Abstract
Acute viral respiratory infections are among the most common causes of human disease. Rapid and accurate diagnosis of viral respiratory infections is important for providing timely therapeutic interventions.
This study evaluated a new multiplex PCR assay (Seegene Inc., Seoul, Korea) for simultaneous detection and identification of 12 respiratory viruses using two primer mixes. The viruses included parainfluenza viruses 1, 2, and 3, human metapneumovirus, human coronavirus 229E/NL63 and OC43, adenovirus, influenza viruses A and B, human respiratory syncytial viruses A and B, and human rhinovirus A.
The analytical sensitivity of the assay was 10–100 copies per reaction for each type of virus. There was no cross-reactivity with common bacterial or viral pathogens. A comparison with conventional viral culture and immunofluorescence was carried out using 101 respiratory specimens from 92 patients. Using viral culture, 57 specimens (56.4%) were positive without co-infection. The same viruses were identified in all 57 specimens using the multiplex PCR. Seven of the 57 specimens (12.3%) were found to be co-infected with other respiratory viruses, and 19 of 44 (43.2%) specimens which were negative by culture were positive by the multiplex PCR.
The Seeplex Respiratory Virus Detection assay represents a significant improvement over the conventional methods for the detection of a broad spectrum of respiratory viruses.
Keywords: Respiratory infection, Viral culture, Multiplex PCR
1. Introduction
Human respiratory tract infections can be caused by many viruses including influenza viruses, parainfluenza viruses (PIV), human respiratory syncytial viruses (RSV), human metapneumovirus (hMPV), human coronaviruses (CoV), human rhinoviruses (HRV), adenoviruses (AdV), and human enteroviruses. These viral infections are often associated with significant morbidity and mortality (Thompson et al., 2003). Therefore, rapid and accurate diagnosis is essential for timely therapeutic interventions (Barenfanger et al., 2000).
Diagnoses of viral respiratory tract infections have been made generally by conventional isolation methods using cell culture and/or detection of antigens. Although these methods are effective and often complementary, they have some disadvantages. Cell culture, which is considered the “gold standard”, takes time, and detection of antigens often lacks sensitivity or specificity. Even when these methods are applied simultaneously, some samples are still found negative despite clinical evidence of a viral respiratory infection (Bellau-Pujol et al., 2005).
Detection of respiratory viruses can be improved with molecular techniques. Many PCR-based methods have been developed and evaluated (Donofrio et al., 1992, Freymuth et al., 1997, Weinberg et al., 2004). However, virus-specific RT-PCR assays which require separate amplification of each virus, are resource intensive. Therefore, multiplex PCR methods were developed with the aim of detecting a panel of viruses simultaneously (Coiras et al., 2004, Grondahl et al., 1999, Li et al., 2007, Templeton et al., 2004).
The Seeplex Respiratory Virus Detection assay (Seegene Inc., Seoul, Korea), based on a multiplex PCR method using dual priming oligonucleotide (DPO) system, has been introduced recently. This assay uses two separate primer mixes, and is capable of detecting 11 types of RNA viruses and 1 type of DNA virus associated commonly with respiratory infections. This study evaluated the performance of the multiplex assay, and compared it with conventional viral culture and immunofluorescence.
2. Materials and methods
2.1. Analytical sensitivity and specificity
AdV strains (serotypes 1,3,8,18,23 and 40), PIV-1, -2, and -3, influenza-A (H3N2 and H1N1) and -B, RSV-A and -B, CoV OC43 and 229E, rhinovirus-A and -B, human herpesvirus 1 and 2, human coxsackievirus, Bordetella pertussis, Haemophilus influenzae, Streptococcus pneumoniae, and Mycoplasma pneumoniae were obtained from the American Type Culture Collection (ATCC, Manassas, VA). An hMPV sample was isolated from a Korean patient, and it was confirmed by direct sequencing to have 99% similarity to the GenBank accession No. DQ023131 hMPV strain.
Viral DNA/RNA was extracted from ATCC freeze-dried cells using Viral Gene-spin™ Kit (iNtRON Biotechnology, Inc., Seoul, Korea). Reverse transcription was performed on 8 μl of the purified RNA in the final reaction volume of 20 μl for 1.5 h at 37 °C, using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Ontario, Canada). A pUC18 vector was used for plasmid DNA preparation.
Tenfold serial dilutions of the prepared plasmid DNA were made from 108 to 10−2 copies per reaction to determine the analytical sensitivity of the assay. ATCC standard organisms (28 species) and human genomic DNA (human adult normal tissue: uterus: cervix, Code number D1234275, Biochain) were used to assess the analytical specificity.
2.2. Respiratory specimens
From April, 2006 to October, 2006, 101 respiratory specimens (nasopharyngeal swab/aspirates and bronchial washing fluids) from 92 patients were tested for respiratory viruses using conventional viral culture and immunofluorescence. The age of patients ranged from 0 to 72, with the mean age of 6.9. There were only six adult patients (6.5%), and the mean age of the other 86 patients was 3.4 years. All the six adult patients had serious underlying diseases such as malignant tumors or advanced pulmonary disease. The medical records of each patient were reviewed at the time of sample collection. All the samples were stored in a freezer at −70 °C until PCR was carried out.
2.3. Viral culture and immunofluorescence
The human laryngeal carcinoma (HEp-2) cell line was used to isolate RSV and AdV. The Madin-Darby canine kidney (MDCK) cell line was used to isolate influenza-A and -B. The monkey kidney (LLC-MK2) cell line was used to isolate PIV-1, PIV-2, and PIV-3. These cell lines were placed in a 24-well tissue culture plates. Each sample was inoculated onto the cells, incubated at 35 °C, and cytopathic effects (CPE) were observed for 10 days. The cells were scraped on day 3 and 10, and an indirect immunofluorescence assay was carried out using Respiratory Panel 1 Viral Screening & Identification Kit (Light Diagnostics, Chemicon, Temecula, CA, USA). Slides were read under a fluorescence microscope.
2.4. Multiplex PCR
Viral DNA/RNA was extracted from 300 μl of each respiratory specimen using Viral Gene-spin™ Kit according to the manufacturer's instructions. Reverse transcription step was carried out using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Ontario, Canada) to synthesize cDNA. Cloned murine leukemia virus reverse transcriptase was employed for this step. Multiplex PCR was then undertaken on 3 μl of synthesized first-strand cDNA. 4 μl of multiplex primer sets, 10 μl of master mix (hot start Taq DNA polymerase and dNTP are included in the reaction buffer), and 3 μl of 8-methoxypsoralen (8-MOP) were added. Internal control (Seegen, Seoul, Korea) was also added at this step to validate the PCR. This internal control is a DNA solution extracted from plants. Specific primer targets for each respiratory virus are listed in Table 1 . 8-MOP, accompanied by UV irradiation for 20 min, prevents amplification of contaminated DNA. For the positive control, a mixture of 12 viral clones was used as a template. Sterile deionized water was used for negative control. The pre-PCR products were stored at −20 °C until use.
Table 1.
Targets for detection of respiratory viruses by the Seeplex Respiratory Virus Detection assay.
| Respiratory virus 1-A set | Target | Accession no. | Amplicon size (bp) |
|---|---|---|---|
| Internal control | rbcL | AJ746297 | 719 |
| Human adenovirus | Pol gene | ADV B: NC_004001 | 534 |
| ADV C: NC_001405 | |||
| ADV E: NC_003266 | |||
| Human metapnemovirus | F gene | NC_004148 | 469 |
| Human coronavirus 229E/NL63 | S gene | 229E: NC_002645 | 375 |
| NL63: NC_005831 | |||
| Human parainfluenzavirus 1 | HN gene | NC_003461 | 324 |
| Human parainfluenzavirus 2 | HN gene | NC_003443 | 264 |
| Human parainfluenzavirus 3 | HN gene | NC_001796 | 219 |
| Respiratory virus 1-B set | Target | Accession no. | Amplicon size (bp) |
|---|---|---|---|
| Internal control | rbcL | AJ746297 | 719 |
| Influenza A virus | segment 7 | NC_007377 | 516 |
| Influenza B virus | segment 1 | NC_002204 | 455 |
| Human respiratory syncytial virus B | F gene | AY353550 | 391 |
| Human rhinovirus A | 5’-NTR | NC_001617 | 337 |
| Human respiratory syncytial virus A | F gene | AY330614 | 273 |
| Human coronavirus OC43 | M gene | NC_005147 | 231 |
After preheating at 95 °C for 15 min, 40 amplification cycles were carried out under the following conditions in a thermal cycler (GeneAmp PCR system 9700, Foster City, CA): 94 °C for 30 s, 60 °C for 1.5 min, and 72 °C for 1.5 min. Amplification was completed at the final extension step at 72 °C for 10 min. The multiplex PCR products were visualized by electrophoresis on an ethidium bromide-stained 2% agarose gel.
Specimen processing, DNA/RNA extraction, PCR amplification, and PCR product analyses were conducted in different rooms. Special care was taken to avoid either contamination with RNAase, or cross-contamination between reactions.
2.5. Comparison of tests
RSV, influenza-A and -B, and PIV-1, PIV-2, PIV-3, and AdV were detected by viral culture. The multiplex PCR, on the other hand, detected four more types of respiratory viruses including rhinovirus, CoV OC43 229E/NL63, and hMPV. It can also distinguish between RSV-A and RSV-B. Therefore, results for the latter viruses were excluded from the comparison.
3. Results
The analytical sensitivity of the multiplex PCR was tested using serial dilutions of plasmid DNA from the ATCC strains. PCR amplifications were observed at 10–100 copies per reaction (Fig. 1 ). The detection limits of single-targeted PCR and multiplex PCR were the same.
Fig. 1.
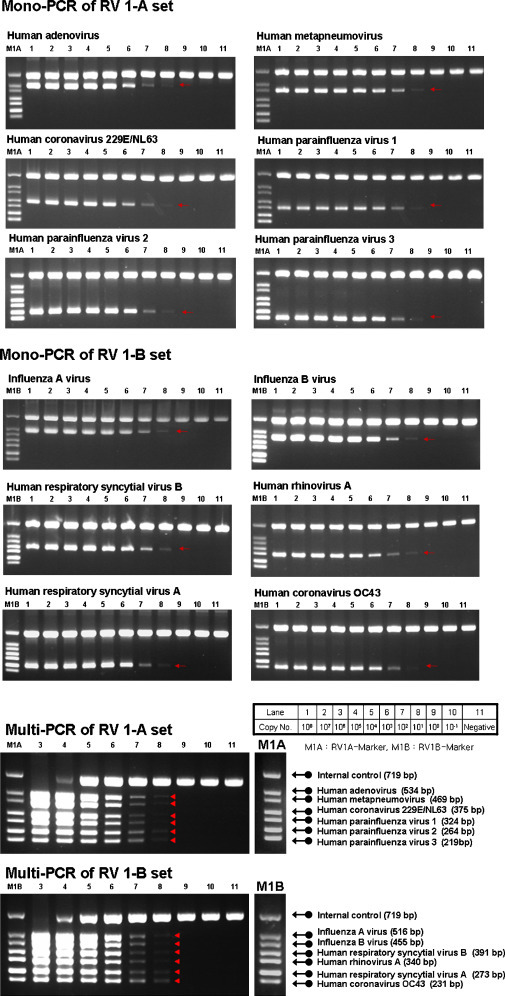
Analytical sensitivity of the multiplex PCR assay. Serially diluted target gene-containing plasmids were used for the analytical sensitivity test.
Fig. 2 shows a summary of the specificity test results. ATCC standard organisms (28 species) and the mixture of 12 respiratory viral clones were used. The multiplex PCR identified each specific respiratory virus. No cross-reactivity with other respiratory viruses or bacteria was observed.
Fig. 2.
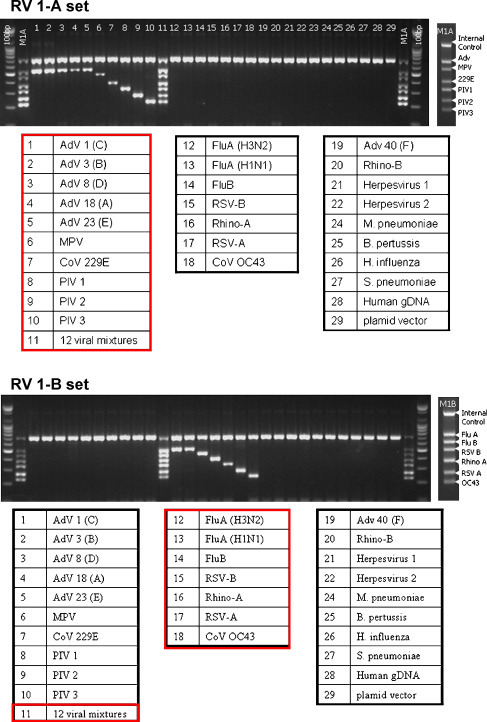
Analytical specificity of the multiplex PCR assay.
A comparison between multiplex PCR and conventional viral culture was made. Among the 101 clinical specimens, 57 samples (56.4%) were positive for the respiratory viruses using viral culture. Isolated viruses were 25 PIV-3, 12 RSV, 8 PIV-1, 8 AdV, 3 influenza-B, and 1 influenza-A. All the culture-positive specimens were also found PCR-positive, without any discrepancy (Fig. 3 ).
Fig. 3.
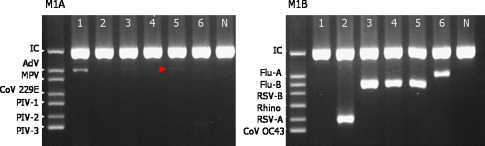
Multiplex PCR assay for respiratory viruses. Six samples were amplified using PCR, and visualized with agarose gel electrophoresis: 1, adenovirus positive. 2, RSV-A positive. 3 and 4, influenza-B positive. 5, adenovirus and influenza-B positive (faint band is observed for adenovirus; arrowhead). 6, influenza-A positive. All samples were tested with a primer mixture A (M1A) and B (M1B). Abbreviations: AdV, adenovirus. CoV, human coronavirus. MPV, human metapneumovirus. PIV, parainfluenza virus. RSV, human respiratory syncytial virus. Rhino, human rhinovirus. Flu, influenza virus. IC, internal control.
The multiplex PCR identified 19 respiratory viruses among 44 culture-negative samples (43.2%). Fifteen of these 19 viruses were 12 RSV, 1 AdV, and 2 co-infections of AdV/PIV-3 and RSV/PIV-1 (Table 2 ), which viral culture was expected to detect.
Table 2.
Viruses isolated by conventional viral culture/immunofluorescence or detected by the multiplex PCR.
| Respiratory viruses isolated by viral culture and immunofluorescence stain | Respiratory viruses isolated by multiplex PCR (n) | Total isolates (%) |
|---|---|---|
| AdV | AdV (7), AdV/RSV-A (1) | 8 (7.9) |
| RSV | RSV-A (12) | 12 (11.9) |
| PIV-1 | PIV-1 (5), PIV-1/RSV-A (1), PIV-1/RSV-B (1), PIV-1/HRV (1) | 8 (7.9) |
| PIV-2 | – | 0 |
| PIV-3 | PIV-3 (21), PIV-3/PIV-2 (1), PIV-3/RSV-A (1), PIV-3/HRV (1), PIV-3/RSV-B/PIV-2 (1) | 25 (24.8) |
| Influenza-A | Influenza-A (1) | 1 (1.0) |
| Influenza-B | Influenza-B (2), Influenza-B/AdV (1) | 3 (3.0) |
| Negative | RSV-A (9), HRV (2), CoV 229E/NL63 (2), AdV (1), AdV/PIV-3 (1), RSV-A/RSV-B (1), RSV-A/PIV-1 (1), RSV-B (1), HRV/RSV-B (1) | 19 (18.8) |
| Negative | Negative | 25 (24.8) |
Excluding the four specimens with rhinoviruses and coronaviruses, which could be detected only by the multiplex PCR, the overall concordance rate was 88.7% (86/97).
In addition, the multiplex PCR detected 13 specimens with co-infections. These included 9 culture-positive specimens and 4 negative ones. After excluding 3 specimens with rhinoviruses and 1 co-infection of RSV-A/RSV-B which could be detected only by the multiplex PCR, the co-infection rate was 12.3% (7/57) of culture-positive specimens and 13.3% (2/15) of culture-negative specimens, respectively. Interestingly, six of the nine co-infections contained RSV. No co-infection was detected by viral culture and immunofluorescence.
At least two times of specimens were collected from six patients (Table 3 ). Follow-up cultures were carried out during the same respiratory event. The interval from collection of the first specimen to the next was no more than 20 days. In the case of Patient 1, the 3rd specimen was culture-negative. However, using the multiplex PCR, the 3rd specimen was positive for RSV-A, as well as the 1st and 2nd specimens. In the cases of Patient 3 and 5, viral infections were detected only by the multiplex PCR. RSV-A was detected repeatedly in Patient 5.
Table 3.
Results of the viral culture/immunofluorescence and multiplex PCR in patients with two or more serial specimens.
| Patient | 1st specimen |
2nd specimen |
3rd specimen |
|||
|---|---|---|---|---|---|---|
| Culture | Multiplex-PCR | Culture | Multiplex-PCR | Culture | Multiplex-PCR | |
| 1 | RSV | RSV-A | RSV | RSV-A | Negative | RSV-A |
| 2 | RSV | RSV-A | RSV | RSV-A | RSV | RSV-A |
| 3 | Negative | Negative | Negative | PIV-1, RSV-A | nd | nd |
| 4 | Negative | Negative | Negative* | Negative* | nd | nd |
| 5 | Negative | RSV-A | Negative | RSV-A | nd | nd |
| 6 | PIV-1 | PIV-1 | PIV-1 | PIV-1, RSV-A | nd | nd |
nd, not done.
All the specimens were bronchial washing fluid, except the 2nd specimen of the Patient 4, which was pleural fluid.
4. Discussion
Viruses causing a variety of respiratory tract infections such as croup, bronchiolitis, and pneumonia in infants and children, can cause also significant morbidity and mortality in elderly and immunocompromised adults (Latham-Sadler and Morell, 1996, Woo et al., 1997). Therefore a sensitive and rapid detection of these viruses is essential for appropriate treatment and reducing nosocomial transmission (Woo et al., 1997).
Conventional viral culture and antigenic detection are often time-consuming with low sensitivity. Therefore, a number of studies aimed to develop and evaluate multiplex PCR for the detection of respiratory viruses. Generally, these studies have confirmed the effectiveness of this technique (Elnifro et al., 2000). However, current multiplex PCR-based assays, such as nested PCR or probe hybridization assay, require further validation due to the high rate of false positives (Bellau-Pujol et al., 2005, Hindiyeh et al., 2005).
Dual priming oligonucleotide (DPO) system is a new molecular technique for PCR, which contains two separate priming regions joined by a polydeoxyinosine linker. These primers allow a wide range of annealing temperatures and provide high specificity which helps to prevent false positive results. Chun et al. (2007) reported that the DPO system prevents non-specific amplification without disrupting efficient amplification of the target sequences. The Seeplex Respiratory Virus Detection Kit is the first multiplex PCR assay based on the DPO system.
This respiratory virus detection kit has been evaluated previously in two studies to assess the diagnostic effectiveness of the kit by comparison with the conventional methods. The results were in agreement between the two methods, although the multiplex PCR was more sensitive for detecting respiratory viruses (Roh et al., 2008, Yoo et al., 2007).
In this study, the kit was evaluated and compared with the conventional methods using 101 respiratory specimens. Multiplex PCR results were superior to those obtained by the conventional methods, as the two previous studies reported. In addition, the analytical sensitivity of the kit was evaluated using multi-step diluted plasmid DNA of ATCC standard organisms, and the minimal number of copies required for the detection of viruses was determined. The kit was highly specific for detecting the target viruses, and no cross-reaction with other microorganisms causing respiratory disease was observed.
The multiplex PCR detected all viruses which were isolated initially using the conventional method. In addition, with the multiplex PCR, at least 15 out of the 44 culture-negative specimens were positive. All these 15 patients had significant respiratory symptoms suggestive of upper or lower respiratory tract infection, and/or showed chest radiograph abnormalities. Bacterial and fungal cultures were all negative in these patients. RSV was the most common pathogen (12/15, 80%). RSV in 3 of these 12 patients was also identified by the direct RSV antigen assay. These respiratory specimens which were multiplex PCR-positive only, contained probably very low virus quantities and/or viruses with decreased viability. Multiplex PCR is reported to be better for the detection of viruses from clinical specimens because it can detect viral genomes at a lower titer as well as viruses that are not replication competent (Liolios et al., 2001).
Several cases of co-infection were detected using the multiplex PCR. Interestingly, most co-infections were due to RSV, which is the most common cause of respiratory tract infections in young children (Hall, 2001). RSV involvement in dual infections has been reported to be associated with an increased severity of illness (Aberle et al., 2005). Although this virus can be detected theoretically by viral culture, multiple infections including those by RSV were detected only by using the multiplex PCR.
Five rhinoviruses and 2 CoV 229E positive samples were detected using the multiplex PCR. These two types of viruses are known to cause common cold, in addition to lower respiratory tract infections in infants and the elderly (Vabret et al., 2003). It has been reported that hMPV is responsible for 5–7% of viral respiratory tract infections in children admitted to hospital (van den Hoogen et al., 2004). However, in this study, hMPV infection was not detected.
When a respiratory virus was detected using viral culture from two or more specimens obtained serially from a patient, the same virus was identified also using the multiplex PCR. Co-infection was also detected. In three patients, only the PCR method revealed the existence and persistence of a viral infection. The multiplex PCR assay therefore provides more consistency, and reflects better infectious status of patients.
In conclusion, the Seeplex Respiratory Virus Detection assay has great potential for the detection and identification of common viral respiratory infections. It can be used as a supplement or alternative to the conventional methods. Furthermore, the results of the multiplex PCR can be obtained within 24 h, while for viral cultures, it takes usually up to a week. Overall, the multiplex PCR assay may help reduce the incidence of nosocomial transmissions, and improve clinical management by earlier treatment after diagnosis.
References
- Aberle J.H., Aberle S.W., Pracher E., Hutter H.P., Kundi M., Popow-Kraupp T. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon-gamma response. Pediatr. Infect. Dis. J. 2005;24:605–610. doi: 10.1097/01.inf.0000168741.59747.2d. [DOI] [PubMed] [Google Scholar]
- Barenfanger J., Drake C., Leon N., Mueller T., Troutt T. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J. Clin. Microbiol. 2000;38:2824–2828. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellau-Pujol S., Vabret A., Legrand L., Dina J., Gouarin S., Petitjean-Lecherbonnier J., Pozzetto B., Ginevra C., Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras M.T., Aguilar J.C., Garcia M.L., Casas I., Perez-Brena P. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 2004;72:484–495. doi: 10.1002/jmv.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donofrio J.C., Coonrod J.D., Davidson J.N., Betts R.F. Detection of influenza A and B in respiratory secretions with the polymerase chain reaction. PCR Methods Appl. 1992;1:263–268. doi: 10.1101/gr.1.4.263. [DOI] [PubMed] [Google Scholar]
- Elnifro E.M., Ashshi A.M., Cooper R.J., Klapper P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymuth F., Vabret A., Galateau-Salle F., Ferey J., Eugene G., Petitjean J., Gennetay E., Brouard J., Jokik M., Duhamel J.F., Guillois B. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin. Diagn. Virol. 1997;8:31–40. doi: 10.1016/s0928-0197(97)00060-3. [DOI] [PubMed] [Google Scholar]
- Grondahl B., Puppe W., Hoppe A., Kuhne I., Weigl J.A., Schmitt H.J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C.B. Respiratory syncytial virus and parainfluenza virus. N. Engl. J. Med. 2001;344:1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- Hindiyeh M., Levy V., Azar R., Varsano N., Regev L., Shalev Y., Grossman Z., Mendelson E. Evaluation of a multiplex real-time reverse transcriptase PCR assay for detection and differentiation of influenza viruses A and B during the 2001–2002 influenza season in Israel. J. Clin. Microbiol. 2005;43:589–595. doi: 10.1128/JCM.43.2.589-595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham-Sadler B.A., Morell V.W. Viral and atypical pneumonias. Prim. Care. 1996;23:837–848. doi: 10.1016/s0095-4543(05)70365-1. [DOI] [PubMed] [Google Scholar]
- Li H., McCormac M.A., Estes R.W., Sefers S.E., Dare R.K., Chappell J.D., Erdman D.D., Wright P.F., Tang Y.W. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 2007;45:2105–2109. doi: 10.1128/JCM.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios L., Jenney A., Spelman D., Kotsimbos T., Catton M., Wesselingh S. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J. Clin. Microbiol. 2001;39:2779–2783. doi: 10.1128/JCM.39.8.2779-2783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh K.H., Kim J., Nam M.H., Yoon S., Lee C.K., Lee K., Yoo Y., Kim M.J., Cho Y. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann. Clin. Lab. Sci. 2008;38:41–46. [PubMed] [Google Scholar]
- Templeton K.E., Scheltinga S.A., Beersma M.F., Kroes A.C., Claas E.C. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial viru, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 2004;42:1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen B.G., Osterhaus D.M., Fouchier R.A. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr. Infect. Dis. J. 2004;23:S25–32. doi: 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- Weinberg G.A., Erdman D.D., Edwards K.M., Hall C.B., Walker F.J., Griffin M.R., Schwartz B. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J. Infect. Dis. 2004;189:706–710. doi: 10.1086/381456. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Chiu S.S., Seto W.H., Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J. Clin. Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.J., Kuak E.Y., Shin B.M. Detection of 12 respiratory viruses with two-set multiplex reverse transcriptase-PCR assay using a dual priming oligonucleotide system. Korean J. Lab. Med. 2007;27:420–427. doi: 10.3343/kjlm.2007.27.6.420. [DOI] [PubMed] [Google Scholar]


