Abstract
A sensitive reporter assay to measure human immunodeficiency virus type 1 (HIV-1) protease (PR) activity is described in this manuscript. This assay measures PR activity as a function of the resonance energy transfer (RET) between a donour molecule [humanized sea pansy Renilla reniformis luciferase (hRLuc)] and an energy acceptor molecule, humanized green fluorescent protein (hGFP2) when expressed in mammalian cells. This is a naturally occurring phenomenon and is an emerging and powerful technology that has significant advantages over alternative in vitro PR assays. The HIV-1 Gag-p2/Gag-p7 (p2/p7) PR site was inserted between hGFP2 and hRLuc. The newly created vector, hRLuc-p2/p7-hGFP2 was co-expressed with an HIV-1 codon-optimized PR+ or PR− Gag/Pol expressor. Expression of the hRLuc-p2/p7-hGFP2 alone or with the PR− Gag-Pol expressor generated a BRET2 indicating that the PR cleavage site was not cleaved, whereas the inclusion of the PR+ Gag-Pol produced a significant reduction in the BRET2. The inclusion of PR inhibitors Saquinavir or Amprenavir, or the expression of a p2/p7 PR substrate mutant also blocked the cleavage to result in a stable BRET2 signal. Because the HIV-1 auxiliary protein Vif has been shown to modulate the HIV-1 p2/p7 cleavage, this assay was then validated in studies in which Vif was expressed. When Vif was overexpressed along with hRLuc-p2/p7-hGFP2 and PR+ Gag-Pol, the decrease in BRET2 was abrogated in a dose-dependent manner, demonstrating that supraphysiologic levels of Vif block p2/p7 cleavage. An accumulation of a Gag processing intermediate was observed, indicating that p2/p7 cleavage was negatively affected. Overexpression of an RNA-binding-defective Staufen protein or a related dsRNA-binding protein TRBP had no effect on PR cleavage activity as shown by Western and BRET2 analyses. The p2/p7 processing data were confirmed by Western blot analyses. BRET is non-invasive and occurs within live cells, is measured in real time, and is not restricted to cellular compartments making it an especially attractive technology to identify small bioactive inhibitory molecules. This PR BRET2 biosensor assay can be adapted for high throughput screening of new HIV-1 PR inhibitors. It can be employed to screen for antiviral compounds that also target the proteases of other viruses.
Keywords: BRET2, HIV-1, Protease, Inhibitor, Vif
1. Introduction
Currently, there are 37 approved drugs for use as antivirals, and upwards of at least 83 new antiviral therapies in clinical trials around the world (The Pharmaceutical Research and Manufacturers of America; De Clercq, 2004). Some of these are new and improved inhibitors of HIV-1 PR. The introduction of the first PR inhibitors in the 1990s was characterized by excitement and hopes for the control of infection. Since most HIV-1 infected individuals rapidly become resistant to these anti-PR drugs, a need to identify new inhibitors became apparent. Whereas current methodologies to screen for these compounds were based largely on electrophoresis and liquid chromatography methods, it was clear that additional screening assays were also needed to respond to this demand. These would ideally be rapid and capable of high throughput screening (HTS) procedures. Initial efforts using fluorogenic and fluorescence quenching substrates for proteolysis enabled higher throughput screens of anti-PR compounds that could readily be adapted to HTS (Matayoshi et al., 1990, McQuade et al., 1990). More recent efforts using identical chemistry (EDANS, Dabcyl) continue to be used in the development of high throughput microplate assays (Bagossi et al., 2004). Other screening assays for anti-PR drugs rely on in vitro analysis of inhibition of certain biochemical reactions resulting in the production of assayable intermediate metabolites. These in vitro estimates are generally followed up with more detailed pharmacological tests and pharmacological screens in various in vitro and in vivo assays. These assays, however, are non-homogeneous and require a synthetic substrate and are executed entirely in in vitro conditions with purified proteins (Baruch et al., 2004). Ideally, initial screening assays should be as close to physiological conditions as possible and even better, be homogeneous assays that test for inhibition of PR in living cells.
In this report, a highly sensitive biosensor assay that can be used to identify HIV-1 PR inhibitors is described. This is based on bioluminescence resonance energy transfer, second generation (BRET2), a naturally occurring phenomenon that relies on the resonance energy transfer (RET) between a donour, hRLuc and an acceptor, hGFP2 molecule. This RET is initiated by the addition of DeepBlue C Coelenterazine that is oxidized by the RLuc resulting in the emission of light at 395 nm. Part of the energy can be transferred non-radiatively to a codon-optimized – humanized – GFP2 (hGFP2) if the hRluc and the hGFP2 are in close proximity (Forster distance being about 10–100 Å). The hGFP2 will then emit light at 510 nm. Both emissions are quantitated by sequential reading using a combined luminometer and fluorescence plate reader and the emission spectra are sufficiently distinct to allow for straight-forward ratiometric analysis of the hGFP2 and hRLuc signals to assess molecular interactions or proximity between hGFP2 and hRLuc in live cells.
BRET2 is an emerging and powerful technology that has significant advantages over alternative in vitro approaches (Baruch et al., 2004). For example, BRET2 is noninvasive and occurs within live cells, is measured in real time, and is not restricted to cellular compartments making it especially attractive to identify small bioactive inhibitory molecules or for use in the study of protein–protein interactions post-translational modification of proteins in live cells (Angers et al., 2000, Bertrand et al., 2002, Germain-Desprez et al., 2003, Issafras et al., 2002, Perroy et al., 2004). The assay described here utilizes this new technology to assay for HIV-1 PR inhibition in live cells. Specifically, the utility of BRET2 in the establishment of a HIV-1 PR biosensor assay is demonstrated and this assay is validated with the use of several known PR inhibitors. Moreover, because virus–host interactions are also critical to all steps of the viral replication cycle, this assay can also be used to probe for the function of viral proteins that can modulate PR activity and may help elucidate the molecular details of PR function and HIV-1 maturation.
PR activity is highly regulated in time with a well-characterized cleavage order (Pettit et al., 1998). Several factors such as RNA, viral proteins and host proteins have been demonstrated to have an impact on PR activity (Akari et al., 2004, Guo et al., 2005, Sheng and Erickson-Viitanen, 1994, Sheng et al., 1997, Zhang and Barklis, 1997). Furthermore, small molecule compounds have been identified recently that act on selective HIV-1 protease sites to delay maturation of the precursor Gag protein (Zhou et al., 2004). It will be interesting in the future to not only identify antiviral compounds that target PR itself, but to identify small molecules that could block PR cleavage by selectively inhibiting cleavage at a particular site (Zhou et al., 2004).
A PR biosensor assay based on BRET2 is described in this manuscript. This assay is validated using commercially available HIV-1 PR inhibitors. The HIV-1 auxiliary protein, Vif, sterically inhibits HIV-1 PR function at the Gag p2/p7 site and the assay is further validated in cells using this information (Akari et al., 2004). The PR biosensor assay is sensitive and provides a measure of absolute PR activity in living cells. This assay can be employed to screen for new PR inhibitors and can be modified to enable HTS of libraries of small molecules that inhibit PR activity and later steps of virus maturation.
2. Materials and methods
2.1. Preparation of the hGFP2-MCS-hRLuc vector and Gag expressors
pCMV-hGFP2-MCS-hRLuc was obtained from Perkin-Elmer/Packard Biosciences (Montreal, PQ) and harbours a multiple cloning site between hGFP2 and RLuc open reading frames. This is engineered to allow insertion of coding sequence in-frame between these two genes. Gag/Pol expression vectors pVRC4200 (PR+) and pVRC4000 (PR−, due to a point mutation in the PR active site) are reported elsewhere (Huang et al., 2001, Huang et al., 1997). Mammalian expression vectors for Staufen, TAR-RNA binding protein (TRBP) and Vif (pcDNA3-hVif; codon-optimized expression vector that allows high-level expression without the requirement for Rev expression) have been reported elsewhere (Chatel-Chaix et al., 2004, Mouland et al., 2000, Nguyen et al., 2004).
2.2. Generation of the pCMV-hGFP2-MCS-hRLuc PR biosensor constructs
The sets of complementary DNA oligomers (AlphaDNA, Montreal, Quebec) that encode the indicated HIV-1 HxBc2 PR cleavage sites (4 amino acids on either side: P-4 to P4) are presented in Table 1 . Each set of oligonucleotides was designed to be directionally cloned into pCMV-hGFP2-MCS-hRLuc in-frame with the hRLuc and hGFP2 open reading frames at the 5′ PstI and 3′ KpnI sites. The PstI site was engineered to be destroyed to allow for screening. The p2/p7 HIV-1 PR cleavage site was introduced into pCMV-hGFP2-MCS-hRLuc. In addition, a p2/p7mut construct was designed to contain 3 random mutations in the PR cleavage site to prevent recognition and cleavage of the PR (Fig. 1 ). A construct containing the p24/p2 PR substrate was generated as well as the p24/p2ext5 construct [to extend the p24/p2 site by five amino acids on both sides since the original p24/p2 PR substrate was incapable of being cleaved in this assay (data not shown]. The parental construct, pCMV-hGFP2-MCS-hRLuc was linearized by digestion with PstI and KpnI and gel-purified using the Qiaquick Gel Purification Kit (Qiagen). The hybridized oligonucleotides above were introduced into pCMV-hGFP2-MCS-hRLuc by standard ligation procedures using an optimal ratio of 0.5:4 of the pCMV-hGFP2-MCS-hRLuc:oligonucleotide duplex. All clones were verified by dideoxynucleotide sequence analysis (Amersham) and in transient expression analyses in 293T cells.
Table 1.
DNA oligonucleotides used for cloning HIV-1 PR substratesa in the parental hGFP2-MCS-hRLuc plasmid
| HIV-1 PR cleavage site | Sense oligonucleotide, antisense oligonucleotide |
|---|---|
| P2/p7 | 5′-CGAATTCAGCTACCATAATGATGCAGAGAGGCAATTTTAGGTAC-3′ |
| 5′-CTAAAATTGCCTCTCTGCATCATTATGGTAGCTGAATTCGTGCA-3′ | |
| P2*P7mut | 5′-CGAATTCAGGTACCATAATTATGCAGAGCGGCAATTTTAGGTAC-3′ |
| 5′-CTAAAATTGCCGCTCTGCATAATTATGGTACCTGAATTCGTGCA-3′ | |
| P24/p2 | 5′-AGAAGGCAAGAGTTTTGGCTGAAGCAATGAGCCAAGGGTAC-3′ |
| 5′-CCTTGGCTCATTGCTTCAGCCAAAACTCTTGCCTTCTTGCA-3′ | |
| P24*P2ext5 | 5′AGGGACCCGGCCATAAGGCAAGAGTTTTGGCTGAAGCAATGAGCCAAGTAACAAATTGGTAC-3′ |
| 5′-CAATTTGTTACTTGGCTCATTGCTTCAGCCAAAACTCTTGCCTTATGGCCGGGTCCCTTGCA-3′ | |
| P17/p24 | 5′-TCAATCAGGTCAGCCAAAATTACCCTATAGTGCAGAACATCCAGGGGTAC-3′ |
| 5′-CCCTGGATGTTCTGCACTATAGGGTAGTTTTGGCTGACCTGATTGATGCA-3′ | |
Amino acid translations are shown in the inset in Fig. 1.
Fig. 1.
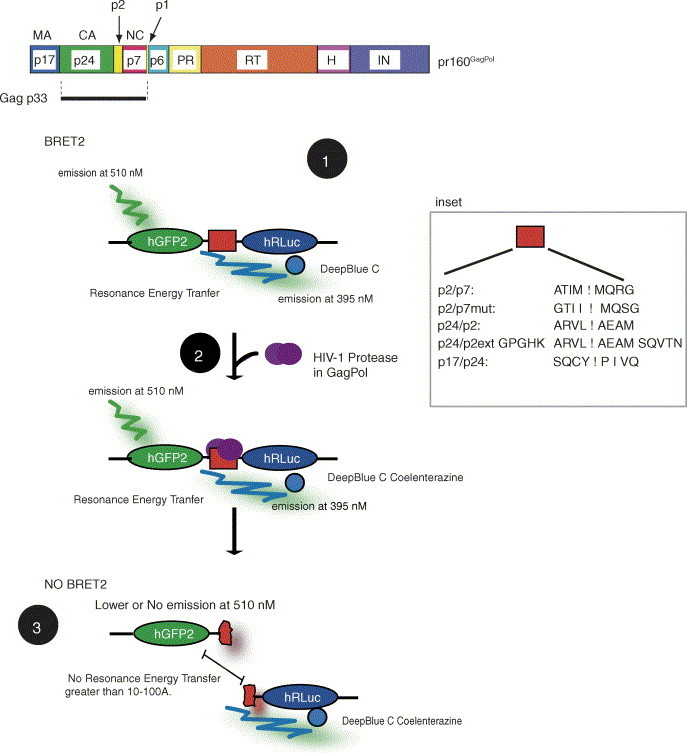
pr160GagPol is cleaved by the viral PR to generate mature porteins indicated in the scheme in A. The pr160GagPol-derived proteins are shown [MA (p17); CA (p24); spacer peptide, p2; NC (p7); spacer peptide, p1; PR, protease; RT, reverse transcriptase; H, RNAseH; IN, integrase). PR cleavage sites are located between each of the pr160GagPol mature proteins listed above. The p2/p7 site is located between p2 and p7 (NC). A scheme of the BRET2 assay is shown in B as described in the text. Briefly, the addition of DeepBlueC Coelenterazine initiates the resonance energy transfer to hGFP2 in the case of a close physical interaction or in the context of a hGFP2-hRLuc fusion protein (Step 1). HIV-1 PR will cleave its substrate (Step 2) to distance the donor and acceptor molecules thereby lowering the RET (Step 3). Inhibitors of PR such as Vif (this manuscript), or PR inhibitors used in HAART will maintain the BRET2 signal because PR activity is inhibited. The inset shows the PR cleavage sites and mutant sites introduced between the donor and acceptor molecules tested in this report. The letter codes for amino acids are indicated.
2.3. Cells lines and cell culture
Human embryonic kidney (HEK)293T cells were cultured in Dulbecco's modified Eagle's medium with the addition of 8% fetal bovine serum and penicillin–streptomycin (Invitrogen). These cells were minimally passaged in order to maintain high transfection efficiencies.
2.4. Transfections and BRET2 analysis
HEK293T cells are plated into NUNC 6-well plates at approximately 300,000 cells/well. Cells were incubated overnight in antibiotic-free DMEM supplemented with 8% FBS before transfection. The optimal amount of DNA for transfection was determined to be 2:2 (μg:μg) of Gag/Pol expressor (pVRC4000 or pVRC4200) (Huang et al., 1997) to pCMV-hGFP2-X-hRLuc vectors (X is the PR cleavage site), with a maximum of 4 μg DNA/well. Lipofectamine 2000 (Invitrogen) was used essentially according to the manufacturer's instructions, at 2.5 μl Lipofectamine/μg of DNA (ratio of 1:2.5) in FBS- and antibiotic-free DMEM. At 6-h post-transfection, the medium of the cells was replaced with fresh DMEM containing 8% FBS. Cells were collected at 40 h post-transfection in 1 ml of phosphate-buffered saline containing 5 mM EDTA and centrifuged 500 × g for 10 min at 4 °C. Cells were then resuspended in PBS to a final concentration of 1 × 106 cells/ml. For experiments in which Vif was overexpressed, a codon-optimized Vif expressor was used (Nguyen et al., 2004). The quantities of DNA in the transfection (0.5, 1.0 or 1.5 μg) as well as the amount of Lipofectamine were adjusted accordingly when viral and cellular proteins were co-expressed.
hGFP2 and hRLuc measurements to calculate the BRET2 ratio were performed essentially as described previously (Germain-Desprez et al., 2003, Mercier et al., 2002) with modifications adapted to the use of the detection instrument as follows. 100 μL of the 293T cell suspension (100,000 cells) was aliquotted into a well of a white, opaque 96-well, flat-bottom microplate (Perkin-Elmer Life Sciences). The DeepBlueC Coelenterazine (BioPackard-Signal Montreal, PQ) cell permeant luciferase substrate was added to each well to obtain a final concentration of 5 μM. Immediately following this addition, five readings for hRLuc and hGFP2 were sequentially recorded using a Fusion ∝-FP apparatus (Perkin-Elmer/Canberra Packard). First the light emitted from the bioluminescence resulting from the Rluc-mediated Coelenterazine degradation at 410 nm (band pass of 80 nm) was detected. The hGFP2 emission (peak 505–508 nm) was then detected using a 515 nm filter (band pass of 30 nm). Readings for this analysis were from the top of the plate. The BRET2 ratio was found to be stable over several readings performed at different times after addition of the substrate (evaluated within 5–8 min). The expression levels of hGFP2 (non RET-induced) and hRLuc for each experimental condition were determined by direct measurements of total hGFP2 and luminescence levels on aliquots of transfected cell samples as follows. The hGFP2 total fluorescence was measured using Fusion ∝-FP apparatus (Perkin-Elmer/Canberra-Packard) with an excitation filter of 400 nm and an emission filter of 510 nm, with the following parameters: gain 1; PMT 900–1100 V; time 1.0 s. After the fluorescence measurement, the same cells were incubated for 10 min with Coelenterazine H (Molecular Probes) at a final concentration of 5 μM and the total luminescence of cells was measured using the same instrument set up for bioluminescence readings with the following parameters: gain 1; PMT 700 V; time 0.5 s. In contrast to DeepBlue C Coelenterazine, Coelenterazine H does not lead to energy transfer to hGFP2 and thus allows the assessment of hRluc expression without loss due to energy transfer to hGFP2. When expressed as a ratio, the total hGFP2:hRluc ratio was found to be identical in each experiment indicating stable levels of expression of each protein.
The median reading was used in the calculation of the BRET2 ratio. The BRET2 ratio was quantified by calculating the RET-induced hGFP2/luminescence ratio. The BRET2 ratio is determined from the following equation: [(emission at 510 nm/emission at 395 nm) in cells expressing the hRluc & hGFP2 fusion proteins)] – [(emission at 510 nm/emission at 410 in cells expressing hRluc alone)] (Chatel-Chaix et al., 2004, Germain-Desprez et al., 2003). The parental construct that contains only the multiple cloning site between the hGFP2 and hRluc cistrons served as a positive control for BRET2.
2.5. Preparation of cell extracts and western blots
Following the BRET2 measurements, cells were centrifuged for 10 min at 500 × g and then were lysed in NP-40 lysis buffer [(100 mM NaCl, 10 mM Tris, 1 mM EDTA, 0.5% NP-40, with 1 mM protease inhibitors (Complete, Mini, EDTA-free protease inhbitor cocktail)] on ice for 45 min. Cell lysates were centrifuged at 14,000 × g for 30 min and the supernatants were transferred to new Eppendorfs. Protein content in the cytosolic extracts was quantitated by the micro-Bradford assay. Gag-Pol, Gag products, and hGFP2 expression were assessed by Western blot analyses using mouse anti-P24 antibody (#11HC25, generously provided by Dr. Grandgenett and the AIDS Research Reference and Reagent Program) or rabbit anti-p24 antibody (ABT-Trinity Biotechnology, CA, USA, catalogue#201), and anti-GFP antibody (RDI, catalogue #R970-01). For Vif, Staufen and TRBP expression in cells, the following antibodies were used: rabbit polyclonal anti-Vif (Beriault et al., 2004, Nguyen et al., 2004), mouse monoclonal anti-Staufen [#CG11 generously provided by L. DesGroseillers, Université de Montréal; Chatel-Chaix et al., 2004], and rabbit anti-TRBP [#617, generously provided by A. Gatignol, McGill University; Mouland et al., 2000]. In some experiments in which Staufen was overexpressed, Staufen was identified in cellular extracts using an anti-HA tag monoclonal (Mouland et al., 2000). Anti-GAPDH antibodies were purchased from RDI, Inc and expression was used as a loading control (Chatel-Chaix et al., 2004).
2.6. Reagents
The HIV-1 PR inhibitors Saquinavir and Amprenavir were obtained from the Division of AIDS, NIH through the NIH AIDS Research Reference and Reagent Program and were used at 1.5 μM. DeepBlue C Coelenterazine was purchased from Perkin-Elmer (Mississauga, ON) and the Coelenterazine H was from Molecular Probes (Eugene, OR).
2.7. Cell cycle and Annexin 5 apoptosis assays
In order to verify that cells were not being negatively affected by the transfection conditions used in the experiments presented in this report, cell cycle and Annexin 5 analyses was performed exactly as described (Mouland et al., 2002, Yao et al., 1998).
3. Results
3.1. Configuration of the BRET2 biosensor assay for PR activity
There are nine PR substrates in the Gag/Pol polyprotein (Fig. 1A) that when processed, give rise to functional mature Gag proteins. The BRET2 assay was developed to identify viral and cellular factors that influence HIV-1 PR activity. Several PR sites were originally tested in this BRET2 assay (inset in Fig. 1). An original goal of this study was to compare two protease cleavage sites, one primary PR site that is processed early following PR activation (p2/p7), and another, a secondary PR site that is processed at a later stage during viral maturation (p24/p2). The rationale was to examine early and late events of maturation and to eventually develop a high throughput screen for compounds (such as small molecules or viral and cellular proteins) that that can inhibit either or both of these cleavage reactions. However, the p24/p2 site was not amenable to cleavage by the co-expressed PR and the other site that was tested, the p17/p24 PR substrate, showed variable expression levels (data not shown and see Section 4). Because it was possible that the p24/p2 PR cleavage substrate was too short an amino acid stretch perhaps preventing the PR from binding, the p24/p2 site was extended by the addition of five amino acids on each end (p24/p2ext; Fig. 1). Preliminary tests showed that this did not improve its ability to be cleaved (data not shown). Based on preliminary findings, the p2/Nucleocapsid (p7) PR substrate site was chosen for all subsequent experimentation. Therefore the p2/p7 site was inserted in frame between the hGFP2 and hRLuc cistrons as shown by the red box in Fig. 1. BRET2 analyses were performed following the addition of the membrane permeable hRLuc substrate, DeepBlueC Coelenterazine. The PR substrate sites that were used for all subsequent experimentation include the p2/p7 site, a p2/p7mut, harbouring three amino acid substitutions in the cleavage site (Fig. 1).
3.2. The p2/p7 PR cleavage site is cleaved efficiently
In order to verify that PR was able to cleave the inserted PR substrate p2/p7, 293T cells were transfected with Gag/Pol PR− or PR+ expression constructs with hGFP2-p2/p7-hRLuc. Because the Gag/Pol vector expresses both Gag and Gag/Pol, Gag/Pol expression levels were first verified in this system. Cells were mock transfected or transfected with either Gag/Pol PR− or PR+. 40 h post transfection, cellular extracts were prepared and the expression pattern of Gag was determined by Western blot analysis. This analysis showed that when the blot was probed with anti-p24, an antibody that recognizes well the mature Gag proteins, expression of PR− showed that the Gag/Pol polyprotein is detected at 160 kDa (pr160GagPol) and is not processed (Fig. 2A). pr55Gag is also detected and is not processed also. The transfection of the Gag/Pol PR+ showed very efficient processing of Gag/Pol, as shown by strong signals for the mature forms of Gag, p25/p24.
Fig. 2.
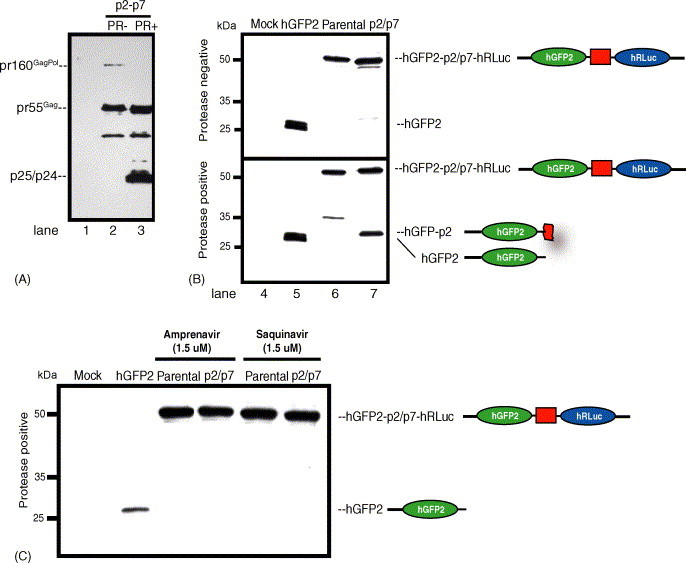
(A). 293T cells were mock transfected (lane 1) or co-transfected with hGFP2-p2/p7-hRLuc (p2/p7) and GagPol [PR− (lane 2) or PR+ (lane 3)]. Extracts were prepared from cells, blotted and subsequently probed for Gag proteins using an anti-p24. Efficient cleavage of the precursor Gag proteins (pr55Gag, pr160GagPol) is observed to generate mature Gag proteins p25/p24 is shown when PR is expressed (lane 3). (B) 293T cells were mock transfected (lane 4), transfected with a hGFP2 expressor (lane 5), or co-transfected with the parental hGFP2-MCS-hRLuc (lane 6) or the hGFP2-p2/p7-hRLuc (p2/p7; lane 7) with either GagPol PR− (top panel) or GagPol PR+ (bottom panel). Cells were processed for Western analyses. The hGFP2-p2 moiety generated by cleavage of the hGFP2-p2/p7-hRLuc fusion protein was observed only in the presence of PR, but not when GagPol PR− was expressed. (C) The addition of PR inhibitors Amprenavir or Saquinavir at 1.5 μM inhibited cleavage of the hGFP2-p2/p7-hRLuc. The parental construct was added as control.
We next determined the expression pattern of co-expressed hGFP2-p2/p7-hRLuc fusion protein. Cell extracts from hGFP2-, parental- (with no insert) and hGFP2-p2/p7-hRLuc-expressing cells in the absence (top) or presence (bottom) of PR were blotted using an anti-GFP antiserum (Fig. 2B). This analysis showed that while there was no detectable cleavage of the parental or hGFP2-p2/p7-hRLuc fusion protein when the Gag/Pol PR− was expressed (top panel), the expression of PR mediated the cleavage of hGFP2-p2/p7-hRLuc fusion protein to yield a hGFP2-p2 signal at about 27 kDa, slightly greater in molecular weight that the hGFP2 signal when expressed alone (bottom panel). These results were also confirmed by blotting with an anti-p24 as shown in Fig. 2A (data not shown). The addition of the HIV-1 PR inhibitors Amprenavir or Saquinavir 12 h before cell harvesting and BRET2 analysis prevented hGFP2-p2/p7-hRLuc cleavage at the p2/p7 scissile bond when PR was expressed (Fig. 2C), to obtain an identical expression pattern as that obtained with the parental vector. Gag and Gag/Pol processing was determined using an anti-p24 antiserum in Western blot analysis. This analysis showed that Gag processing was blocked in the presence of PR inhibitors as evidenced by the absence of mature Gag proteins (data not shown).
3.3. Validation experiments: effects of Vif on PR activity
Recent data have demonstrated that the HIV-1 auxiliary protein Vif selectively blocks processing at the CA-NC boundary through steric interference. This results in an accumulation of Gag processing intermediates, p33/p34, consisting of CA and NC and one or both of the spacer peptides, p1 and p2 (Akari et al., 2004). These recent data were then considered in order to validate this BRET2 PR biosensor assay. Using the construct shown in Fig. 1 and the GagPol PR+, increasing concentrations of Vif were overexpressed from a humanized Vif expression plasmid that does not require Rev for expression (Nguyen et al., 2004). This resulted in a detectable increase in Vif expression in these cells with increasing DNA in the transfection (Fig. 3A, top panel). Supraphysiological expression of Vif inhibited p2/p7 cleavage as indicated by the dose-dependent accumulation of the hGFP2-p2/p7-hRLuc fusion protein and the diminishing abundance of the hGFP2-p2 moiety (Fig. 3A, middle panel). Furthermore, using an anti-p24 or anti-NC (generously provided by Dr. Robert Gorelick), the accumulation of one of the Gag processing intermediates was identified in Western blots (likely p33; Fig. 1, top) when Vif was expressed (not shown). While earlier work showed that Vif mediated the accumulation of p33 and p34 Gag processing intermediates in the context of proviral gene expression, only one of these immunoreactive Gag processing intermediates was detected. This may be due to the conditions of GagPol Pr+ expression used in this manuscript or the low abundance of this Gag species.
Fig. 3.
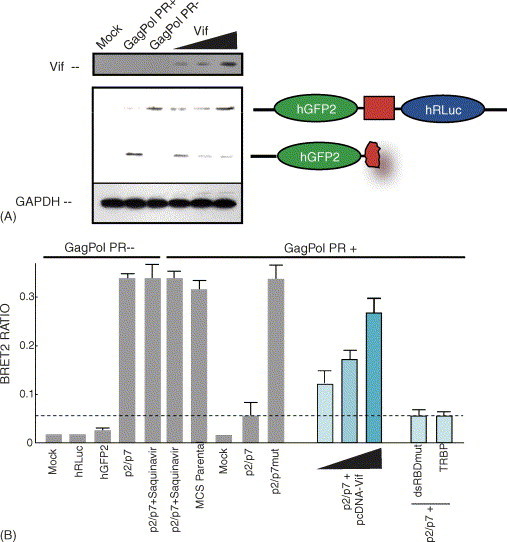
BRET2 analyses. (A) 293T cells were mock transfected or co-transfected with GagPol PR+ or GagPol PR− and hGFP2-p2/p7-hRLuc. Increasing concentrations (0.5, 1, and 1.5 μg) of a Vif expressor were included with GagPol PR+ and hGFP2-p2/p7-hRLuc (last three lanes). Cells were harvested and blotted for hGFP2 and Gag (not shown). Overexpression of Vif caused a dose-dependent accumulation of hGFP2-p2/p7-hRLuc fusion protein precursor indicating an inhibition by Vif on PR activity at the p2/p7 site. Vif expression inhibited PR activity as determined by BRET2 (see B) to almost 85% that obtained in the PR− context. (B) 293T cells were transfected as above except several controls were included (hRLuc and hGFP2 expression alone). The transfection conditions are indicated above and below the histogram. The threshold BRET2 value is shown by the dashed line and the error bars represent the mean ± S.E.M. from at least five independent experiments, except for the last two conditions when the Staufen double stranded RNA-binding mutant protein (dsRBDmut) and TRBP proteins are expressed (1.5 μg) for which the error bar represents the standard deviation calculated from two independent experiments.
3.4. BRET2 analyses
BRET2 analyses were performed as described in Section 2 at 40-h post transfection and the results are presented in Fig. 3B. The BRET2 ratio is a direct measure of the degree to which the p2/p7 is cleaved by the coexpressed GagPol (PR+ or PR−). The BRET2 ratio was determined when cells were mock-transfected or transfected with hRLuc or hGFP2 alone in each experiment, in the presence of PR expression or not. There was no BRET2 signal under any of these control transfection conditions (Fig. 3B). Expression of hGFP2-p2/p7-hRLuc in the absence of PR (i.e. in the context of Gag/Pol PR−) showed a strong BRET2 signal, similar to the levels found when the cells were treated with Saquinavir (with or without HIV-1 PR), Amprenavir or Nelfinavir (data not shown), or transfected with the parental construct with no PR cleavage site (with HIV-1 PR+). hGFP2-p2/p7-hRLuc was efficiently cleaved in the presence of the HIV-1 PR, while a mutant that harbours three amino acid substitutions (Fig. 1) was not cleaved. BRET2 analyses were performed in experiments in which Vif was overexpressed. When Vif was incrementally overexpressed in 293T cells (as in Fig. 3A), the BRET2 analyses reveal that Vif is inhibitory to p2/p7 cleavage and this is reflected in a dose-dependent increase in BRET2 reaching 85% that obtained with PR−. The data in Fig. 3 might suggest that overexpression of any protein will decrease PR activity non-specifically (Akari et al., 2004). Thus, in order to rule this out, either a double-stranded RNA binding mutant (dsRBDmut) form of Staufen or a related dsRNA-binding protein, TRBP was expressed as a control for BRET2 analyses. While a 4- to 8-fold increase in expression of these proteins was achieved as determined by Western blot analyses (data not shown), overexpression of these proteins resulted in a constant BRET2 ratio and did not have an impact on PR activity or the cleavage of the p2/p7 PR site. These overexpression results indicate that Vif's effect on PR action is selective. The histogram presented in Fig. 3B expresses the calculated averages (±S.E.M.) of the BRET2 analyses from at least five experiments per experimental condition.
3.5. Effects of Gag/Pol expression on apoptosis and cell cycle
Because the expression of HIV-1 PR has been associated to apoptosis and cell cycle when expressed (Strack et al., 1996, Wallin et al., 1990, Shoeman et al., 1990), cell cycle and apoptosis Annexin 5 analyses were performed using FACS. These analyses did not reveal any effects of PR expression on cell cycle or apoptosis levels using the transient conditions described in the assays shown here perhaps because the PR is expressed from a precursor protein, a context which has already been shown to prevent the cytotoxicity of HIV-1 PR (Lindsten et al., 2001 and data not shown).
4. Discussion
This report describes a novel biosensor assay for HIV-1 PR activity. This assay is homogeneous in that it is performed in live cells and there is no need for preparing cell extracts. A simple addition of a membrane-diffusible and non-toxic substrate to initiate the biochemical reaction is required and the readout is RET-induced hGFP2 emission that is readily detectable by a fluorescence plate reader (Fig. 1). The assay also relies on a straight-forward ratiometric calculation between the light emitted caused by interaction of hRLuc with its substrate and the resultant emission from hGFP2, in the case of a RET between these two proteins. This experimental system was first tested using the bicistronic construct in both PR+ and PR− backgrounds. The results demonstrated appropriate Gag processing by the PR, and a specific cleavage that resulted in a decreased BRET2 ratio in the PR+ context only (Fig. 2, Fig. 3). Mutated PR substrates were then introduced between the hRLuc and hGFP2 open reading frames or, alternatively, potent anti-PR drugs Amprenavir and Saquinavir were employed to block PR activity in cells. These control experiments demonstrated the sensitivity of this approach and represent the first validation experiments.
One aim of this study was to test a PR site that was cleaved early (e.g., p2/p7) during maturation and one that was cleaved late (e.g., p24/p2). The inability to efficiently cleave the p24/p2 site, even with an extended PR substrate (p24/p2ext, Fig. 1), suggests that this site must be in a contextually correct state to be a substrate for PR. This does not appear to be the case for the p2/p7 cleavage, however, since this was cleaved efficiently in the minimal context (Fig. 2). Nevertheless, because the p24/p2 site is estimated to be cleaved at about 0.0025% that of the p2/p7 (Pettit et al., 1998), other determinants are required that are not recapitulated in the experimental system used here. Previous reports have demonstrated that correct Gag/Pol dimerisation is necessary for proper assembly. Alternatively, other factors including viral proteins, the context of assembly and budding and the presence of genomic RNA that was shown to be important for HIV-1 and other retroviral PRs might also influence the context (Sheng and Erickson-Viitanen, 1994, Sheng et al., 1997, Zhang et al., 2000, Zhang and Barklis, 1997). It is noteworthy here that earlier reports of fluorescence-based PR in vitro screening assays never employed the Gag p24/p2 PR site indicating that it may not be amenable to this type of analysis. Whereas, these assays have used the p17/p24 PR cleavage site in their in vitro PR assays in which small peptide substrates are employed, the expression of the hGFP2-p17/p24-hRLuc construct that was prepared was too low to be employed in this study (K.H. and A.J.M., data not shown). Reconstitution of the missing factors may be helpful in the identification of selective PR inhibitors of this site.
Recent results demonstrate that the overexpression of the HIV-1 auxiliary protein Vif sterically inhibits processing at the p2/p7 PR substrate site. This information was used to validate the PR biosensor assay described in this report. The results shown in Fig. 3A and B confirmed that supraphysiologic expression of Vif, a HIV-1 auxiliary protein, inhibited p2/p7 cleavage and resulted in a stable BRET2 signal. Vif overexpression also resulted in an accumulation of a Gag processing intermediate (p33) (Fig. 1A). The data presented here are consistent with earlier work in that during proviral gene expression, an accumulation of Gag processing intermediates was found when Vif was overexpressed (Akari et al., 2004).
Recent work has characterized an important virus–host interaction between Lysyl tRNA synthetase (LysRS) and the main structural protein pr55Gag of HIV-1 (Javanbakht et al., 2003). The value of this PR biosensor assay has been demonstrated recently and has identified LysRS as a cellular mediator of HIV-1 PR activity (Guo et al., 2005). These data support the notion that LysRS, that is part of the HIV-1 Gag assembly complex (Kleiman and Cen, 2004), can dramatically impact on Gag processing and maturation. The impact of Vif on virus maturation has been derived from earlier reports that demonstrate that Vif associates with Gag (Bouyac et al., 1997) and to genomic RNA (Zhang et al., 2000), has effects on core stability (Ohagen and Gabuzda, 2000), and directly interacts or interferes with HIV-1 PR (Baraz et al., 2002, Bardy et al., 2001). Finally, Vif has been shown to be incorporated into budding virions (Kao et al., 2003). Several of these characteristics are shared between LysRS and Vif. Vif, however, has been shown to selectively influence p2/p7 cleavage by steric interference lending to the idea that it may somehow regulate the timing of PR activity during viral maturation. Processing of the p2/p7 PR site also has an influence on genomic RNA dimer maturation (Shehu-Xhilaga et al., 2001) and future work will have to address how both viral and cellular protein function to coordinate HIV-1 maturation during the budding process.
BRET was first exploited to study the dimerization of circadian proteins in bacteria (Xu et al., 1999) and the association of seven-membrane spanning G protein coupled receptors with their coreceptor or cofactors during signaling reactions (Angers et al., 2000). It was extended to examine tyrosine kinase receptor activation (Boute et al., 2001), transcription factor associations (Germain-Desprez et al., 2003) and more recently BRET2 has been used in several studies on virus-host interactions and HIV-1 coreceptor dynamics in the membrane (Issafras et al., 2002, Babcock et al., 2003, Chatel-Chaix et al., 2004, Guo et al., 2005, Percherancier et al., 2005). Recent work has also employed BRET2 in an in vivo ubiquitination assay (Perroy et al., 2004). BRET is considered a valuable approach and is at the forefront of HTS to identify new and desperately needed pharmacological drugs (Roda et al., 2003). Because small molecule inhibitors that target specific PR sites in HIV-1 have already been identified (Zhou et al., 2004), the assay reported here should prove useful to identify sequence specific inhibitors for HIV-1 PR sites in living cells. This BRET2 PR biosensor assay could also be useful to screen for small molecules that remain active due to drug resistance mutations that can occur in or around the PR sites in Gag (Feher et al., 2002). Finally, this BRET2-based assay can also be adapted to HTS of small molecule inhibitors in living cells by generating a stable indicator cell line that expresses all the necessary components of this assay.
In sum, the development of this novel and sensitive biosensor assay for HIV-1 PR activity provides an essential benefit over other reported assays in that it can be performed in live cells and is amenable to HTS of new anti-HIV-1 compounds without any apparent toxicity. This assay also has applications in the identification of small molecules that inhibit the essential function of proteases of other emerging viruses like the coronavirus that causes severe acute respiratory syndrome (SARS-CoV) and Hepatitus C (Blanchard et al., 2004, Lee et al., 2004).
Acknowledgements
We thank Billy Breton for advice on the BRET2 analyses and the NIH AIDS Research Reference and Reagent Program, Division of Acquired Immunodeficiency Syndrome (DAIDS, NIH), Duane Grandgenett, Stuart LeGrice, Robert Gorelick, Paul Spearman, Luc DesGroseillers and Anne Gatignol for supplying antibodies, reagents and genetic clones. J.F.C. was a recipient of an Fonds pour la recherche en santé du Québec Studentship, M.B. is a recipient of a Canada Research Chair in Signal Transduction and Molecular Pharmacology, and A.J.M. is a recipient of a CIHR New Investigator Award. This work was supported by grants from the Canadian Foundation for Innovation to A.J.M., the Canadian Institutes of Health Research to A.J.M, L.K. and M.B. and from the NIH to L.K. and K.S.
References
- The Pharmaceutical Research and Manufacturers of America (PhRMA), http://www.phrma.org/newmedicines/aids/.
- Akari H., Fujita M., Kao S., Khan M.A., Shehu-Xhilaga M., Adachi A., Strebel K. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 2004;279:12355–12362. doi: 10.1074/jbc.M312426200. [DOI] [PubMed] [Google Scholar]
- Angers S., Salahpour A., Joly E., Hilairet S., Chelsky D., Dennis M., Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc. Natl. Acad. Sci. U.S.A. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Farzan M., Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J. Biol. Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- Bagossi P., Kadas J., Miklossy G., Boross P., Weber I.T., Tozser J. Development of a microtiter plate fluorescent assay for inhibition studies on the HTLV-1 and HIV-1 proteinases. J. Virol. Methods. 2004;119:87–93. doi: 10.1016/j.jviromet.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Baraz L., Hutoran M., Blumenzweig I., Katzenellenbogen M., Friedler A., Gilon C., Steinitz M., Kotler M. Human immunodeficiency virus type 1 Vif binds the viral protease by interaction with its N-terminal region. J. Gen. Virol. 2002;83:2225–2230. doi: 10.1099/0022-1317-83-9-2225. [DOI] [PubMed] [Google Scholar]
- Bardy M., Gay B., Pebernard S., Chazal N., Courcoul M., Vigne R., Decroly E., Boulanger P. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing. J. Gen. Virol. 2001;82:2719–2733. doi: 10.1099/0022-1317-82-11-2719. [DOI] [PubMed] [Google Scholar]
- Baruch A., Jeffery D.A., Bogyo M. Enzyme activity—it's all about image. Trends Cell Biol. 2004;14:29–35. doi: 10.1016/j.tcb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Beriault V., Clement J.F., Levesque K., Lebel C., Yong X., Chabot B., Cohen E.A., Cochrane A.W., Rigby W.F., Mouland A.J. A late role for the association of hnRNP A2 with the HIV-1 hnRNP A2 response elements in genomic RNA, Gag, and Vpr localization. J. Biol. Chem. 2004;279:44141–44153. doi: 10.1074/jbc.M404691200. [DOI] [PubMed] [Google Scholar]
- Bertrand L., Parent S., Caron M., Legault M., Joly E., Angers S., Bouvier M., Brown M., Houle B., Menard L. The BRET2/arrestin assay in stable recombinant cells: a platform to screen for compounds that interact with G protein-coupled receptors (GPCRS) J. Recept. Signal Transduct. Res. 2002;22:533–541. doi: 10.1081/rrs-120014619. [DOI] [PubMed] [Google Scholar]
- Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N., Pernet K., Issad T. Monitoring the activation state of the insulin receptor using bioluminescence resonance energy transfer. Mol. Pharmacol. 2001;60:640–645. [PubMed] [Google Scholar]
- Bouyac M., Courcoul M., Bertoia G., Baudat Y., Gabuzda D., Blanc D., Chazal N., Boulanger P., Sire J., Vigne R., Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Clement J.F., Martel C., Beriault V., Gatignol A., DesGroseillers L., Mouland A.J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Antiviral drugs in current clinical use. J. Clin. Virol. 2004;30:115–133. doi: 10.1016/j.jcv.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Feher A., Weber I.T., Bagossi P., Boross P., Mahalingam B., Louis J.M., Copeland T.D., Torshin I.Y., Harrison R.W., Tozser J. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. Eur. J. Biochem. 2002;269:4114–4120. doi: 10.1046/j.1432-1033.2002.03105.x. [DOI] [PubMed] [Google Scholar]
- Germain-Desprez D., Bazinet M., Bouvier M., Aubry M. Oligomerization of transcriptional intermediary factor 1 regulators and interaction with ZNF74 nuclear matrix protein revealed by bioluminescence resonance energy transfer in living cells. J. Biol. Chem. 2003;278:22367–22373. doi: 10.1074/jbc.M302234200. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gabor, J., Cen, S., Hu, K., Mouland, A.J., Kleiman, L., 2005. Stabilization of HIV-1 GagPol by Lysyl-tRNA Synthetase. J. Biol. Chem., in press. [DOI] [PubMed]
- Huang Y., Kong W.P., Nabel G.J. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 2001;75:4947–4951. doi: 10.1128/JVI.75.10.4947-4951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang J., Shalom A., Li Z., Khorchid A., Wainberg M.A., Kleiman L. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J. Virol. 1997;71:726–728. doi: 10.1128/jvi.71.1.726-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issafras H., Angers S., Bulenger S., Blanpain C., Parmentier M., Labbe-Jullie C., Bouvier M., Marullo S. Constitutive agonist-independent CCR5 oligomerization and antibody-mediated clustering occurring at physiological levels of receptors. J. Biol. Chem. 2002;277:34666–34673. doi: 10.1074/jbc.M202386200. [DOI] [PubMed] [Google Scholar]
- Javanbakht H., Halwani R., Cen S., Saadatmand J., Musier-Forsyth K., Gottlinger H., Kleiman L. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 2003;278:27644–27651. doi: 10.1074/jbc.M301840200. [DOI] [PubMed] [Google Scholar]
- Kao S., Akari H., Khan M.A., Dettenhofer M., Yu X.F., Strebel K. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 2003;77:1131–1140. doi: 10.1128/JVI.77.2.1131-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman L., Cen S. The tRNALys packaging complex in HIV-1. Int. J. Biochem. Cell Biol. 2004;36:1776–1786. doi: 10.1016/j.biocel.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Lee J.C., Chang C.F., Chi Y.H., Hwang D.R., Hsu J.T. A reporter-based assay for identifying hepatitis C virus inhibitors based on subgenomic replicon cells. J. Virol. Methods. 2004;116:27–33. doi: 10.1016/j.jviromet.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Lindsten K., Uhlikova T., Konvalinka J., Masucci M.G., Dantuma N.P. Cell-based fluorescence assay for human immunodeficiency virus type 1 protease activity. Antimicrob. Agents Chemother. 2001;45:2616–2622. doi: 10.1128/AAC.45.9.2616-2622.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matayoshi E.D., Wang G.T., Krafft G.A., Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- McQuade T.J., Tomasselli A.G., Liu L., Karacostas V., Moss B., Sawyer T.K., Heinrikson R.L., Tarpley W.G. A synthetic HIV-1 protease inhibitor with antiviral activity arrests HIV-like particle maturation. Science. 1990;247:454–456. doi: 10.1126/science.2405486. [DOI] [PubMed] [Google Scholar]
- Mercier J.F., Salahpour A., Angers S., Breit A., Bouvier M. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 2002;277:44925–44931. doi: 10.1074/jbc.M205767200. [DOI] [PubMed] [Google Scholar]
- Mouland A.J., Coady M., Yao X.J., Cohen E.A. Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology. 2002;292:321–330. doi: 10.1006/viro.2001.1261. [DOI] [PubMed] [Google Scholar]
- Mouland A.J., Mercier J., Luo M., Bernier L., DesGroseillers L., Cohen E.A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K.L., llano M., Akari H., Miyagi E., Poeschla E.M., Strebel K., Bour S. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology. 2004;319:163–175. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Ohagen A., Gabuzda D. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 2000;74:11055–11066. doi: 10.1128/jvi.74.23.11055-11066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percherancier Y., Berchiche Y., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J. Biol. Chem. 2005 doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- Perroy J., Pontier S., Charest P.G., Aubry M., Bouvier M. Real-time monitoring of ubiquitination in living cells by BRET. Nat. Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- Pettit S.C., Sheng N., Tritch R., Erickson-Viitanen S., Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- Roda A., Guardigli M., Pasini P., Mirasoli M. Bioluminescence and chemiluminescence in drug screening. Anal. Bioanal. Chem. 2003;377:826–833. doi: 10.1007/s00216-003-2096-6. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga M., Kraeusslich H.G., Pettit S., Swanstrom R., Lee J.Y., Marshall J.A., Crowe S.M., Mak J. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 2001;75:9156–9164. doi: 10.1128/JVI.75.19.9156-9164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N., Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J. Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng N., Pettit S.C., Tritch R.J., Ozturk D.H., Rayner M.M., Swanstrom R., Erickson-Viitanen S. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 1997;71:5723–5732. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeman R.L., Honer B., Stoller T.J., Kesselmeier C., Miedel M.C., Traub P., Graves M.C. Human immunodeficiency virus type 1 protease cleaves the intermediate filament proteins vimentin, desmin, and glial fibrillary acidic protein. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6336–6340. doi: 10.1073/pnas.87.16.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack P.R., Frey M.W., Rizzo C.J., Cordova B., George H.J., Meade R., Ho S.P., Corman J., Tritch R., Korant B.D. Apoptosis mediated by HIV protease is preceded by cleavage of Bcl-2. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9571–9576. doi: 10.1073/pnas.93.18.9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin M., Deinum J., Goobar L., Danielson U.H. Proteolytic cleavage of microtubule-associated proteins by retroviral proteinases. J. Gen. Virol. 1990;71:1985–1991. doi: 10.1099/0022-1317-71-9-1985. [DOI] [PubMed] [Google Scholar]
- Xu Y., Piston D.W., Johnson C.H. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.J., Mouland A.J., Subbramanian R.A., Forget J., Rougeau N., Bergeron D., Cohen E.A. Vpr stimulates viral expression and induces cell killing in human immunodeficiency virus type 1-infected dividing Jurkat T cells. J. Virol. 1998;72:4686–4693. doi: 10.1128/jvi.72.6.4686-4693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Pomerantz R.J., Dornadula G., Sun Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Yuan X., Dismuke D., Forshey B.M., Lundquist C., Lee K.H., Aiken C., Chen C.H. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 2004;78:922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


