Abstract
A TaqMan probe-based real-time RT-PCR assay was developed for simultaneous detection of RNA of transmissible gastroenteritis virus (TGEV) in pig fecal samples and RNA of enhanced green fluorescent protein (EGFP) added exogenously as an internal amplification control. The TGEV primers and probe were designed to be specific to a portion of the S gene sequence conserved in all TGEV isolates, but absent in the closely related porcine respiratory coronaviruses. The optimized TaqMan assay detected a minimum of 2.8 copies of in vitro transcribed RNA of the target S gene and RNA extracted from 1 TCID50/ml of TGEV. Using 113 clinical samples received at our diagnostic laboratory over a 4-year period, the performance of the assay was tested and compared with that of a previously described nested RT-PCR assay. All the fecal samples which tested positive for TGEV by the nested RT-PCR assay also tested positive by the TaqMan assay. However, approximately 9% of the samples that tested negative by the nested RT-PCR assay tested positive by the TaqMan assay. These results indicate that the developed TaqMan assay is a highly sensitive diagnostic test for rapid detection of TGEV in pig fecal samples.
Keywords: Transmissible gastroenteritis virus, TaqMan assay, Real-time RT-PCR, Pig fecal samples, Internal amplification control
1. Introduction
Transmissible gastroenteritis (TGE) is an acute enteric viral disease of pigs of all ages (Saif and Wesley, 1999). The disease is caused by TGE virus (TGEV), an enveloped virus with a large, single positive-strand RNA genome that belongs to Coronaviridae family. TGEV replicates in intestinal enterocytes and shed in the feces of infected pigs. The infection is transmitted by fecal–oral route. Clinical signs of the disease include watery diarrhea, vomiting, dehydration and high mortality in unweaned piglets (Saif and Wesley, 1999). TGEV infections can cause severe economic losses to swine industry. Because of the highly contagious nature of TGE, availability of rapid diagnostic methods that are highly specific and sensitive for detection of TGEV in fecal samples is very useful for timely implementation of disease management practices.
Natural mutants of TGEV with deletions in spike (S) gene, known as porcine respiratory coronavirus (PRCV), show tropism towards respiratory tissue and cause mild or subclinical respiratory infections (Rasschaert et al., 1990, Kim et al., 2000b). Some pigs infected with PRCV can shed the virus in their feces (Saif and Wesley, 1999, Kim et al., 2000a, Costantini et al., 2004). Therefore, any diagnostic assay for TGE should be able to differentiate it from PRCV. The conventional diagnostic methods used for TGE include, cell culture isolation of the virus, identification of coronavirus in feces by electron microscopy, fluorescent antibody test on cryosections of intestines, antigen-capture enzyme-linked immunosorbent assay (ELISA) for detection of TGEV in feces, and detection of serum antibodies to TGEV by virus neutralization assay or ELISA (Dulac et al., 1977, van Nieuwstadt et al., 1988a, van Nieuwstadt et al., 1988b, Sirinarumitr et al., 1997, Saif and Wesley, 1999, Carman et al., 2002). These methods are not suitable for the rapid detection of clinical TGE because of either their low specificity and sensitivity or the prolonged time required to perform the assay. A previously developed nested reverse transcriptase-polymerase chain reaction (nRT-PCR) that can differentially detect TGEV and PRCV has been very useful for the diagnostic testing of TGE cases (Kim et al., 2000a). However, this assay is also time-consuming because of the need to perform two PCR amplifications and detect the amplified products by agarose gel electrophoresis. In addition, nested PCR assays are generally more prone to give false positive results because of cross-contamination of the amplified DNA products, especially when processing large number of samples (Aslanzadeh et al., 1996, Belák and Thorén, 2001, Khlif et al., 2009).
Probe-based real-time PCR assays are better suited for rapid, accurate and sensitive detection of pathogens in clinical samples (Espy et al., 2006, Belák, 2007). This technology also makes it easier to carryout high-throughput testing of samples. In this study, a TaqMan probe-based real-time RT-PCR was developed for specific detection of TGEV in pig fecal samples. The assay included an internal amplification control to monitor the presence of PCR inhibitors in RNA extracted from pig feces. Performance of the assay was tested with field clinical samples and compared with that of nRT-PCR.
2. Materials and methods
2.1. Primers and probe design
In comparison to TGEV, all PRCV strains have a deletion of 621–681 nucleotides within the amino terminus encoding part of the S gene (Kim et al., 2000b). Multiple alignment of the TGEV and PRCV S gene sequences showed that a 561 nucleotide-long region, corresponding to sequence between nucleotides 105 and 666 of the TGEV S gene open reading frame, is absent from all PRCV strains (Kim et al., 2000b). Nucleotide sequences of S gene of 15 TGEV strains available in the databases at GenBank (China strain TH-98, AF494337; China strain TS, AY335549; Japan strain TO14, AF302263; Korea strain KT2, AF481360; Russia strain PPS, Y15449; Spain strain NEB72-RT, M94099; Taiwan strain TFI, Z35758; The Netherlands isolate, M21950; UK isolate TGEV FS772/70, X53128; UK strain 96-1933, AF104420; USA isolate BW021898B, AF179882; USA isolate CHV-TGE, U26215; USA strain Miller, S51223; USA strain PUR46-MAD, AJ271965; USA strain Purdue-115, X05695) were retrieved and aligned using ClustalW multiple alignment algorithm (MegAlign program, LaserGene sequence analysis software, DNASTAR, Inc., Madison, WI) to identify the conserved sequences within the region that is absent in S gene of PRCV. A set of primers (forward primer: 5′-TCTGCTGAAGGTGCTATTATATGC-3′; reverse primer: 5′-CCACAATTTGCCTCTGAATTAGAAG-3′) and a TaqMan probe (5′-C/TAAGGGCTCACCACCTACTACCACCA-3′) targeting a conserved 145 bp region (corresponding to the region between nucleotides 370 and 515 of the TGEV S gene open reading frame) were designed using Beacon Designer 3 software (PREMIER Biosoft International, Palo Alto, CA). In silico analysis was performed to confirm the TGEV-specificity of the primers and probe by BLAST search of the nucleotide sequences present in databases at Genbank. The TGEV TaqMan probe contained the fluorescent dye 6-carboxyfluorescein (FAM) at the 5′-end and a non-fluorescent quencher (Black Hole Quencher 1, BHQ1) at the 3′-end. Previously described primers and probe were used for detection of synthetic RNA of enhanced green fluorescent protein (EGFP) gene (forward primer: 5′-GACCACTACCAGCAGAACAC-3′; reverse primer: 5′-GAACTCCAGCAGGACCATG-3′; probe: 5′-AGCACCCAGTCCGCCCTGAGCA-3′) (Hoffmann et al., 2006). The EGFP TaqMan probe contained hexachlorofluorescein (HEX) at the 5′-end and BHQ1 at the 3′-end. The primers and probes were custom synthesized at a commercial source (Operon Biotechnologies, Inc., Huntsville, AL).
2.2. Viruses and fecal samples
Cell culture grown TGEV (Purdue reference strain) with 50% tissue culture infectious dose (TCID50)/ml of 2.4 × 107 was obtained from National Veterinary Services Laboratory, Ames, IA. Field isolates of TGEV, PRCV, bovine coronavirus (BCV) and feline infectious peritonitis virus (FIPV) were obtained from our virology culture collection. Fecal samples collected from 24 healthy 10-week-old pigs of an unrelated experimental study were used as true negative samples in the development of the assay. Some of the fecal samples from pigs with diarrhea submitted over a 4-year period (2003–2006) to our Animal Disease Diagnostic Laboratory for detection of diarrheogenic pathogens (viral and bacterial) were used to evaluate the performance of the TaqMan assay.
2.3. Synthetic RNA
A 874 bp DNA product encompassing the TGEV-specific S gene sequence that was the target for the TaqMan assay was amplified by RT-PCR using previously described F2 and R2 primers (Kim et al., 2000a), cloned in pCR2.1 vector (Invitrogen, Carlsbad, CA), and sequenced to confirm the nucleotide sequences. The insert was excised from pCR2.1 with XbaI and BamHI digestion and cloned into the same sites of pCDNA3.1 (Invitrogen). For an internal control, EGFP gene from pEGFP-N1 (Clontech, Mountain View, CA) was excised with XbaI and BamHI digestion and cloned into the same sites of pCDNA3.1. The pCDNA3.1 plasmids with the inserts were linearized with BamHI and in vitro transcribed from the Sp6 promoter using RNAMaxx High Yield Transcription kit (Stratagene, Cedar Creek, TX). The transcribed RNA was extracted using TRIzol reagent (Invitrogen) and digested with RNase-free DNase to remove any contaminating DNA, and the RNA was purified again using RNeasy column as per the RNeasy kit protocol (Qiagen, Valencia, CA). Quant-iT RiboGreen RNA assay kit (Invitrogen) was used to quantify the purified RNA transcripts. RNA copy numbers were calculated based on the exact length and nucleotide composition of the RNA molecules. Tenfold serial dilution of the transcripts were prepared in TE buffer (10 mM Tris and 1 mM EDTA, pH 8.0) with 0.5 U/μl of RNasin (Promega, Madison, WI) and stored at −20 °C.
2.4. RNA extraction from feces
After initial evaluation of different RNA extraction methods, a hybrid protocol that uses TRIzol LS reagent (Invitrogen) and RNeasy spin column (Qiagen) was developed to reduce the time required for RNA extraction with no reduction in the RNA yield. A 250 μl of 20% (w/v) fecal suspension in DNase- and RNase-free water was added to a 1.5 ml microcentrifuge tube with 750 μl of TRIzol LS reagent, mixed thoroughly by vortexing for 15 s, and incubated at room temperature for 5 min. To this, 200 μl chloroform was added, vortexed briefly and incubated for 3 min at room temperature. The tube was centrifuged at 12,000 × g for 10 min and 600 μl of the aqueous phase was transferred to a clean tube containing 600 μl of 70% ethanol. After mixing by inverting, the entire contents, 600 μl at a time, were passed through a RNeasy spin column following the RNeasy kit protocol (Qiagen). The spin column was washed, once with 700 μl of RW1 buffer and twice with 500 μl of RPE buffer, and the RNA was eluted using 30 μl of DNase- and RNase-free water. The extracted RNA samples were stored at −20 °C until use.
2.5. TaqMan assay
Reagents of the One-Step RT-PCR kits from either Qiagen or Invitrogen were used for the assay. The assays were performed in a 25 μl reaction volume using SmartCycler II (Cepheid, Sunnyvale, CA). The assays were performed in a single-target format for detection of TGEV only and in a double-target (duplex) format for detection of TGEV and simultaneous amplification of the EGFP RNA, the internal amplification control. The assays were optimized for primer, probe and MgCl2 concentrations as well as the thermal cycling parameters. The final optimized duplex assay contained 3 mM MgCl2, 0.2 mM dNTP mix, 13 U of RNasin (Promega), 0.4 μM of TGEV F primer, 1.2 μM of TGEV R primer, 0.3 μM of TGEV probe, 0.4 μM each of EGFP primers, 0.3 μM of EGFP probe and 10 fg of EGFP RNA template. In the single-target format assay for detection of TGEV, the EGFP-specific primers, probe and the synthetic RNA template were omitted. In both assay formats, 5 μl of RNA extracted from samples or serially diluted TGEV RNA or the synthetic transcripts was used as template; DNase- and RNase-free water was used in the no template controls. The amplification parameters were: reverse transcription at 50 °C for 30 min followed by denaturation at 95 °C for 15 min. Next, 50 cycles of amplification were performed with each cycle containing denaturation at 94 °C for 15 s, annealing at 56 °C for 30 s and extension at 72 °C for 15 s. The data were acquired during the annealing step and analyzed by using Cepheid Smart Cycler software (version 2.0b). The threshold cycle (Ct) values were determined by primary curve analysis program with the threshold limit set at 30 fluorescent units.
2.6. Nested RT-PCR
A gel-based, nRT-PCR assay, described previously by Kim et al. (2000a), was performed with slight modifications for the detection of TGEV. One-Step RT-PCR reagents from Invitrogen were used for the first-step RT-PCR amplification. Five microliters of the RNA sample was used as template. The tubes were incubated at 50 °C for 30 min, followed by 95 °C for 5 min and 35 cycles of 94 °C for 1 min, 60 °C for 1.5 min and 72 °C for 2.5 min. The second-step, nested PCR was performed with NovaTaq polymerase (Novagen, Madison, WI) using 2 μl of the RT-PCR product as the template and 35 cycles with same parameters as above. Both rounds of the amplification were performed in 25 μl reaction volumes. Twelve microliters of the products of the nested PCR were separated by electrophoresis on 0.8% agarose gel and the DNA fragments were detected by staining the gel with ethidium bromide.
2.7. PCR quality control measures
Except for the generation of synthetic RNA transcripts, all other procedures were performed in our molecular diagnostics laboratory that is setup to carryout PCR-based diagnostic assays following the necessary procedural precautions to prevent nucleic acid and nuclease contamination of the samples and reagents (Dragon et al., 1993).
3. Results
3.1. Evaluation of the TaqMan assay in single-target format
The TaqMan assay in single-target format was performed with different copies of the TGEV S gene synthetic RNA transcripts as the template. As shown in Fig. 1 , the TaqMan assay detected a range of 2.8–2.8 × 108 target copies per reaction in a linear manner.
Fig. 1.
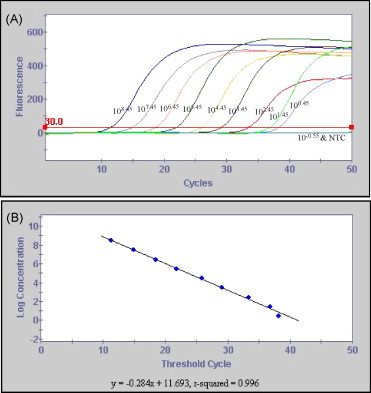
Analytical sensitivity of the TGEV TaqMan assay with the 10-fold serially diluted synthetic RNA as template. The amplification (A) and the associated standard curve (B) graphs are shown. In panel A, the numbers on each line represent the target RNA copies per reaction. The horizontal line above the X-axis is the threshold line set at 30 fluorescent units. NTC: no template control.
To compare the performance of the TaqMan assay with that of the nRT-PCR assay, aliquots of a pooled fecal sample negative for TGEV were spiked with different concentrations of cell culture grown TGEV and RNA extracted from the spiked samples was used as template in the PCR assays. The TaqMan assay detected a range of 107–1 TCID50/ml of TGEV in the fecal suspensions in a linear manner (Fig. 2 ). In the nRT-PCR assay, specific amplification was detected when the samples contained TGEV in the range of 107–10 TCID50/ml; no specific amplification was detected in the sample with 1 TCID50/ml concentration of the virus (Fig. 3 ). To assess the inter-assay variability, the TaqMan assay was performed on 3 separate days using the same spiked fecal samples. The inter-assay variability was found to be very low with coefficient of variation (CV) ranging from 0.2% to 1.8%.
Fig. 2.
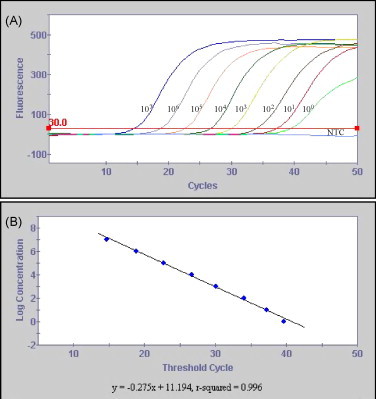
Analytical sensitivity of the TGEV TaqMan assay with fecal samples spiked with different concentrations of the virus. The amplification (A) and the associated standard curve (B) graphs are shown. In panel A, the numbers on each line represent the virus concentration, in TCID50/ml, in the fecal sample. The horizontal line above the X-axis is the threshold line set at 30 fluorescent units. NTC: no template control.
Fig. 3.
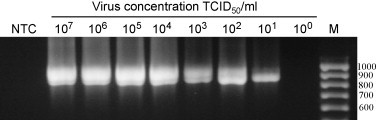
Detection of TGEV by the nested RT-PCR assay in fecal samples spiked with different concentrations of the virus. NTC: no template control. M: molecular size maker. Numbers at the right indicate the molecular size of the corresponding DNA fragment in base-pairs.
When RNA of PRCV, FIPV, and BCV was used as template, no specific amplification was detected in the TaqMan assay. In the nRT-PCR, a DNA fragment of similar length as that for TGEV (874 bp) was amplified from FIPV, but not BCV, RNA; as expected a 201 bp fragment was amplified from PRCV RNA (data not shown). Both the assays readily detected the four TGEV field isolates tested.
3.2. Evaluation of the TaqMan assay with the internal amplification control
The TGEV-specific TaqMan was modified into a duplex assay by adding 10 fg of EGFP transcript RNA and the optimal concentrations of corresponding primers and probe (see Section 2.5) to the reaction mixture. The 10 fg amount of the EGFP transcript RNA was selected so that the amplification of EGFP target sequences did not affect the efficiency of amplification of the TGEV target sequences. The performances of the duplex and the single-target assays were compared using the spiked fecal samples with different concentrations of TGEV and a set of 7 fecal samples from pigs previously confirmed to be TGEV positive. As shown in Table 1 , for each sample, the Ct values for TGEV were very similar between the two assay formats. In the duplex assay, no EGFP-specific amplification was detected when the Ct values for the TGEV were low (Table 1).
Table 1.
Performance of the TGEV TaqMan assay in duplex format with internal amplification control.
| Sample | Single target assay Ct for TGEV | Duplex assay |
|
|---|---|---|---|
| Ct for TGEV | Ct for EGFP | ||
| Spiked fecal samples | |||
| 107 TCID50/ml | 14.29 | 14.66 | 0.00a |
| 106 TCID50/ml | 18.75 | 18.82 | 27.75 |
| 105 TCID50/ml | 22.35 | 22.74 | 26.66 |
| 104 TCID50/ml | 26.41 | 26.62 | 26.51 |
| 103 TCID50/ml | 29.68 | 30.05 | 27.21 |
| 102 TCID50/ml | 34.00 | 34.07 | 27.39 |
| 101 TCID50/ml | 36.65 | 37.21 | 27.42 |
| 100 TCID50/ml | 38.84 | 39.63 | 26.69 |
| No template control | 0.00 | 0.00 | 25.63 |
| Field fecal samples | |||
| 1 | 24.37 | 24.95 | 25.58 |
| 2 | 32.37 | 32.86 | 26.65 |
| 3 | 15.59 | 15.80 | 0.00 |
| 4 | 14.75 | 14.88 | 0.00 |
| 5 | 30.96 | 31.12 | 29.30 |
| 6 | 12.34 | 12.19 | 0.00 |
| 7 | 26.10 | 26.37 | 28.60 |
| No template control | 0.00 | 0.00 | 25.56 |
No Ct value—no target amplification detected.
3.3. Evaluation of the TaqMan assay with field fecal samples
The TaqMan and the nRT-PCR assays were used to test 113 fecal samples from different field diarrhea cases. All 61 samples that tested negative by the nRT-PCR also tested negative for TGEV by the TaqMan assay (Table 2 ). However, 6 of the samples that tested negative by the nRT-PCR assay were positive for TGEV in the TaqMan assay (Table 2). The Ct values of these 6 samples were 25.58, 23.68, 30.14, 33.94, 19.21 and 18.82. The TGEV Ct values for all 52 positive samples ranged from 9.2 to 33.94, and 75% of the samples had Ct values below 26 (data not shown). The EGFP Ct values for the TGEV positive samples varied between 0 and 35.
Table 2.
Comparison of the TaqMan and the gel-based nested RT-PCR assays carried out on pig fecal samples from field cases. Numbers indicate the samples positive (+) or negative (−) for TGEV.
| TaqMan assay |
||
|---|---|---|
| + | − | |
| Nested RT-PCR | ||
| + | 46 | 0 |
| − | 6 | 61 |
The EGFP Ct values were within the expected values (26.63 ± 0.8) for most of the TGEV negative samples, but for 7 samples the EGFP Ct values were above 29. When the RNA of these samples were diluted 1 in 5 with TE buffer and then used as template in the assay, the EGFP Ct values decreased to the expected values, suggesting that the presence of potential PCR inhibitors (data not shown). These samples, however, tested negative for TGEV even when the diluted RNA was used as the template.
4. Discussion
PCR-based assays are becoming the diagnostic method of choice for rapid and sensitive detection of pathogens in fecal samples. Previous studies show that RT-PCR assays are superior to other diagnostic methods for detection of TGEV in fecal samples (Paton et al., 1997, Kim et al., 2000a). The TaqMan RT-PCR assay reported in this study showed a broad dynamic range and high sensitivity (Fig. 1, Fig. 2). The lower limit of detection for the assay was determined to be 2.8 target RNA copies per reaction or RNA extracted from fecal samples with 1 TCID50/ml of the virus. The lower limit of detection for the nRT-PCR assay was 10 TCID50/ml of the virus, indicating that this assay was less sensitive than the TaqMan assay.
The lack of any amplification with RNA of PRCV, FIPV and BCV suggests that the TaqMan assay is highly specific to TGEV. We have not examined the assay specificity with the RNA of porcine epidemic diarrhea virus (PEDV), another porcine coronavirus that is of concern in parts of Europe and Asia, but not in North or South America (Pensaert, 1999). However, our in silico analysis showed that the primers and probe used for TGEV TaqMan assay did not share sequence identity with the PEDV genes, strongly suggesting that the TaqMan assay cannot detect the presence of PEDV in fecal samples.
Nucleic acids extracted from fecal samples may contain unknown additional molecules, usually referred to as PCR inhibitors, which can interfere with the PCR amplification (Dye et al., 2008, Scipioni et al., 2008, Oikarinen et al., 2009). Presence of PCR inhibitors can lead to false negative results. The TGEV-specific real-time PCR assays reported previously do not detect the presence of PCR inhibitors (Chen et al., 2004, Kim et al., 2007). An effective way to monitor for the presence of PCR inhibitors is to include an internal amplification control in the PCR assay. We employed the EGFP RNA-based universal internal amplification control described previously for this purpose (Hoffmann et al., 2006). The EGFP Ct values in most of the TGEV negative samples were within the expected range, suggesting that RNA extracted from these fecal samples did not contain PCR inhibitors. The EGFP internal amplification control detected the presence of potential PCR inhibitors in RNA of 7 of the TGEV negative samples. In case of TGEV positive samples, the EGFP Ct values were highly variable (in the range of 0–35). In samples where the TGEV Ct values were low (<15, indicating high viral concentration in the sample), no amplification of the EGFP internal control occurred (Ct value 0). This could be mainly because of the competitive advantage of the high concentration of the TGEV target sequences for reagent utilization during PCR amplification. On the other hand, the high EGFP Ct values (>29) in the TGEV positive samples suggest the presence of PCR inhibitors; though the relevance of such finding has no effect on the diagnostic interpretation of the sample (i.e. positive for TGEV). In none of the tested samples there was complete inhibition of both the EGFP and TGEV targets, suggesting that the PCR inhibitors present in the fecal RNA extractions were not at high enough levels to cause 100% RT-PCR inhibition. Overall, these results suggest that the developed TaqMan assay can effectively monitor the presence of PCR inhibitors, and along with the RNA extraction method used in this study, the assay provides a reliable means to detect TGEV in the fecal samples.
The TGEV Ct values of 75% of the positive field samples were below 26. Calculations using the standard curves of synthetic RNA transcripts and spiked fecal samples (Fig. 1, Fig. 2) suggest that in most of the clinical cases tested the excretion of TGEV was higher than 105 TCID50 or 9.6 × 106 target copies per gram of feces; the highest detected viral concentration was 1010 TCID50 per gram of feces. The time of sample collection during the course TGEV infection and the virulence of the infecting viral strain are two obvious factors that can affect the viral load of a fecal sample (van Nieuwstadt et al., 1988b, Kim et al., 2000b, Kim et al., 2007). In the present study, in most of the cases, each tested sample was a pooled fecal sample from pigs of a herd with diarrhea. Therefore, the calculated viral loads do not represent the level of shedding by an individual pig.
Six samples were negative for TGEV by the nRT-PCR but were positive by the TaqMan assay. When 1 in 10 dilution (with TE buffer) of the RNA extracted from these samples was used as template in the nRT-PCR, 2 samples with higher viral load (Ct values of 18.82 and 19.21) showed a low level amplification of the specific DNA product; no detectable amplification was observed with the other 4 samples. In addition, even with some positive field samples, only a low level of amplification was detected in the nRT-PCR even though the TGEV Ct values of the same samples indicated very high viral loads. A probable reason for this discrepancy could be that the nRT-PCR was more prone to PCR inhibition than the TaqMan assay, similar to the observations reported for norovirus detection in fecal and environmental samples (Trujillo et al., 2006, Scipioni et al., 2008).
Effective management of TGE outbreaks in pig herds requires quick and accurate detection of the virus in the feces. Compared to the nested RT-PCR assay, the main advantages of the TaqMan assay are the less labor-intensive nature and the rapid turn-around time. Employing the TaqMan assay developed in this study should permit diagnostic laboratories to provide the results of TGE suspect cases within a few hours of receiving the samples. Moreover, the TaqMan assay can also be used for research purposes where rapid quantification of TGEV in fecal or other samples is needed.
Acknowledgement
We thank Barb Million for her excellent technical assistance.
References
- Aslanzadeh J., Padilla B.B., Shanley J.D. Evaluation of PCR and nested PCR for laboratory diagnosis of hepatitis C virus infection. Mol. Cell Probes. 1996;10:173–178. doi: 10.1006/mcpr.1996.0024. [DOI] [PubMed] [Google Scholar]
- Belák S. Molecular diagnosis of viral diseases, present trends and future aspects: a view from the OIE collaborating centre for the application of polymerase chain reaction methods for diagnosis of viral diseases in veterinary medicine. Vaccine. 2007;25:5444–5452. doi: 10.1016/j.vaccine.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belák S., Thorén P. Molecular diagnosis of animal diseases: some experiences over the past decade. Expert Rev. Mol. Diagn. 2001;1:434–443. doi: 10.1586/14737159.1.4.434. [DOI] [PubMed] [Google Scholar]
- Carman S., Josephson G., McEwen B., Maxie G., Antochi M., Eernisse K., Nayar G., Halbur P., Erickson G., Nilsson E. Field validation of a commercial blocking ELISA to differentiate antibody to transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus and to identify TGEV-infected swine herds. J. Vet. Diagn. Invest. 2002;14:97–105. doi: 10.1177/104063870201400202. [DOI] [PubMed] [Google Scholar]
- Chen R., Huang W., Lin Z., Zhou Z., Yu H., Zhu D. Development of a novel real-time RT-PCR assay with LUX primer for the detection of swine transmissible gastroenteritis virus. J. Virol. Methods. 2004;122:57–61. doi: 10.1016/j.jviromet.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini V., Lewis P., Alsop J., Templeton C., Saif L.J. Respiratory and fecal shedding of porcine respiratory coronavirus (PRCV) in sentinel weaned pigs and sequence of the partial S-gene of the PRCV isolates. Arch. Virol. 2004;149:957–974. doi: 10.1007/s00705-003-0245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon E.A., Spadoro J.P., Madej R. Quality control of polymerase chain reaction. In: Persing D.H., Smith T.F., Tenover F.C., White T.J., editors. Diagnostic Molecular Microbiology—Principles and Applications. American Society for Microbiology; Washington, DC: 1993. pp. 160–168. [Google Scholar]
- Dulac G.C., Ruckerbauer G.M., Boulanger P. Transmissible gastroenteritis: demonstration of the virus from field specimens by means of cell culture and pig inoculation. Can. J. Comp. Med. 1977;41:357–363. [PMC free article] [PubMed] [Google Scholar]
- Dye C., Helps C.R., Siddell S.G. Evaluation of real-time RT-PCR for the quantification of FCoV shedding in the faeces of domestic cats. J. Feline Med. Surg. 2008;10:167–174. doi: 10.1016/j.jfms.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A., Yao J.D., Wengenack N.L., Rosenblatt J.E., Cockerill F.R., 3rd, Smith T.F. Real-time PCR in clinical microbiology: application for routine laboratory testing. Clin. Microbiol. Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Depner K., Schirrmeier H., Beer M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods. 2006;136:200–209. doi: 10.1016/j.jviromet.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Khlif M., Mary C., Sellami H., Sellami A., Dumon H., Ayadi A., Ranque S. Evaluation of nested and real-time PCR assays in the diagnosis of candidaemia. Clin. Microbiol. Infect. 2009;15:656–661. doi: 10.1111/j.1469-0691.2009.02762.x. [DOI] [PubMed] [Google Scholar]
- Kim L., Chang K.-O., Sestak K., Parwani A., Saif L.J. Development of a reverse transcription-nested polymerase chain reaction assay for differential diagnosis of transmissible gastroenteritis virus and porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J. Vet. Diagn. Invest. 2000;12:385–388. doi: 10.1177/104063870001200418. [DOI] [PubMed] [Google Scholar]
- Kim L., Hayes J., Lewis P., Parwani A.V., Chang K.O., Saif L.J. Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch. Virol. 2000;145:1133–1147. doi: 10.1007/s007050070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kim I.J., Pyo H.M., Tark D.S., Song J.Y., Hyun B.H. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J. Virol. Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikarinen S., Tauriainen S., Viskari H., Simell O., Knip M., Virtanen S., Hyöty H. PCR inhibition in stool samples in relation to age of infants. J. Clin. Virol. 2009;44:211–214. doi: 10.1016/j.jcv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Paton D., Ibata G., Sands J., McGoldrick A. Detection of transmissible gastroenteritis virus by RT-PCR and differentiation from porcine respiratory coronavirus. J. Virol. Methods. 1997;66:303–309. doi: 10.1016/S0166-0934(97)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B. Porcine epidemic diarrhea. In: Straw B.E., D’Allaire S., Mengeling W.L., Taylor D.I., editors. Diseases of Swine. 8th ed. Iowa State University Press; Ames: 1999. pp. 179–185. [Google Scholar]
- Rasschaert D., Duarte M., Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71:2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- Saif L.J., Wesley R.D. Transmissible gastroenteritis and porcine respiratory coronavirus. In: Straw B.E., D’Allaire S., Mengeling W.L., Taylor D.I., editors. Diseases of Swine. 8th ed. Iowa State University Press; Ames: 1999. pp. 295–325. [Google Scholar]
- Scipioni A., Bourgot I., Mauroy A., Ziant D., Saegerman C., Daube G., Thiry E. Detection and quantification of human and bovine noroviruses by a TaqMan RT-PCR assay with a control for inhibition. Mol. Cell Probes. 2008;22:215–222. doi: 10.1016/j.mcp.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Sirinarumitr T., Paul P.S., Halbur P.G., Kluge J.P. An overview of immunological and genetic methods for detecting swine coronaviruses, transmissible gastroenteritis virus, and porcine respiratory coronavirus in tissues. Adv. Exp. Med. Biol. 1997;412:37–46. doi: 10.1007/978-1-4899-1828-4_4. [DOI] [PubMed] [Google Scholar]
- Trujillo A.A., McCaustland K.A., Zheng D.P., Hadley L.A., Vaughn G., Adams S.M., Ando T., Glass R.I., Monroe S.S. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 2006;44:1405–1412. doi: 10.1128/JCM.44.4.1405-1412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nieuwstadt A.P., Cornelissen J.B., Vreeswijk J. Solid phase immune electron microscopy for diagnosis of transmissible gastroenteritis in pigs. Res. Vet. Sci. 1988;44:286–294. doi: 10.1016/S0034-5288(18)30859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nieuwstadt A.P., Cornelissen J.B., Zetstra T. Comparison of two methods for detection of transmissible gastroenteritis virus in feces of pigs with experimentally induced infection. Am. J. Vet. Res. 1988;49:1836–1843. [PubMed] [Google Scholar]


