the specific function of viral genes associated with PHEV replication and may have potential therapeutic applications.
Keywords: PHEV, shRNA, N gene, Inhibition
Porcine hemagglutinating encephalomyelitis coronavirus (PHEV) is a member of the Coronaviridae family; it is an enveloped virus containing a non-segmented, single-stranded, positive-sense RNA genome of approximately 30 kb that codes for 5 major proteins: Hemagglutinin-esterase protein (HE), Spike glycoprotein (S), Envelope protein (E), Membrane protein (M), and Nucleocapsid protein (N). NS4.9 and NS12.7, however, are non-structural proteins. The genes that encode these proteins occur in the order 5′-HE-S-NS4.9-NS12.7-E-M-N-3′ (GenBank accession no.: NC_007732). PHEV was first isolated in 1962 in Canada from suckling piglets with encephalomyelitis and has since been isolated from swine worldwide. It was first isolated in primary cultures of pig kidney (PK-15) cells through visible cytopathic effects (CPE) and infectivity assays (Greig et al., 1962, Mengeling et al., 1972). When piglets younger than 3 weeks are infected with PHEV, their mortality rates range from 20 to 100% (Pensaert, 1989).
RNA interference (RNAi) is an accurate and potent gene silencing method that uses double-stranded RNA (dsRNAs) molecules consisting of 19–27 nucleotides (nt) (Jana et al., 2004). Subsequent RNAi studies have used synthetic small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) extensively to study the interference of viral replication. Thus far, the replication of various viruses, including many coronaviruses, has been effectively inhibited in vitro and in vivo (Galan et al., 2009, Lambeth et al., 2009, Lan et al., 2011, Wu et al., 2005, Zhao et al., 2005, Zhou et al., 2007, Zhou et al., 2010, Zhuang et al., 2009); however, no reports have shown that the replication of PHEV in cell culture could be disrupted by shRNAs targeting the N gene of PHEV.
N is an extensively phosphorylated, highly basic, vital structural protein; its primary function is to form a helical ribonucleoprotein complex with viral RNA (vRNA) (Wang et al., 2010). N plays an important and necessary role as an enhancer of coronavirus replicon activity (Almazan et al., 2004, Chang and Brian, 1996). Here, we constructed a single short hairpin RNA (shRNA) plasmid expression system that targeted the N gene and investigated whether shRNA-mediated RNA interference could inhibit PHEV replication in PK-15 cells.
The HEV-67N strain (GenBank accession no.: AY078417) was replicated in PK-15 cells (Mengeling et al., 1972). Prior to being infected with PHEV, the cells were maintained in MEM supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 μg/ml streptomycin and 100 U/ml penicillin) in a 37 °C, 5% CO2 incubator overnight. When 70% of the virus-infected cells showed cytopathic effects (CPE), the cultures were collected, purified by sucrose density gradient centrifugation, and stored at −80 °C until use.
Based on recent research (Elbashir et al., 2002) and the experience of researchers from the Ambion Corporation (Jacque et al., 2002) using GenBank sequences (GenBank accession no.: AY078417, NC_007732) for HEV-67N and VW527, the conserved areas were selected, and Ambion's online siRNA target design tool was utilized to choose the two best target sequences for targeting N. BLASTN searches were conducted on all sequences to ensure gene specificity. The targeted oligonucleotides were inserted into the pGPU6/GFP/Neo plasmid vectors using the BbsI and BamHI restriction sites to produce shN1 and shN2 (sequences shown Table 1 ); the negative control shRNA (shNC), which targeted GTTCTCCGAACGTGTCACGT sequences and did not match any gene, was purchased from Shanghai Genepharma Co, Ltd (Shanghai, China).
Table 1.
List of shRNA sequences in this study.
| Target name | Sequences of shRNAa |
|---|---|
| shN1 (nt7004–7024) | Sense: 5′-caccGGTACTGGTACAGACACAACAttcaagagaTGTTGTGTCTGTACCAGTACCTTTTTTg-3′ Antisense: 5′-gatccAAAAAAGGTACTGGTACAGACACAACAtctcttgaaTGTTGTGTCTGTACCAGTACC-3′ |
| shN2 (nt7218–7238) | Sense: 5′-caccGGCTATTCCGACTAGGTTTCCttcaagagaGGAAACCTAGTCGGAATAGCCTTTTTTg-3′ Antisense: 5′-gatccAAAAAAGGCTATTCCGACTAGGTTTCCtctcttgaaGGAAACCTAGTCGGAATAGCC-3′ |
The underlined sequences targeted the N gene, and the bold italic letters indicate the loop sequence. Near the end of all shRNA sense templates was a 6-nt poly(T) tract that is recognized as a termination signal by RNA pol III, which terminated shRNA synthesis. The 5′ ends of the two oligonucleotides were non-complementary and formed the BbsI and BamHI restriction site overhangs that facilitated efficient directional cloning into the pGPU6/GFP/Neo plasmid vector.
PK-15 cells were seeded in 24-well plates and incubated for 24 h at 37 °C in a 5% CO2 atmosphere. When the cells were 70–80% confluent, they were washed and overlaid with transfection complexes containing 1.5 μg of shN1, 1.5 μg of shN2, or 1.5 μg of shNC, in 100 μL of MEM medium mixed with Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The transfection complexes were completely removed after incubating for 6 h, and the cells were infected with 400 TCID50 (104.49) of PHEV. Non-transfected cells were used as a control.
To study the inhibitory effects of RNA interference on PHEV replication, the level of viral antigen produced in the PK-15 cells after shRNA transfection and viral infection was examined 48 h post-infection by an indirect immunofluorescence assay (IFA) using anti-PHEV serum. Infected cells were visualized by IFA, as has been described previously (Xu et al., 2008). Positive anti-PHEV serum and negative control serum were prepared, as described previously (Yu et al., 2001). Cells were washed three times with PBS and incubated with Cy3-conjugated rabbit anti-pig IgG (Sigma, St. Louis, MO, USA; 1:60) for 1 h. After washing, the cells were photographed and examined with an Olympus FV 1000 laser scanning confocal microscope. At 48 h post-PHEV infection, the majority of the non-transfected cells and the cells transfected with shNC exhibited red fluorescence in the cytoplasm, indicating that these cells were virus-producing (Fig. 1A and C). By contrast, only a few cells in wells treated with shN1 and shN2 transfection displayed red fluorescence (Fig. 1D and E), indicating that most of these cells were effectively protected by shRNAs and resisted viral infection.
Fig. 1.
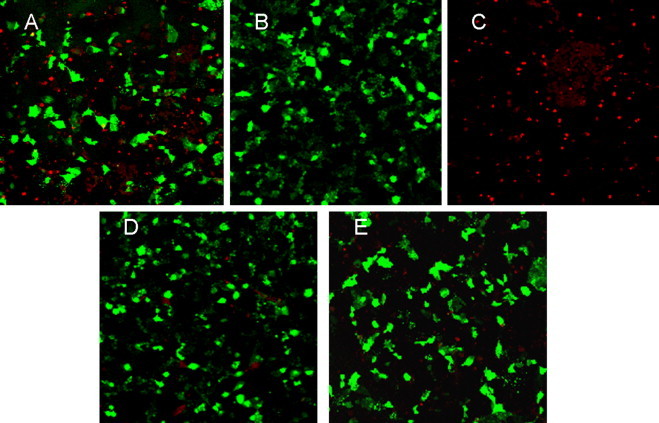
Protective effect of shRNA against PHEV infection. The viral infection in PK-15 cells was examined by IFA. (A) shNC transfection (stained with PHEV-positive serum); (B) shNC transfection (stained with PHEV-negative serum); (C) non-transfection (stained with PHEV-positive serum); (D) shN1 transfection; (E) shN2 transfection.
Oligonucleotides for the quantitation of PHEV by TaqMan real-time RT-PCR were designed using the corresponding GenBank sequences (GenBank accession no.: AY078417, NC_007732) for HEV-67N and VW527; the conserved areas were selected, and primers were designed for PHEV using Primer express 2.0. Three primers were synthesized (JiKang, Shanghai, China) for quantification of the PHEV genome by real-time PCR: 5′-AGCGATGAGGCTATTCCGACTA-3′ (NFP primer), 5′-TTGCCAGAATTGGCTCTACTACG-3′ (NRP primer), and 5′-FTTC-CGCCAGGCACGGTACTCCCP-3′ (TaqMan probe). The target region for real time RT-PCR spanned nucleotides 7210–7348 of the N protein gene of PHEV (GenBank accession no.: AY078417). Total RNA (100 ng) was extracted and purified using the Trizol LS Reagent (Invitrogen, Carlsbad, CA, USA). PCR was used to amplify the N gene with the NFP and NRP primers under conditions of 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 60 s, with a final extension at 72 °C for 7 min. The PCR product was gel-purified using an Agarose Gel DNA Extraction Kit (Axygen, Union, CA, USA) and then cloned into the pGEM-18T vector (Promega, Madison, WI, USA). The resulting plasmid, pT-N, for which the correct sequence was confirmed by direct sequencing, was selected as a quantitative standard for the determination of the viral RNA copy number. Real-time PCR was performed with the ABI PRISM® 7000 Sequence Detection System using a QuantiTect™ Probe PCR Kit (Qiagen, Hilden, Germany) under the conditions of 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing, and extension at 60 °C for 30 s. The quantitative standard curve for the determination of the PHEV genome copy number was created by real-time PCR of standard plasmid pT-N preparations at serial dilutions of 103, 104, 105, 106 and 107 copies/μL. The specificity of the real-time PCR was confirmed by sequencing the product.
To quantify the effect of shRNA on viral replication at 48 h post-viral infection, the viral genome copy number was determined by real-time PCR, using the serially diluted plasmid pT-N as a standard. The R 2 value of the standard curve was 0.99, and the average amplification efficiency E was 0.993. Sequencing showed that the amplified fragment contained the expected portion of the PHEV genome, thereby demonstrating the specificity and reliability of the analysis. As shown in Fig. 2 , the copy number of the viral genome per nanogram of total RNA calculated from the standard curve was 8.79 × 104 copies/ng of viral genome in the total RNA from the shNC-treated cells, which was similar to that observed in the mock transfected cells (non-transfected cells), whereas there were 2.67 × 104 and 1.60 × 104 copies/ng of the viral genome in total RNA from cells treated with shN1 and shN2, respectively. These values correspond to reductions of 70% and 82%, respectively.
Fig. 2.
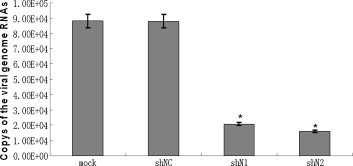
Real-time quantitative RT-PCR results indicate a significant decrease of N gene mRNA expression level by shN1 and shN2 at 48 h after virus infection. The values are shown as means of triplicates and three independent experiments. *Significant difference from shNC groups (P < 0.05).
To substantiate further the inhibitory effect of shN1 and shN2, the viral 50% tissue culture infectious dose (TCID50) was used to titrate PHEV at 48 h post-infection. Briefly, the cultures were collected, subjected to 3 freeze–thaw cycles, serially diluted 10-fold from 10−1 to 10−8, and added to PK-15 cells in 96-well plates. TCID50 was calculated using the Reed and Muench method (Reed and Muench, 1938). The results (Fig. 3 ) showed that in control cells transfected with shNC, the PHEV TCID50 reached 104.30 at 48 h post-infection, which was similar to that observed in non-transfected cells. In contrast, the titers at 48 h post-infection were 102.45 and 102.03 for cells transfected with shN1 and shN2, respectively, which corresponded to 71- and 186-fold reductions compared to shNC-transfected cells. Our results demonstrated that shN1 and, especially, shN2 both blocked viral replication with very high efficiency, but the effects were transient (data not shown).
Fig. 3.
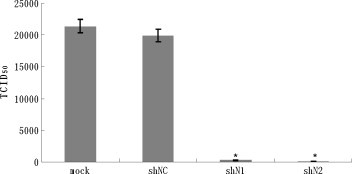
Transfection with shN1 and shN2 reduces the viral titer in the PK-15 cells after 48 h of virus infection. At 48 h after virus infection, cell culture supernatants were collected for viral titration. All the data were obtained by experiments performed in triplicates and repeated three times. *Significant difference from shNC group (P < 0.05).
To conform further the effect of using RNA interference (RNAi), stably transfected lines were created. For shN2 transfection, 105 cells per well were plated in six-well plates and transfected with 4 μg of shRNA duplex using Lipofectamine 2000 according to the manufacturer's instructions. After 48 h of incubation, the medium was replaced with MEM containing 1300 mg/ml G418. After maintenance for 2 weeks in selection media, resistant cell clones, named shN2-1, shN2-2 and shNC-1, were selected, cultured, infected with 400 TCID50 (104.49) of PHEV per well in 24-well plates, and incubated at 37 °C for the various time periods indicated. To study PHEV-induced CPE (Fig. 4 ) at 12 h post-infection, cells were examined by phase-contrast microscopy every 4 h. The infected shNC-1 cells and non-transfected cells exhibited obvious morphological changes starting from the brims of the wells at 56 h post-infection, such as loss of the monolayer as well as rounding and shrinking of the cells. Analysis of cell morphology indicated that the negative control (shNC-1) had no inhibitory effect on PHEV-induced CPE compared with the mock control (non-transfected cells). In contrast, the trial group (shN2-1 and shN2-2) effectively blocked CPE in the cell cultures up to 120 h (data not shown).
Fig. 4.
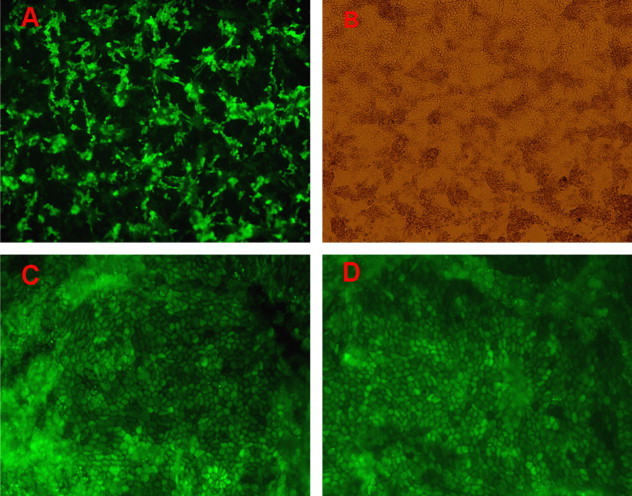
Effect of siRNAs on PHEV-induced CPE in PK-15 cells. Virus-produced CPE was observed at 56 h after virus incubation. (A) shNC-1; (B) non-transfection; (C) shN2-1; (D) shN2-2.
In addition, infectious virus production was determined by a TCID50 assay. Fig. 5 shows that titers of PHEV reached 104.31, 104.46, and 104.48 TCID50/mL at 48 h, 56 h and 72 h post-infection, respectively, in shNC-1, followed by a gradual decrease in titer from 72 to 120 h post-infection. In contrast, titers at 48 h post-infection were 102.55 and 102.14 TCID50/mL in shN2-1 and shN2-2, corresponding to 57- and 147-fold reductions, respectively. Titers at 72 h post-infection were 102.79 and 102.65 TCID50/mL in shN2-1 and shN2-2, respectively, corresponding to 49- and 66-fold reductions. At 120 h post-infection, the viral titers were still 11- and 25-fold lower in shN2-1 and shN2-2, respectively, compared to that of shNC-1. The data indicate that the shRNA stably expressing cells experienced markedly inhibited infectious virus production, but the level of inhibition decreased as time progressed.
Fig. 5.
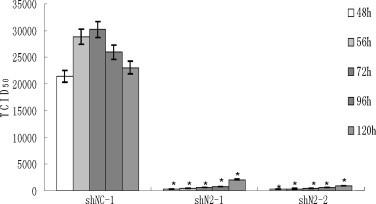
Viable viral production in two siRNA stably expressing PK-15 cell clones (shN2-1, shN2-2). TCID50 are the means of three repeat titrations at the time points indicated. *P < 0.05 vs. shNC stably expressing PK-15 cells (shNC-1).
To confirm the specificity of the RNAi effect and the absence of interferon activity in stably transfected lines, shN2-1, shN2-2 and shNC-1 were also challenged with 200 TCID50 (103.38) of Pseudorabies virus (PRV) JL strain in a plaque counting assay, as has been described previously (Ferrari et al., 2000). No significant difference in plaque number and plaque size between mock control (non-transfected cells) and stably transfected lines was observed from 0 to 48 h after PRV infection (shown in Fig. 6 ).
Fig. 6.
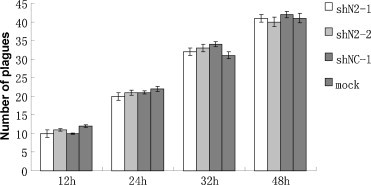
The stably transfected lines (shN2-1, shN2-2, shNC-1) failed to alter significantly plaque number or size (not shown) from 0 to 48 h after 200 TCID50 of PRV incubation. At 12, 24, 36 and 48 h after virus infection, respectively, the medium was removed, cells were fixed and stained, and the number of plaques was counted. All data were collected in triplicate wells and in three independent experiments.
PHEV is the only known CoV that is neurotropic in pigs. Recently, infection rates of PHEV have increased in some countries (Gao et al., 2011, Rho et al., 2011); thus, pigs without antibodies to PHEV, such as SPF pigs, were threatened by a high risk of infection by PHEV (Quiroga et al., 2008). The study of PHEV replication and of the control or inhibition of this infection is of great significance because, currently, no effective preventative measures or cures exist for this disease.
RNAi provides effective antiviral defenses in plants and other organisms; so several studies have focused on harnessing RNAi to inhibit viral infection (Leonard and Schaffer, 2006). However, the effectiveness of siRNA that has been transfected transiently into cells is restricted because of low transfection efficiency and the short-term persistence of silencing effects. Recently, the vector-based approach of shRNA interference has been developed to achieve stable, long-term, and highly specific suppression of gene expression in mammalian cells (Li et al., 2011, Siolas et al., 2005). To explore the possibility of interrupting PHEV replication with siRNAs or shRNAs, siRNA expression plasmids targeting the N gene in PHEV have been synthesized.
N plays an important role in viral packaging. In the process of viral packaging, N acts as a bridge between the M protein and the viral RNA. The interaction of N and mRNA may affect on viral RNA transcription and replication. In addition, N is highly conserved among coronaviruses, is expressed during viral infection and has a strong immunogenicity. These features make the N gene coding region a good RNAi target site. Our results have shown that the replication of PHEV in cell culture can be disrupted by shRNAs targeting the N gene of PHEV.
It is clear from this study that the DNA vector-based shRNA approach, that is, the use of RNAi expression plasmids directed against the PHEV N gene, could effectively block expression of the viral target gene and inhibit viral replication. In addition, siRNA therapy can be directed at the different levels of viral function, such as transcription or translation. Based on the present data and the advantages of siRNA technology, we believe that siRNA is a potential therapeutic agent against chronic PHEV infection.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Nos. 31072134, 30871849, 30671551, 31172291), the Young Scientist Program of Science and Technology Development Plan in Jilin Province (No. 20090154), the Natural Science Foundation in Jilin Province (No. 201115040), and the International Cooperation Program of Science and Technology Development Plan in Jilin Province (No. 20080722).
References
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R.Y., Brian D.A. cis Requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 1996;70:2201–2207. doi: 10.1128/jvi.70.4.2201-2207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Ferrari M., Mettenleiter T.C., Romanelli M.G., Cabassi E., Corradi A., Dal Mas N., Silini R. A comparative study of pseudorabies virus (PRV) strains with defects in thymidine kinase and glycoprotein genes. J. Comp. Pathol. 2000;123:152–163. doi: 10.1053/jcpa.2000.0406. [DOI] [PubMed] [Google Scholar]
- Galan C., Sola I., Nogales A., Thomas B., Akoulitchev A., Enjuanes L., Almazan F. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Zhao K., Zhao C., Du C., Ren W., Song D., Lu H., Chen K., Li Z., Lan Y., Xie S., He W., Gao F. Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin, China. Virol. J. 2011;8:130. doi: 10.1186/1743-422X-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig A.S., Mitchell D., Corner A.H., Bannister G.L., Meads E.B., Julian R.J. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 1962;26:49–56. [PMC free article] [PubMed] [Google Scholar]
- Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Chakraborty C., Nandi S., Deb J.K. RNA interference: potential therapeutic targets. Appl. Microbiol. Biotechnol. 2004;65:649–657. doi: 10.1007/s00253-004-1732-1. [DOI] [PubMed] [Google Scholar]
- Lambeth L.S., Zhao Y., Smith L.P., Kgosana L., Nair V. Targeting Marek's disease virus by RNA interference delivered from a herpesvirus vaccine. Vaccine. 2009;27:298–306. doi: 10.1016/j.vaccine.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Lan Y., Lu H., Zhao K., He W., Chen K., Wang G., Song D., Gao F. In vitro inhibition of porcine hemagglutinating encephalomyelitis virus replication with siRNAs targeting the spike glycoprotein and replicase polyprotein genes. Intervirology. 2011 doi: 10.1159/000323523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard J.N., Schaffer D.V. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo H., Shi Z., Tu C. In vitro inhibition of CSFV replication by retroviral vector-mediated RNA interference. J. Virol. Methods. 2011;169:316–321. doi: 10.1016/j.jviromet.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengeling W.L., Boothe A.D., Ritchie A.E. Characteristics of a coronavirus (strain 67N) of pigs. Am. J. Vet. Res. 1972;33:297–308. [PubMed] [Google Scholar]
- Pensaert M. Virus infections of porcines. Vet. Q. 1989;2:142–148. [Google Scholar]
- Quiroga M.A., Cappuccio J., Pineyro P., Basso W., More G., Kienast M., Schonfeld S., Cancer J.L., Arauz S., Pintos M.E., Nanni M., Machuca M., Hirano N., Perfumo C.J. Hemagglutinating encephalomyelitis coronavirus infection in pigs, Argentina. Emerg. Infect. Dis. 2008;14:484–486. doi: 10.3201/eid1403.070825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H.A. A simple method of estimating fifty percent end points. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Rho S., Moon H.J., Park S.J., Kim H.K., Keum H.O., Han J.Y., Van Nguyen G., Park B.K. Detection and genetic analysis of porcine hemagglutinating encephalomyelitis virus in South Korea. Virus Genes. 2011;42:90–96. doi: 10.1007/s11262-010-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D., Lerner C., Burchard J., Ge W., Linsley P.S., Paddison P.J., Hannon G.J., Cleary M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Wang Q., Li C., Zhang Q., Wang T., Li J., Guan W., Yu J., Liang M., Li D. Interactions of SARS coronavirus nucleocapsid protein with the host cell proteasome subunit p42. Virol. J. 2010;7:99. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Huang H.W., Liu C.Y., Hong C.F., Chan Y.L. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Guo H., Xiao C., Zha Y., Shi Z., Xia X., Tu C. In vitro inhibition of classical swine fever virus replication by siRNAs targeting Npro and NS5B genes. Antiviral Res. 2008;78:188–193. doi: 10.1016/j.antiviral.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Yu X., Tu C., Li H., Hu R., Chen C., Li Z., Zhang M., Yin Z. DNA-mediated protection against classical swine fever virus. Vaccine. 2001;19:1520–1525. doi: 10.1016/s0264-410x(00)00334-0. [DOI] [PubMed] [Google Scholar]
- Zhao P., Qin Z.L., Ke J.S., Lu Y., Liu M., Pan W., Zhao L.J., Cao J., Qi Z.T. Small interfering RNA inhibits SARS-CoV nucleocapsid gene expression in cultured cells and mouse muscles. FEBS Lett. 2005;579:2404–2410. doi: 10.1016/j.febslet.2005.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Huang F., Hua X., Cui L., Zhang W., Shen Y., Yan Y., Chen P., Ding D., Mou J., Chen Q., Lan D., Yang Z. Inhibition of porcine transmissible gastroenteritis virus (TGEV) replication in mini-pigs by shRNA. Virus Res. 2010;149:51–55. doi: 10.1016/j.virusres.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.F., Hua X.G., Cui L., Zhu J.G., Miao D.N., Zou Y., He X.Z., Su W.G. Effective inhibition of porcine transmissible gastroenteritis virus replication in ST cells by shRNAs targeting RNA-dependent RNA polymerase gene. Antiviral Res. 2007;74:36–42. doi: 10.1016/j.antiviral.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T., Ling H., Yang B., Saitoh H., Zhang L., Qin C., Sugamura K., Hattori T. Procyanidins and butanol extract of Cinnamomi cortex inhibit SARS-CoV infection. Antiviral Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


