Highlights
► A novel homogenous method to quantify limited virus induced CPE was developed. ► As a model system we used human coronavirus NL63 in Vero118 cell cultures. ► The method confirms the antiviral activity of an Anti-ACE-2 polyclonal antiserum. ► Repeated testing results in Z’ > 0.5 indicating potential use for screening.
Keywords: Antiviral assay, Fluorescent protease substrate, High-throughput screening, Human coronavirus HCoV-NL63, CPE quantitation
Abstract
For antiviral screenings purposes, infection of cell cultures with the virus under study, should ideally result in the induction, within just a few days, of (nearly) complete CPE and allow the calculation of acceptable Z’ factors (>0.5). The human Corona virus NL63 (HCoV-NL63) causes only limited CPE on different cell lines (Schildgen et al., 2006). Following infection of Vero118 cells, virus-induced CPE was too low to allow readout based on classical colorimetric methods (such as the MTS assay), even following prolonged incubation times (>7 days). To develop an antiviral screenings-assay against HCoV-NL63, we explored whether a dead-cell protease substrate could be used instead. The substrate used is a quenched peptide (bis-AAF-R110) that releases a fluorophore upon proteolytic-cleavage by proteases; the latter released from dead cells. Following different rounds of optimization a screening protocol was developed: Vero118 cells in 96-well plate format were infected with HCoV-NL63 (MOI = 0.01; 200 μL cell culture; 2.104 cells/mL, IMDM 5% FBS medium). Cultures were subsequently incubated for 5 days at 35 °C after which 20 μL of the peptide solution was added. Fluorescence was quantitated 2 hr after incubation at 37 °C. A roughly 3-fold increase in fluorescence intensity in the infected cultures was observed as compared to the uninfected cultures with a low well-to-well variability. Z’ factors calculated from different experiments were in the range of 0.6–0.8, indicating excellent assay quality. An anti-ACE2 polyclonal antiserum (that prevents coronavirus infection in cell cultures) was used as a positive control and allowed to validate the assay for antiviral screening purposes. In conclusion, in conditions where a viability staining is inadequate to quantitate virus-induced CPE, a novel simple and convenient method that detects cell-death and that is suitable for high-throughput screening purposes can be employed.
1. Introduction
Viral infections remain a major burden for human health world-wide. While it is difficult to estimate the total number of disease causing viral infections, the mortality caused by some specific viral pathogens is well described. Today, antiviral drugs are only available for the treatment of infections with (i) HIV, (ii) some DNA viruses (herpes viruses, hepatitis B virus, poxviruses) and (iii) a small number of RNA viruses (HCV, RSV, and influenza virus). Decades of rigorous effort were needed to develop these drugs and specific therapy is still lacking for the treatment of infections by all other RNA viruses, leaving supportive care as the only option.
A major strategy to identify novel antivirals is to screen compound libraries using phenotypic assays. As in other disease areas the contribution of phenotypic screening to the discovery of first-in-class small-molecule drugs exceeds that of target-based approaches (Swinney and Anthony, 2011). The main phenotype used in antiviral assays is the development of a virus induced cythopathic effect (CPE). If a virus infection renders sufficient CPE in cell culture then standard viability assays (ex. MTS reduction) are feasible to detect viral and antiviral activity. However if the CPE is limited than more complicated and expensive tools are needed (ex ATP quantitation).
Previously a method was reported to quantify cell toxicity. This method uses a “dead-cell protease substrate”, which contains a fluorophore (R110) quenched by the covalent linkage of two Ala-Ala-Phe peptides (bis-AAF-R110). The method can be used to detect cell death since proteolytic-cleavage by proteases, released from dead cells, restores the fluorophore (Niles et al., 2007) (Fig. 1 ). This method was developed further for the quantitation of limited CPE. The read-out is homogeneous and rather inexpensive and therefore amendable to medium- or high-throughput screening campaigns. In this paper the method is described as a solution to screen for inhibitors of human coronavirus. The human Coronavirus NL63 is used as a model system since it uses the same cellular receptor as the SARS-virus (ACE2). HCoV-NL63 causes only limited CPE on different cell lines (Schildgen et al., 2006) making CPE quantitation troublesome with classical viability staining (ex MTS reduction).
Fig. 1.

A novel assay principle to quantitate virus induced CPE.
2. Materials and methods
The bis-AAF-R110 peptide was purchased from Eurogentec (Liège, Belgium). Both the human coronavirus NL63 strain and the Vero118 cell line were kindly provided by Ron A. Fouchier (Erasmus Medical Center, Rotterdam, The Netherlands). The HCoV-NL63 strain was isolated from an 8 months old boy suffering from pneumonia (Fouchier et al., 2004). Vero-118 cells are a subclone of Vero-WHO cells (Kuiken et al., 2004), and are cultured in Iscove's Modified Dulbecco's Medium (Life Technologies, Gent, Belgium – cat no 21980-032) with 10% FBS, 100 IU penicillin/mL and 100 μg streptomycin/mL. Cells are split ¼ twice weekly. Anti-ACE-2 IgG and polyclonal goat IgG control were obtained from R&D systems (Abingdon, United Kingdom) (cat no AF933 and AB-108-C), solubilized in DPBS at 0.2 mg/mL and stored at −20 °C until use.
After different rounds of optimization the following antiviral protocol was developed: Vero-118 cells in 96-well plate format are infected with HCoV-NL63 (MOI = 0.01, 200 μL cell culture, 20,000 cells/well, IMDM 5% FBS medium). Cultures are incubated subsequently for 5 days at 35 °C after which 20 μL of the peptide solution (16 μM final concentration) is added. Fluorescence is measured after 2 h incubation at 37 °C using parameters for rhodamine fluorescence read-out.
For the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) reduction assay an MTS/Phenazine methosulphate (PMS) stock solution (2 mg/mL MTS (Promega, Leiden, The Netherlands) and 46 μg/mL PMS (Sigma–Aldrich, Bornem, Belgium) in PBS at pH 6–6.5) was diluted 1/20 in MEM (Life Technologies, Gent, Belgium – cat no 21090-022). Medium was aspirated from wells and 70 μL of MTS/PMS solution was added. After 1–2 h incubation at 37 °C absorbance was measured at 498 nm.
3. Results
In a first experiment a virus titration of two different virusstocks of HCoV-NL63 was carried out and on day 6 p.i. read-out was performed by (i) microscopic scoring of the CPE and (ii) by using the CytoTox-Fluor™ Cytotoxicity Assay (Promega, Leiden, the Netherlands – cat no G9260). This CytoTox-Fluor™ assay has been developed to quantitate cell-death based on the cleavage of the bis-AAF-R110 peptide by proteases released from dead cells (Niles et al., 2007). In this experiment the assay was used according to the manufacturers’ instructions. Results are presented in Fig. 2 . For both virusstocks a very good correlation between the observed levels of CPE and the increased levels of fluorescence are observed. This indicates that the release of proteases by cultures that develop CPE and the subsequent cleavage of the bis-AAF-R110 peptide to its fluorescent substrate can be used to quantitate CPE. When a similar experiment was carried out using other methods to detect cell viability such as the MTS reduction method (Fig. 3 ) or the quantitation of LDH release (data not shown), no significant difference between infected and non-infected cell cultures could be observed.
Fig. 2.
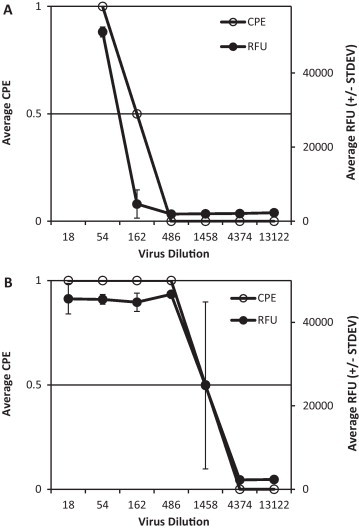
Two different virusstocks (A and B) were titrated in a 96-well plate using the standard protocol. At day 6 p.i. titrations were read by microscopic scoring and with the CytoTox-Fluor™ Cytotoxicity Assay. CPE scoring was done on a scale from 0 to 1 where 0 indicates no CPE and 1 indicates CPE. Each measurement was performed in duplicate.
Fig. 3.
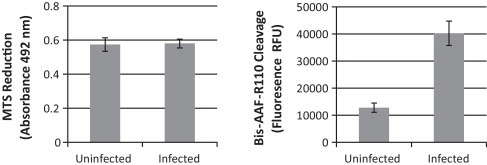
Comparing MTS Reduction with bis-AAF-R110 cleavage. An infectivity assay was carried out using the same protocol as the antiviral assay described in the methods. Five days p.i. the CPE was quantitated using either MTS reduction (Left) or bis-AAF-R110 cleavage (CytoTox-Fluor™ assay) (Right). Only the bis-AAF-R110 cleavage assay allows quantitation of the limited amount of CPE.
Next experiments were designed to optimize the assay for screening purposes. The optimal condition obtained was: 20,000 cells/well of Vero-118 cells infected with HCoV-NL63 (MOI = 0.01 CCID50/cell) in IMDM medium supplemented with 5% FBS medium. Upon infection cultures are maintained for 5 days at 35 °C after which 20 μL of the peptide solution (16 μM final concentration) is added. Fluorescence is measured after 2 hr incubation at 37 °C using parameters for rhodamine fluorescence read-out. The amount of peptide used under these conditions is 3x lower than proposed by the manufacturer of the Cytotox-Fluor™ assay. Using lower levels of peptide results in a decreased signal but the signal-to-noise ratio remains sufficient high to be used under screening conditions (see below). The main reason to lower the amount of peptide is to make a screening campaign affordable.
A batch of bis-AAF-R110 peptide was prepared by a commercial supplier. The peptide (MW 910 g/mol) was obtained as a light yellow powder, with >95% purity as determined by HPLC and was stored at −20 °C until use. Peptide stock solutions were prepared in DMSO at 100 mM and again stored at −20 °C until use.
In a subsequent experiment the performance of the custom synthesized peptide stock was compared with that of the CytoTox-Fluor™ kit. In addition the antiviral activity of a known SARS inhibitor, that is, Anti-ACE2 IgG was evaluated. This antibody inhibits SARS virus replication by binding ACE2 and preventing the virus to enter the cell (Li et al., 2003, Ivens et al., 2005). The experiment was set-up as a standard antiviral experiment in which the antibody was diluted in medium at different concentrations in duplicate after which virus and cells were added. In the same way a polyclonal goat IgG was used as a control. Untreated infected (virus controls), and treated uninfected (cell controls) cultures were included on each plate (6 wells each). On day 5 p.i. one plate was analyzed using the CytoTox-Fluor™ kit according to manufacturers’ protocol (except that the peptide was used at a 3× lower concentration). In brief the peptide solution was diluted 1/600 in assay buffer (as supplied with the kit) and 20 μL/well was added to the 96-well plate. The other plate was analyzed in parallel using the custom made peptide. To this end a 100 mM DMSO stock of the peptide was diluted 1/600 in assay medium and 20 μL/well was added (final peptide concentration 16 μM). Plates were incubated at 37 °C for 90′ and following thorough shaking the plates were read. Results are presented in Fig. 4 . The performance of the custom prepared peptide batch was comparable to that of the CytoTox-Fluor™ kit. A Z’ factor of 0.73 and 0.70 was calculated in this experiment for the custom prepared and commercial peptide respectively. A Z’ > 0.5 indicates sufficient quality for screening (Zhang et al., 1999). In addition the results show that the Anti-ACE2 IgG inhibits in vitro HCoV-NL63 replication with an EC50 ∼ 0.3 μg/mL. The polyclonal goat IgG, used as a negative control, showed an EC50 > 10 μg/mL (Fig. 4). It has been reported previously that the anti-ACE2 IgG is not toxic at concentrations up to 125 μg/mL (Li et al., 2003, Ivens et al., 2005). Also in the experiment above no effect of the anti-ACE2 IgG on cellular morphology could be observed microscopically (data not shown). The antiviral activity found is ∼3× more pronounced than the activity against the SARS coronavirus (Li et al., 2003, Ivens et al., 2005). This is not unexpected since both viruses are known to bind the ACE2 receptor differently and have evolved independently in their ability to use it for cellular entry (Wu et al., 2011).
Fig. 4.

Assaying Z’ factors in virus infected Vero118 cells using either (A) the Cytotox-Fluor™ kit or (C) a custom-prepared bis-AAF-R110 peptide. Data are means and ±SD for 6 wells/condition. Panels B and D demonstrate the effect of Anti-ACE-2 IgG (●) or polyclonal goat IgG (○) on the replication of HCoV-NL63 using the Cytotox-Fluor™ kit or the custom-prepared bis-AAF-R110 peptide respectively.
Finally two independent antiviral assays were carried out using the custom prepared peptide. In each assay four 96-well plates were used with 6-infected and 6-uninfected wells in each plate. Fluorescence was quantified on day 5 p.i. and resulted in a Z’-factor of 0.63 (Fig. 5 ).
Fig. 5.

Assay quality under HTS conditions using the custom prepared bis-AAF-R110 peptide. Two independent antiviral experiments were performed each using four 96-well plates. The individual results of the infected and uninfected control wells (total number of wells/condition is 48) as well as the average and Z’ are presented.
4. Discussion
We demonstrate that a method, previously validated for quantitation of cell toxicity (Niles et al., 2007), can also be used to quantitate virus induced CPE and antiviral activity under conditions suitable for screening. The assay principle relies on the detection of proteases released from dead cells by using the bis-AAF-R110 peptide, which upon cleavage, turns fluorescent. The main advantage of this method, in comparison to cell viability measurements such as the MTS reduction or LDH release methods, is the sensitive detection of limited virus induced CPE. This allows its use in antiviral screening campaigns with viruses that cause only limited levels of CPE. In addition the assay is highly homogenous (no washing steps) and affordable. In our hands the cost of substrate is ∼€0.03/well. Another advantage is the non-destructive nature of the method, allowing microscopic inspection and simultaneous or sequential use of other assay methods in the same well (for example to evaluate toxic or anti-proliferative effects of the compounds). Because the method is non-destructive the samples remain infectious. In general this is not a limitation as fluorescence can be measured in most plate readers with the plate lid covering the plate.
Acknowledgments
We are very grateful to Ron A. Fouchier and Bernadette van den Hoogen from Erasmus Medical Center (Rotterdam, the Netherlands) for providing us with the HCoV-NL63 virus stock and the necessary input to set up the cell culture system. We thank Tom Bellon for excellent technical assistance. This work was funded by EU FP7 project SILVER (260644).
References
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens T., Van den Eynde C., Van Acker K., Nijs E., Dams G., Bettens E., Ohagen A., Pauwels R., Hertogs K. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome-associated coronavirus. Journal of Virological Methods. 2005;129:56–63. doi: 10.1016/j.jviromet.2005.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., van den Hoogen B.G., van Riel D.A., Laman J.D., van Amerongen G., Sprong L., Fouchier R.A., Osterhaus A.D. Experimental human metapneumovirus infection of cynomolgus macaques (Macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. American Journal of Pathology. 2004;164:1893–1900. doi: 10.1016/S0002-9440(10)63750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles A.L., Moravec R.A., Eric Hesselberth P., Scurria M.A., Daily W.J., Riss T.L. A homogeneous assay to measure live and dead cells in the same sample by detecting different protease markers. Analytical Biochemistry. 2007;366:197–206. doi: 10.1016/j.ab.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Schildgen O., Jebbink M.F., de Vries M., Pyrc K., Dijkman R., Simon A., Müller A., Kupfer B., van der Hoek L. Identification of cell lines permissive for human coronavirus NL63. Journal of Virological Methods. 2006;138:207–210. doi: 10.1016/j.jviromet.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D.C., Anthony J. How were new medicines discovered? Nature Reviews Drug Discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Wu K., Chen L., Peng G., Zhou W., Pennell C.A., Mansky L.M., Geraghty R.J., Li F. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. Journal of Virology. 2011;85:5331–5337. doi: 10.1128/JVI.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of Biomolecular Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]


