Abstract
Two sensitive and specific RT-PCR assays were standardised for testing the presence of human metapneumovirus. A total of 300 nasopharyngeal aspirates collected from infants suffering from bronchiolitis since October 2000 to June 2003 and shown previously as negative to common respiratory viruses were examined. Matrix and polymerase viral genes, which show a low rate of variation, were chosen to design amplification assays to ensure that any genotype of the human metapneumovirus could be detected. A RT-PCR followed by a reverse line blotting hybridisation was developed for viral polymerase gene. For the matrix gene, after the RT-PCR assay, a subsequent nested PCR was carried out. Both assays had similar sensitivity, equivalent to 0.1 TCID50 of human metapneumovirus strain NL/1/99 which was used as the positive control. The human metapneumovirus was present in 16.6% of the specimens studied. The approaches described below are not only a robust method for rapid diagnosis of the human metapneumovirus, but also to establish an etiological surveillance tool for epidemiological studies. Based on the results obtained, human metapneumovirus infections in Madrid followed a seasonal pattern, with most of the infections occurring between February and April.
Keywords: RT-heminested-PCR, Reverse line blotting hybridisation, Human metapneumovirus, Bronchiolitis, Respiratory viruses
1. Introduction
Respiratory infections are the most common diseases in the world being the origin of considerable morbidity and mortality specially in infants and elderly (Monto, 2002). Viruses causing acute respiratory infections include influenza viruses, respiratory syncytial virus, parainfluenza viruses, adenoviruses, coronaviruses, enteroviruses and rhinoviruses (Mackie, 2003). Among these, respiratory syncytial virus is a major pathogen of the lower respiratory tract infections in infants and young children, causing bronchiolitis and pneumonia with an important rate of hospitalizations (McCarthy and Hall, 2003).
Another recently discovered paramyxovirus, named human metapneumovirus, has also been associated with respiratory disease in children (van den Hoogen et al., 2001). It causes infections similar to those produced by respiratory syncytial virus, with symptoms ranging from wheeze and fever to severe cough, bronchiolitis or even pneumonia. Assisted ventilation is required in the most severe cases (Boivin et al., 2003, Esper et al., 2003). Since it was discovered in 2001 in The Netherlands, the virus has also been detected in specimens from adults, elderly and immunocompromised patients suffering from acute respiratory infections (Boivin et al., 2002, Pelletier et al., 2002).
Studies in different countries indicate the worldwide circulation of the human metapneumovirus in respiratory specimens from North (Peret et al., 2002) and South America (Galiano et al., 2004); China (Peiris et al., 2003a); Israel (Wolf et al., 2003); Australia (Nissen et al., 2002). The Netherlands (van den Hoogen et al., 2001) and other European countries (Maggi et al., 2003, Nicholson et al., 2003, Viazov et al., 2003, Vicente et al., 2003, Xepapadaki et al., 2004). Two main human metapneumovirus genetic lineages have been described and cocirculate during different years in several geographical areas (Biacchesi et al., 2003, Viazov et al., 2003).
The role of the human metapneumovirus in the paediatric acute respiratory infections is frequently decieved due to the difficulty to detect this virus by traditional diagnostic methods such as isolation in cell culture, immunofluorescence assay or enzyme immunoassay. Isolation in cell cultures is inefficient and monoclonal antibodies for immunofluorescence assay or enzyme immunoassay are not easily available and further testing is required to ensure reliability. Consequently, specific and sensitive molecular diagnostic methods are essential to achieve the accurate detection of human metapneumovirus in clinical specimens. This diagnosis may help the clinical managment of patients and may establish the role of this pathogen in the respiratory infection and its epidemiological behaviour.
In the present study, the convenience of using two RT-PCR based assays, designed in two different highly conserved genes (van den Hoogen et al., 2002), which codify the matrix and the polymerase viral proteins have been evaluated. These genes were selected to ensure the detection of human metapneumovirus independently of its genetic variation. The assays, a RT-heminested-PCR and a RT-PCR followed by a reverse line blotting hybridisation assay, had high specifity and sensitivity. Their inclusion as routine diagnosis methods can reduce the number of negative results. The presence of human metapneumovirus was evaluated in 300 respiratory secretions collected from children with respiratory disease from October 2000 to June 2003 in Madrid (Spain).
2. Material and methods
2.1. Clinical specimens
A total of 747 nasopharyngeal aspirates from young infants and children presenting with acute respiratory infections were collected from October 2000 to June 2003 at the Hospital Severo Ochoa Paediatric Unit (Madrid, Spain). Isolation in cell culture, indirect immunofluorescenee assays and/or multiplex RT-nested-PCR (Coiras et al., 2003) were attempted with every sample in order to detect influenza viruses, respiratory syncytial virus, parinfluenza viruses, adenovirus or enteroviruses. The presence of human metapneumovirus was checked in 300 aspirates, which were proven to be negative for the main respiratory viruses. All specimens were collected in 3 ml of virus transport medium (MEM, Gibco-BRL, Life Technologies, Paisley, Scotland; penicillin 200 U/ml, and streptomycin 200 μg/ml, BioWhittaker, MA; mycostatin 200 U/ml, Sigma; bovine serum albumin 0.25%, Merck, Darmstadt, Germany). These specimens were analysed at the Respiratory Virus Laboratory, in the National Centre for Microbiology (ISCIII, Madrid, Spain) to test for the presence of respiratory viruses. The specimens were frozen and stored at −70 °C until testing.
2.2. Virus isolation
Rhesus monkey kidney (LLC-MK2) cell line was maintained in DMEM (BioWhittaker, Belgium) supplemented with 5% foetal bovine serum and 1% penicillin–streptomycin (ICN Biomedicals, Ohio). Human metapneumovirus strain NL/1/99 was inoculated in LLC-MK2 in the presence of 5 μg/ml of trypsin in order to prepare positive controls for the amplification assays.
2.3. Nucleic acid extraction
Total nucleic acids were extracted from 200 μl aliquots from nasopharyngeal aspirates and also from the supernatant of infected cell cultures that were used as positive controls. Extraction was made using the guanidinium thiocyanate method described by Casas et al. (1995). Negative controls, consisting on RNase free sterile water were also submitted to the same extraction process. Lysis buffer including 100 molecules of a cloned amplified product of the RNA control supplied by the Promega RT-PCR system kit (Promega), was used as internal control of the extraction step and the following PCR process (Coiras et al., 2003).
2.4. Primer design for the matrix gene detection
Specific primer pairs were designed to amplify highly conserved regions of the human metapneumovirus gene that codes for matrix (M) protein. All the human metapneumovirus sequences available in GeneBank database were used to perform computer-assisted alignments using Macaw 2.0.5 program (Multiple Alignment Construction and Analysis Workbench, NCBI, Bethesda, MD). These primers were evaluated to ensure they fitted essential criteria for optimal PCR primers (Dieffenbach et al., 1993). The G + C contents, melting temperatures and lengths of the primers chosen, were analysed by using PrimerSelect v3.04a (DNAstar Inc., Madin, Wisconsin).
Primers used to amplify a part of the M gene by a RT-heminested-PCR assay were MIS (5′-GAGTCCTACCTAGTAGACAC-3′) and MIA (5′-TTGTYCCTTGRTGRCTCCA-3′) for RT-PCR and MIS and M2A (5′-TCTTGCAKATYYTRCTKATGCT-3′) for the subsequent heminested amplification reaction.
2.5. Primer and probe design for the polymerase gene detection
A conserved fragment of the polymerase gene was amplified using L6 and L7 oligonucleotides (van den Hoogen et al., 2003). L6 olignonucleotide was biotin-labelled at 5′ end to permit the detection of PCR product by chemiluminiscence. The specific probe (5′-CTGTTAATATCCCACACCA-3′), designed to be used in the reverse line blotting hybridisation with the PCR product, was 5′ end amino labelled.
2.6. RT-heminested-PCR assay for matrix gene detection
Reverse transcription PCR amplification for M gene detection was carried out in a single tube, using a commercial kit (Qiagen OneStep RT-PCR Kit, Qiagen, California). To each reaction tube was dispensed a mixture which contained 10 μl of buffer 5× with 25 mM MgCl2, 0.1 mM dNTPs, 0.1 μM of specific primers MIS and MIA, 0.1 μM of specific primers for internal control amplification, 2 μl of QIAGEN OneStep RT-PCR Enzyme Mix, according to manufacture's instructions, and 5 μl of nucleic acid extracted for a final volume of 50 μl. Thermal cycling conditions were as followed: for the RT proccess an initial cycle of 45 °C for 45 min, and 95 °C for 5 min; cycling conditions for the PCR were 45 cycles of 95 °C for 30 s, 53 °C for 2 min, 68 °C for 30 s and finally 10 min incubation at 68 °C for 10 min.
The heminested-PCR reaction contained 5 μl of buffer II 5× (Applied Biosystems, New Jersey), 2 mM MgCl2 (Applied Biosystems), 0.2 mM of each dATP, dGTP, dCTP and dTTP (Amersham Pharmacia Biotech, Piscataway, New Jersey), 0.8 μM of specific primers MIS and M2A, 0.2 μM specific primers for internal control amplification and 2.5 U Taq DNA polymerase (Amplitaq DNA polymerase, Applied Biosysterns). A total of 2 μl from the first reaction products was added for a final volume of 50 μl. Before heminested PCR, samples were heated to 95 °C for 5 min. Cycling conditions were 35 cycles: 95 °C for 30 s, 55 °C for 2 min, 72 °C for 3 min and a final incubation at 72 °C for 10 min.
The heminested PCR products were analysed by electrophoresis on 2% Seakem agarose gel, containing 5 μg/ml of ethidium bromide, in 0.5× Tris-borate buffer. The specific sizes of PCR products were 887 bp for the internal control and 718 bp for human metapneumovirus.
The procedures for preparing and amplifying specimens included all conventional recommendations to avoid contamination with PCR amplification products or positive controls (Kowk and Higuchi, 1989).
2.7. RT-PCR followed by RLB hybridisation assay for polymerase gene detection
The RT-PCR assay for L gene amplification was carried out in a single tube using the Qiagen OneStep RT-PCR Kit (Qiagen) as above described except for dNTPs final concentration, which was 0.2 mM. Ten picomole of each specific oligonucleotide was included to obtain a 170 pb sized amplified product. Antisense primer was labelled with biotin at 5′ end to get a biotin-labelled PCR amplification product.
Thermal cycling conditions were the following: an initial cycle of 42 °C for 45 min, and 95 °C for 5 min, followed by 45 cycles of 95 °C for 30 s, 52 °C for 1 min, 68 °C for 30 s and a final incubation of 68 °C for 7 min.
The reverse line blotting hybridisation procedure was performed according to Coiras et al. (2005). Briefly, the carboxyl groups of a Biodine C blotting membrane (Pall Biosupport) were activated with EDAC (l-ethyl-3-(3-dimethylaminopropyl)carbodiimide; Pall Gelman Laboratory, Michigan) and the membrane was placed in a MN45 miniblotter (Biometra, Germany). The specific probe for human metapneumovirus L gene, diluted to a final concentration of 0.8 pmol/μl in 500 mM NaHCO3 (pH 8.4), was covalently linked to the activated membrane. The remaining active esters on the membrane were hydrolysed by incubation with 0.1N NaOH. The membrane was then washed with 2× SSPE-0.1% SDS and placed in the miniblotter rotated 90 °C from the previous position. The slots were filled with 40 μl of denatured biotin-labelled PCR products diluted with 2× SSPE-0.5% SDS. The samples were incubated for 2 h at 48 °C. After washing, the membrane was treated with streptavidin–peroxidase conjugate (Roche, Indiana). Finally, the membrane was washed with 2× SSPE and the hybridised PCR products were detected by chemiluminiscence using ECL detection reagents (Amersham Pharmacia Biotech, England) and visualised by exposure to a light sensitive film (Hyperfiltn ECL; Amersham).
2.8. Sequence analyses for confirmation of the results
Positive specimens for human metapneumovirus M and L genes were purified with QIAquick PCR purification kit (Qiagen). Sequenced were obtained by an automatic DNA sequencer (ABI Prism 3700; Applied Biosystems) using Big Dye Terminator Cycle Sequencing kit Version 3.1 (Applied Biosystems). Nucleotide sequences obtained were aligned with sequences published previously using Macaw 2.0.5 program.
3. Results
The primers designed for the RT-PCR based assays amplify specifically partial sequences of the human metapneumovirus genes which encode for the matrix and the polymerase viral proteins. Primer concentrations and annealing temperatures, thermocycling parameters, concentration of each reaction component and probe hybridisation, both temperature and concentration, were standardised by experimentation. After optimizing the assay conditions, both assays showed concordant results with serial 10-fold dilutions of the human metapneumovirus strain NL/1/99 used as positive control (Fig. 1 ). The efficient limit of detection was 0.1 TCID50 on each assay. Specificity of the method was assayed by using RNA extracted from different respiratory viruses (influenza A, B and C, respiratory syncytial virus A and B, parainfluenza 1–3, adenoviruses and enteroviruses).
Fig. 1.
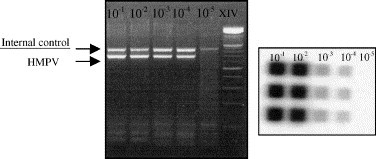
Sensitivity limits of RT-PCR assays evaluated by serial dilutions from 10−1 to 10−5, correspond to 102 to 10−2 TCID50 of the prototype strain of human metapneumovirus grown in MK2 cells. In the left panel can be visualised the RT-heminested-PCR assay for matrix gene in a 2% agarose gel in which both internal control and specific band are indicated. The right panel shows a sensitive film for polymerase RLB assay in which specific probe is used in triplicate for every virus dilution. In both assays, the detection limit reached was the dilution 10−4 which corresponds to 10−1 TCID50.
The reverse line blotting procedure for polymerase gene and the RT-heminested-PCR for the matrix gene were validated by using a total of 300 clinical nasopharyngeal aspirates taken from young infants. The number of specimens collected per season is summarized in Table 1 . Out of 300 specimens, a total of 50 resulted positive for the presence of human metapneumovirus (16.6%) by both RT-PCR assays. Nasopharyngeal aspirates were considered positive for the presence of human metapneumovirus only when a positive result for this virus was obtained in both assays. When enough amount of sample was available, positive human metapneumovirus results were confirmed by sequence analysis.
Table 1.
Number of human metapneumovirus infections detected in the nasopharingeal aspirates studied during three consecutive seasons
| 2000–2001 | 2001–2002 | 2002–2003 | Total | |
|---|---|---|---|---|
| Number of samples analysed | 100 | 93 | 107 | 300 |
| Number of positives to HMPV | 14 (14.00%) | 26 (27.9%) | 10 (9.3%) | 50 (16.6%) |
The percentage of human metapneumovirus infections in every season is presented in brackets.
The number of positive human metapneumovirus samples found was higher in the 2001–2002 season than in the other two seasons, although the number of samples analysed in the three periods was rather similar (Table 1). The annual distribution of human metapneumovirus positive specimens, summarized in Fig. 2 , showed a peak in the viral detection between February and April, that corresponded to a maximum in February and March in both 2000–2001 and 2001–2002 seasons, and another between March and April in the 2002–2003 season. Although most of the positive nasopharyngeal aspirates were collected between February and April, human metapneumovirus infection was also detected in specimens collected in January, May, June, October and December at a lower rate (nine positive specimens in total).
Fig. 2.
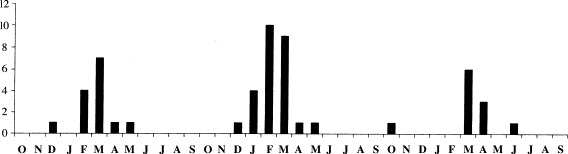
The graphic represents the number of positive samples found out of 300 specimens studied which were collected from October 2000 to June 2003.
All nasopharyngeal aspirates, collected from one hospital during the three consecutive seasons, had been tested previously for the main respiratory viruses (influenza A–C, respiratory syncytial virus groups A and B, parainfluenza 1–3, adenoviruses and enteroviruses) using RT-PCRs assays (Coiras et al., 2003), indirect immunofluorescence assay and/or isolation in cell culture. The number of positives for each virus and the percentage obtained is summarized in Table 2 and in Fig. 3 . As shown, the percentage of human metapneumovirus, referred to the total 747 nasopharyngeal aspirates, is at least 6.7%, due to the fact that only negative samples have been analysed and so double infections, if any, could not been found.
Table 2.
Respiratory viruses detected in the specimens collected during the three consecutive seasons (2000–2003)
| Season | Type of respiratory viruses |
|||||||
|---|---|---|---|---|---|---|---|---|
| ADV | EV | IV | HMPV | PIV | RSV | Negative | Total | |
| 2000–2001 | 5 | 4 | 5 | 14 | 1 | 162 | 86 | 277 |
| 2001–2002 | 12 | 1 | 8 | 26 | 4 | 148 | 67 | 266 |
| 2002–2003 | 20 | 1 | 11 | 10 | 0 | 65 | 97 | 204 |
| Totals | 37 (4.95%) | 6 (0.8%) | 24 (3.2%) | 50 (6.7%) | 5 (0.67%) | 375 (50.2%) | 250 (33.5%) | 747 |
The total percentage of each virus infection is presented in brackets: ADV, adenovirus; EV, enterovirus; IV, influenza viruses; HMPV, human metapneumovirus; PIV, parainfluenza viruses; RSV, respiratory syncytial viruses; negative, specimens in which no respiratory virus was found.
Fig. 3.
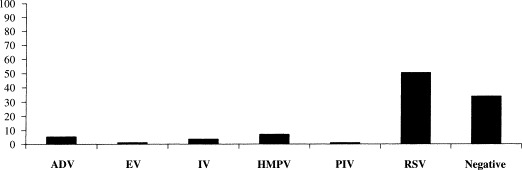
Percentage of the different viruses detected in positive samples during the consecutive seasons studied: ADV, adenovirus; EV, enterovirus; IV, influenza virus; HMNV, human metapneumovirus; PIV, parainfluenzavirus; RSV, respiratoty syncytial virus.
4. Discussion
Acute respiratory infection comprises a wide range of clinical pictures and it is caused by a heterogeneous group of viruses. Thus, there is a need for specific detection of the virus involved in each particular case, including those discovered more recently such as human metapneumovirus, human coronavirus NL63 (van der Hoek et al., 2004) or SARS coronavirus (Drosten et al., 2003). As in the case of other new viruses, the human metapneumovirus is specially difficult to grow in cell cultures. For this reason, molecular amplification methods are a valuable tool to detect the viral RNA present in the respiratory secretions (Kahn, 2003).
Two highly specific and sensitive RT-PCR based assays for the detection of human metapneumovirus RNA in nasopharyngeal aspirates were designed and can be performed rapidly and easily. In order to identify genetic variants of the main human metapneumovirus lineages, the design of amplification assays was focused on fragments of two different genes highly conserved. The use of two alternative RT-PCR assays presents the advantage of confirming the positive results in two independent genes and it also guarantees a decrease in the number of false positive results. The RT-heminested-PCR for the matrix gene is easy to perform and do not need high technology requirements. The reverse line blotting protocol for the viral polymerase gene is a safe procedure since non-radioactive reactives are used for labelling the probe and has the advantage of being suitable for large-scale epidemiological screening. The usage of the probe increases the specificity of the assay. In our experience, both methods were reliable and reproducible. Contamination with amplified products is prevented by careful separation of each mixture preparation, extraction, addition of the sample RNA, amplification and postamplification areas. Different sets of laboratory coats, pipettes and filtered pipette tips are used in each different area.
The assays were validated by examinining 300 nasopharyngeal aspirates from a complete cohort of 747 collected from infant population, less than 2 years old, suffering from acute respiratory infections such as bronchiolitis, pneumonia and influenza like illnesses, attending the Paediatric Unit of the Hospital Severo Ochoa in Madrid (Spain). Nasopharyngeal aspirate was a suitable type of specimen for the specific diagnosis of infections due to respiratory viruses. The specimens were collected from 2000 to 2003, not only during the respiratory virus circulation season but also during the rest of the year. The results provide further evidence that human metapneumovirus accounts for many cases of acute respiratory infection in children of this age, as 16.6% positives were detected in the 300 nasopharyngeal aspirates studied. The analysis of these results suggests that paediatric respiratory disease due to human metapneumovirus may have been underestimated due to the lack of rapid, sensitive and reliable diagnostic methods in most paediatric units. Most of the incidence studies associated with human metapneumovirus reported a lower rate of infection, which corresponded to values between 5 and 10% (Esper et al., 2004, Ijpma et al., 2004, Mackay et al., 2003, van den Hoogen et al., 2003). The percentage obtained in our study would be similar to that reported by Maggi et al. (2003) in Italy and by Viazov et al. (2003) in Germany, who also studied specimens from infants less than 2 years old. Other researchers, who evaluated children older than ours (Boivin et al., 2003, Esper et al., 2003, Peiris et al., 2003b, Vicente et al., 2003) or a wider age group ranging from children to adults (Stockton et al., 2002) found a lower incidence of human metapneumovirus. Therefore, our results support the views that human metapneumovirus is, after respiratory syncytial virus, an important cause of viral acute respiratory infections in children under 5 years.
On the other hand, the human metapneumovirus was detected in the present study as a unique pathogen because all 300 samples tested were found negative previously for the presence of the most common respiratory viruses. This type of samples had also been tested in other studies (Boivin et al., 2002, Freymouth et al., 2003, Peret et al., 2002). However, when co-infections were studied, some of the researchers concluded that they rarely occurred (Stockton et al., 2002, Vicente et al., 2003). Co-infections would be more numerous if the diagnosis techniques used for common respiratory viruses, mainly cellular cultures and indirect immunofluorescence assays, were as sensitive as the PCR assays are at the moment. Further studies are necessary to obtain a more real analysis of the respiratory virus infections.
Although this is a retrospective study, some epidemiological conclusions may be drawn. Human metapneumovirus positive specimens were collected mainly during the late winter and early spring months. Some positive cases were detected during the rest of the year. In contrast to other studies, no alternative years of high and low incidence were observed (Maggi et al., 2003). The peak of virus detection was found between February and April on the 3 years period from 2000 to 2003. Besides, the onset of respiratory syncytial virus disease was determined between December and January, just before the peak of human metapneumovirus detection, during the same seasons (data not shown). These results confirm that human metapneumovirus circulates primarily during the late winter and early spring, at the end of the circulation of respiratory syncytial virus (Falsey et al., 2003, Maggi et al., 2003, von Linstow et al., 2004). However, positive specimens for human metapneumovirus were also found during the late spring and summer months. Peiris et al. (2003b) reported human metapneumovirus activity in these months too. Nevertheless, few data related to summer months are available, since most of the studies only examined samples collected during winter season. In relation to the age of the children studied, most of the human metapneumovirus infections affected children between 6 and 12 months of age (data not shown), in agreement with previous data (van den Hoogen et al., 2003). Further studies will be needed to determine the real incidence of human metapneumovirus in community-acquired acute respiratory infections in young children in Spain.
In conclusion, the methods developed in this study and the results obtained permit the detection of human metapneumovirus, indicating its important role as a pathogen causing bronchiolitis in young infants. Diagnosis of human metapneumovirus is essential to reduce the number of respiratory infections in which the etiologic agent is not recognized. It should be noted that human metapneumovirus was detected in 16.6% of the subgroup with negative results out of the total 747 nasopharyngeal aspirates, the use of these specific molecular assays have increased at least in 6.7% the number of specimens in which a viral agent could have been determined. The results obtained indicate that human metapneumovirus should be taken into account in the differential diagnosis of acute respiratory infections, mainly to distinguish it from respiratory syncytial virus infections. The RT-PCR based assays described above are specific and sensitive for a rapid and reliable diagnosis of human metapneumovirus in nasopharyngeal aspirates.
Acknowledgements
M.R. López-Huertas is a predoctoral fellow funded by de Instituto de Salud Carlos III (03/0028). The authors acknowledge skillful technical assistance from Manuela López-Valero and Nieves Cruz in the Respiratory Viruses Laboratory, CNM, ISCIII, Madrid, Spain. The authors thank Dr. Osterhaus at the Erasmus Medical Centre Rotterdam for kindly supplying human Metapneumovirus strain NL/1/99 used as positive control.
References
- Biacchesi S., Skiadopoulos M.H., Boivin G., Hanson C.T., Murphy B.R., Collins P.L., Buchholz U.J. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315(1):1–9. doi: 10.1016/s0042-6822(03)00528-2. [DOI] [PubMed] [Google Scholar]
- Boivin G., Abed Y., Pelletier G., Ruel L., Moisan D., Cote S., Peret T.C., Erdman D., Anderson L.J. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 2002;186(9):1330–1334. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- Boivin G., De Serres G., Cote S., Gilca R., Abed Y., Rochette L., Bergeron M.G., Déry P. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 2003;9(6):634–640. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas I., Powell L., Klapper P.E., Cleator G.M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods. 1995;53(1):25–36. doi: 10.1016/0166-0934(94)00173-e. [DOI] [PubMed] [Google Scholar]
- Coiras M.T., Pérez-Breña P., Garcia M.L., Casas I. Simultaneous detection of influenza A, B and C viruses, respiratory syncytial virus and adenoviruses in clinical samples by multiplex reserve transcription nested-PCR assay. J. Med. Virol. 2003;69:32–44. doi: 10.1002/jmv.10255. [DOI] [PubMed] [Google Scholar]
- Coiras M.T., López-Huertas M.R., López-Campos G., Aguilar J.C., Pérez-Breña P. Oligonucleotide array for simultaneous detection of respiratory viruses using a reserve-line blot hybridisation assay. J. Med. Virol. 2005;76:256–264. doi: 10.1002/jmv.20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieffenbach C.W., Lowe T.M., Dveksler G.S. General concepts for PCR primer design. PCR Methods Appl. 1993;3(3):S30–S37. doi: 10.1101/gr.3.3.s30. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Esper F., Boucher D., Weibel C., Martinello R.A., Kahn J.S. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics. 2003;111:1407–1410. doi: 10.1542/peds.111.6.1407. [DOI] [PubMed] [Google Scholar]
- Esper F., Martinello R.A., Boucher D., Weibel C., Ferguson D., Landry M.L., Kahn J.S. A 1-year experience with human metapneumovirus in children aged <5 years. J. Infect. Dis. 2004;189(8):1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Erdman D., Anderson J.L., Walsh E.E. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 2003;187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Freymouth F., Vabret A., Legrand L., Eterradossi N., Lafay-Delaire F., Brouard J., Guillois B. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr. Infect. Dis. J. 2003;22(1):92–94. doi: 10.1097/00006454-200301000-00024. [DOI] [PubMed] [Google Scholar]
- Galiano M., Videla C., Puch S.S., Martinez A., Echavarria M., Carballal G. Evidence of human metapneumovirus in children in Argentina. J. Med. Virol. 2004;72(2):299–303. doi: 10.1002/jmv.10536. [DOI] [PubMed] [Google Scholar]
- Ijpma F.F., Beekhuis D., Cotton M.F., Pieper C.H., Kimpen J.L., van den Hoogen B.G., van Doornum G.J., Osterhaus D.M. Human metapneumovirus infection in hospital referred South African children. J. Med. Virol. 2004;73(3):486–493. doi: 10.1002/jmv.20116. [DOI] [PubMed] [Google Scholar]
- Kahn J.S. Human metapneumovirus, a newly emerging respiratory virus. Pediatr. Infect. Dis. J. 2003;22(10):923–924. doi: 10.1097/01.inf.0000091347.27554.ff. [DOI] [PubMed] [Google Scholar]
- Kowk S., Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- Mackay I.M., Jacob K.C., Woolhouse D., Waller K., Syrmis M.W., Whiley D.M., Siebert D.J., Nissen M., Sloots T.P. Molecular assays for detection of human metapneumovirus. J. Clin. Microbiol. 2003;41(1):100–105. doi: 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie P.L. The classification of viruses infecting the respiratory tract. Paediatr. Respir. Rev. 2003;4(2):84–90. doi: 10.1016/S1526-0542(03)00031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F., Pifferi M., Vatteroni M., Fornai C., Tempestini E., Anzilotti S., Lanini L., Andreoli E., Ragazzo V., Pistello M., Specter S., Bendinelli M. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 2003;41(7):2987–2991. doi: 10.1128/JCM.41.7.2987-2991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C.A., Hall C.B. Respiratory syncytial virus: concerns and control. Pediatr. Rev. 2003;24(9):301–309. doi: 10.1542/pir.24-9-301. [DOI] [PubMed] [Google Scholar]
- Monto A.S. Epidemiology of viral respiratory infections. Am. J. Med. 2002;112:4S–12S. doi: 10.1016/s0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- Nicholson K.G., McNally T., Silverman M., Simons P., Zambon M.C. Influenza-related hospitalizations among young children in Leicestershire. Pediatr. Infect. Dis. J. 2003;22(10):S28–S30. doi: 10.1097/01.inf.0000092193.91306.2f. [DOI] [PubMed] [Google Scholar]
- Nissen M.D., Siebert D.J., Mackay I.M., Sloots T.P., Withers S.J. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 2002;176(4):188. doi: 10.5694/j.1326-5377.2002.tb04354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Tang W.H., Chan K.H., Khong P.L., Guan Y., Lau Y.L., Chiy S.S. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg. Infect. Dis. 2003;9(6):628–633. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G., Dery P., Abed Y., Boivin G. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 2002;8(9):976–978. doi: 10.3201/eid0809.020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret T.C., Boivin G., Li Y., Couillard M., Humphrey C., Osterhaus A.D., Erdman D.D., Anderson L.J. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 2002;185(11):1660–1663. doi: 10.1086/340518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton J., Stephenson I., Fleming D., Zambon M. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 2002;8(9):897–901. doi: 10.3201/eid0809.020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyre K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A., Osterhaus A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001;7(6):719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., Bestebroer T.M., Osterhaus A.D., Fourchier R.A. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295(1):119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- van den Hoogen B.G., van Doornum G.J., Fockens J.C., Cornelissen J.J., Beyer W.E., Groot Rd R., Osterhaus A.D., Fouchier R.A. Incidence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 2003;188(10):1571–1577. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- Viazov S., Ratjen F., Scheidhauer R., Fiedler M.L., Roggendorf M. High incidence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 2003;41(7):3043–3045. doi: 10.1128/JCM.41.7.3043-3045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente D., Cilia G., Montes M., Perez-Trallero E. Human metapneumovirus and community-acquired respiratory illness in children. Emerg. Infect. Dis. 2003;9(5):602–603. doi: 10.3201/eid0905.020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Linstow M.L., Larsen H.H., Eugen-Olse J., Koch A., Winther T.N., Meyer A.N., Westh H., Lundgren B., Melbye M., H∅gh B. Human metapneumovirus and resporatory syncytial virus in hospitalized Danish children with acute resporatory tract infection. Scand. J. Infect. Dis. 2004;36:578–584. doi: 10.1080/00365540410018166. [DOI] [PubMed] [Google Scholar]
- Wolf D.G., Zakay-Rones Z., Fadeela A., Greenberg D., Dagan R. High seroprevalence of human metapneumovirus among young children in Israel. J. Infect. Dis. 2003;188(12):1865–1867. doi: 10.1086/380100. [DOI] [PubMed] [Google Scholar]
- Xepapadaki P., Psarras S., Bossios A., Tsolia M., Gourgiotis D., Liapi-Adamidou G., Constantopoulos A.G., Kafetzis D., Papadopoulos N.G. Human metapneumovirus as a causative agent of acute bronchiolitis in infants. J. Clin. Virol. 2004;30(3):267–270. doi: 10.1016/j.jcv.2003.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


