Abstract
RIG-I and MDA5 receptors are key sensors of pathogen-associated molecular pattern (PAMP)-containing viral RNA and transduce downstream signals to activate an antiviral and immunomodulatory response. Fifteen years of research have put them at the center of an ongoing hunt for novel pharmacological pan-antivirals, vaccine adjuvants, and antitumor strategies. Current knowledge testifies to the redundant, but also distinct, functions mediated by RIG-I and MDA5, opening opportunities for the use of specific and potent nucleic acid agonists. We critically discuss the evidence and remaining knowledge gaps that have an impact on the choice and design of optimal RNA ligands to achieve an appropriate immunostimulatory response, with limited adverse effects, for prophylactic and therapeutic interventions against viruses and cancer in humans.
Keywords: RIG-I, MDA5, antiviral, vaccine, cancer, immunotherapy
The Immunomodulatory Functions of RIG-I-Like Receptors (RLRs): Therapeutic Opportunities and Challenges
Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) (see Glossary) are pattern-recognition receptors (PRRs) that play a key role in the cytosolic sensing of RNA viruses through recognition of viral RNA or viral replication intermediates to initiate the innate immune response [1]. The RLR family is composed of three ubiquitously expressed members, RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). All three RLRs possess a DExD/H-box RNA helicase domain and a C-terminal domain (CTD), while only RIG-I and MDA5 harbor N-terminal caspase-recruitment domains (CARDs) that are necessary for signaling (Figure 1 ) [2]. Upon ligand recognition and binding, RIG-I and MDA5 undergo a conformational change that renders the CARD domains available for homotypic interaction with the membrane-associated common adaptor mitochondrial antiviral-signaling (MAVS) protein anchored in the membranes of mitochondria, peroxisomes, and mitochondrial-associated endoplasmic reticulum membranes (MAMs) [3]. MAVS activation results in the coordinated activation of the transcription factors nuclear factor κ/light-chain enhancer of activated B cells (NF-κB) and interferon regulatory factor (IRF)3 through the IκB kinase (IKK) complex (α/β/γ) and IKK-related tank-binding kinase (TBK) 1/IKKε kinases, respectively. These events culminate in the transcriptional regulation of type I and III interferons (IFNs) and proinflammatory cytokines, thereby establishing an antiviral and immunoregulatory state 1, 3 (Figure 2 , Key Figure).
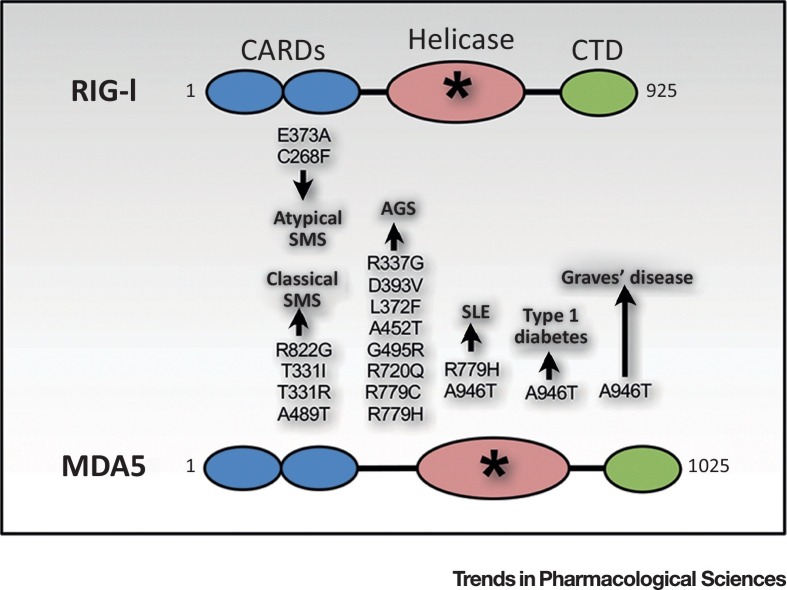
Figure 1.
Structure and Associated Polymorphisms in RIG-I and MDA5. Both RIG-I and MDA5 possess N-terminal caspase-recruitment domains (CARDs, in blue), a helicase domain (in pink), and a C-terminal domain (CTD, in green). Single-nucleotide polymorphisms (SNPs) in DDX58 and IFIH, genes encoding RIG-I and MDA5 respectively, result in gain-of-function mutations in the helicase domain (depicted by the asterisk) that are associated with various autoimmune and inflammatory diseases such as Singleton–Merten syndrome (SMS), Aicardi–Goutières syndrome (AGS), systemic lupus erythematosus (SLE), type 1 diabetes, and Graves’ disease (Box 1).
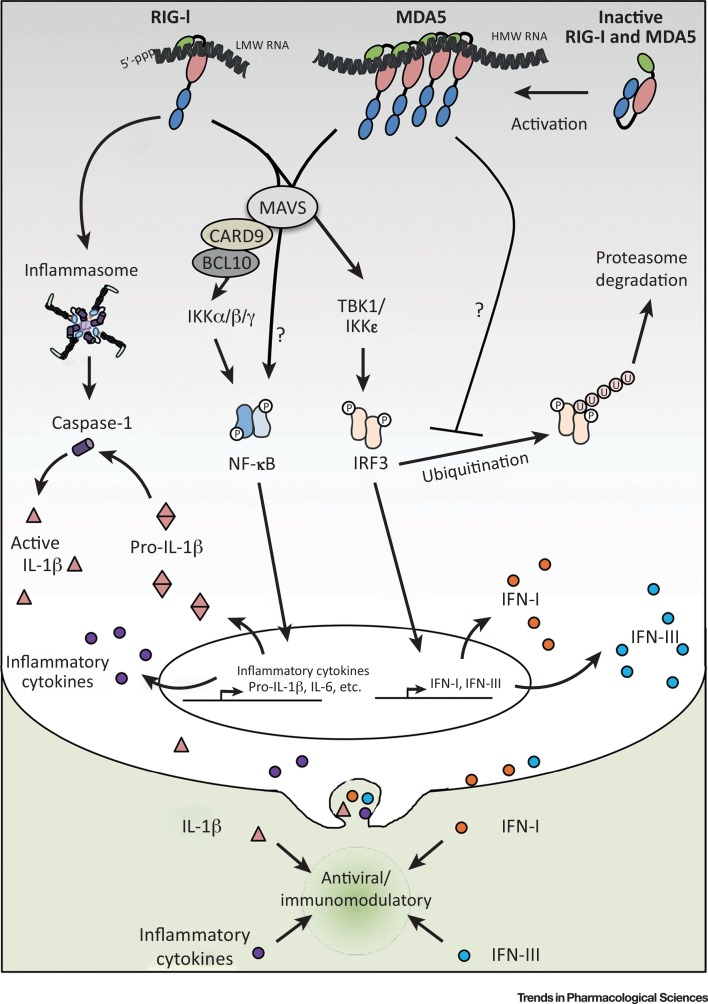
Figure 2.
Key Figure: RIG-I- and MDA5-Dependent Immunomodulatory Pathways
The RIG-I and MDA5 sensors are respectively activated by 5′ppp low molecular weight (LMW) and high molecular weight (HMW) viral RNA or viral intermediates containing double-stranded (ds) RNA structures. Ligand binding results in CARD-mediated interactions with the mitochondrial antiviral-signaling (MAVS) adaptor. MAVS activation results in the coordinated activation of the NF-κB and IRF3 transcription factors. NF-κB activation through the IKK α/β/γ kinase complex is in part mediated by a CARD9–BCL10-dependent pathway. IRF3 activation is dependent on the IKK-related TBK1/IKKε kinases. These in turn regulate the transcription of type I (orange spheres) and type III interferons (IFNs) (blue spheres), as well as of proinflammatory cytokines (purple spheres) including pro-IL-1β (double inverted pink triangle). RIG-I, but not MDA5, also mediates the activation of the inflammasome and subsequent caspase-1 activity (purple cylinder) that leads to the production of mature IL-1β (pink triangle). MDA5 is also involved in the regulation of the non-canonical NF-κB pathway and in the sustained activity of IRF3 by interfering with its proteasome-mediated degradation. Mechanisms currently uncharacterized are depicted by question marks. Abbreviations: P, phosphorylation; U, ubiquitination.
Initial attempts to use synthetic RNA as immunostimulants to improve human health were performed using polyinosinic:polycytidylic acid [poly(I:C)] or derivatives, but with limited success [4]. With the advancement of knowledge about the mechanisms of ligand recognition and function of RLRs, the possibility of using RLR ligands as pan-antivirals, vaccine adjuvants, and antitumor agents has gained much attention [5]. While studies using RIG-I RNA agonists are accumulating, MDA5-targeted strategies suffer from the limited availability and diversity of pharmacologically usable ligands. However, important functional divergences between RIG-I and MDA5 have recently been reported, potentially providing new therapeutic opportunities. We address here the recent concepts and remaining gaps in our knowledge regarding the similarities and differences of RIG-I and MDA5 ligands and their signaling and biological functions. We highlight how such knowledge should be considered in the choice and design of specific and potent nucleic acid agonists, and discuss options for targeting RIG-I and/or MDA5 to achieve the best antiviral and immunomodulatory response for therapeutic and vaccine adjuvant development.
RIG-I and MDA5 Ligand-Binding Specificities: What Do We Know?
Both RLRs recognize RNA species carrying pathogen-associated molecular patterns (PAMPs). The characteristics of selective RIG-I and MDA5 ligands have recently been reviewed 1, 2. Thus, we will only highlight the recent advances and concepts relevant to the design of selective and potent RIG-I and MDA5 agonists. Using in vitro transcribed (ivt) viral RNA analogs, it was determined that the consensus minimum RNA size required for RIG-I recognition and signal transduction is as short as 10 bp, but the maximum size over which binding to RIG-I is suppressed remains unknown 6, 7, 8, 9. By contrast, it is generally accepted that MDA5 senses high molecular weight (HMW) RNA species 7, 10. Although the minimum length of MDA5 ligand remains unresolved, length-dependent induction of IFNs by ivt double-stranded (ds) RNA in RIG-I-deficient murine embryonic fibroblasts (MEFs) suggests that ligands as short as 100 bp can activate MDA5 [10]. Elucidation of the structural mechanisms of RLR activation has provided key information that has helped explain RIG-I and MDA5 ligand length discrimination. Long dsRNA species serve as a scaffold to facilitate the cooperative assembly of MDA5 into long polymers, thereby facilitating optimal interaction of the exposed multiple CARDs with MAVS 11, 12, 13, 14. On the other hand, RIG-I forms shorter signaling-competent ligand-bound filaments 15, 16. In vitro and biochemical analyses indicate that MDA5 filaments nucleate from an internal region and propagate toward the ends of dsRNA, while RIG-I exhibits an end-capping pattern before oligomerization into shorter filaments 13, 15, 16, 17. Although these observations support the hypothesis that filament dynamics explain the difference in RIG-I and MDA5 oligomerization capacities and signaling from RNA duplexes of different lengths, this might be overly simplistic. Considering that filament formation is ATP-dependent and that binding of a monomer of RIG-I, but not of MDA5, to the 5′-triphosphate (5′ppp) terminal region of short ivt dsRNA is sufficient for optimal signal transduction, it can be speculated that RIG-I filament formation on HMW dsRNA would be energy-demanding and less efficient 8, 13, 15.
In addition to ligand length, RNA structure and modifications must also be taken into consideration for the synthesis of specific and high-affinity RIG-I and MDA5 agonists. The salient chemical features for RIG-I engagement consist of higher-order structure, in the form of an RNA duplex or panhandles, and a 5′ppp moiety 18, 19, 20. Although ivt single-stranded (ss) RNA synthesized using bacteriophage T7 polymerase activates RIG-I-dependent IFN production, it is important to consider that phage polymerases produce transcripts containing 5′ppp and regions of higher-order structure resulting from a ‘copy-back’ mechanism [21]. dsRNA structures of ligands are also essential for MDA5 activation, but 5′ppp is not required 7, 10, 22. Furthermore, research aimed at understanding the mechanism of selective recognition of non-self versus self-RNA by RIG-I has also provided insights into the impact of RNA capping. 2′-O-methylation of cap-1, but not N-7-methylation of cap-0, is crucial to abolish the binding of RIG-I to self-RNA 6, 23. 2′-O-methylation of mRNA also prevents detection by MDA5, a mechanism used by coronaviruses to evade MDA5 recognition [24]. Comparison of unmodified RNA with RNA containing modified nucleotides that are present in the human transcriptome, such as N-6-methyladenosine, 5-methylcytidine, pseudouridine, N-1-methylpseudouridine, and 2′-fluorodeoxyribose, showed that modified nucleotides suppress RIG-I binding and signaling, although through different mechanisms [25]. However, it remains to be determined if and how these modifications alter MDA5 activation.
An additional open question concerns the contribution of nucleotide composition to the specificity and efficiency of RIG-I and MDA5 binding and activation. The first suggestion of a role of the nucleotide sequence came from the observation that the poly(U/UC) region of the 3′-untranslated region (UTR) of hepatitis C virus (HCV) [26] viral RNA, and of the corresponding poly-A-containing RNA replication intermediate, confers the capacity to functionally bind to RIG-I, but not to MDA5 27, 28, 29. Reduction of the A/U content of the RNA impairs RIG-I activation [27]. Similarly, modification of the nucleotide sequence of ivt 5′ppp RNA can potentiate the activation of RIG-I-dependent antiviral and inflammatory activities without altering ligand specificity [30]. In a measles virus infection, deep sequencing revealed that RNA species crosslinked to MDA5 or RIG-I were enriched in AU-rich RNA species, and the AU content positively correlated with their immunostimulatory capacity [31]. Although this supports the importance of the RNA sequence in RLR activation, it remains unclear if the nucleotide sequence impacts on specific binding to RIG-I and/or MDA5. Overall, ligand length remains the only discriminating parameter that can currently be used to design specific RIG-I and MDA5 agonists for pharmacological applications. It seems clear, however, that future developments will need to take into account not only length but also higher-order structure, sequence, and RNA modifications. Future work should aim at characterizing the impact of each, and combinations thereof, on RIG-I and MDA5 binding, as well as the ability to engage downstream signaling.
Redundancy and Divergence of RIG-I and MDA5 Functions
The rationale behind the search for specific ligands relies on accumulating evidence of striking functional differences between RIG-I and MDA5. The first observation of a major difference arose from the attempts to generate RLR-specific knockout mice in the C57BL/6 mice strain, which is the most commonly used to study the antiviral immune response 32, 33. Whereas MDA5-deficient mice were healthy without noticeable defects, RIG-I-deficient mice exhibited high embryonic mortality, growth retardation, with death within 3 weeks of birth or fetal liver degeneration 32, 33. Generation of a viable RIG-I-deficient mouse model required backcrossing onto the 129S1 strain, which is widely used for its genetic flexibility [34]. These RIG-I-deficient mice spontaneously develop pathological phenotypes leading to colitis [33]. None of these phenotypes have been reported in MDA5-deficient mice.
Single-nucleotide polymorphisms (SNPs) in DDX58 and IFIH1, encoding RIG-I and MDA5, respectively, are associated with various clinical inflammatory and autoimmune presentations, suggesting important functional redundancies but also likely differences (Figure 1). Several gain-of-function (GOF) mutations in DDX58 and IFIH1 are associated with Singleton–Merten syndrome (SMS) (Box 1 ), although with variable clinical features, as recently reviewed [35]. Mutations c.2465G > A [p.R822G], c.992C > T [p.T331I] and c.992C > G [p.T331R], and c.1465G > A [p.A489T] in IFIH1 were found in patients with classical SMS, whereas c.1118A > C [p.E373A] and c.803G > T [p.C268F] in DDX58 were associated with atypical SMS [35]. Although currently known missense mutations of DDX58 are restricted to SMS phenotypes, additional IFIH1 GOF polymorphisms, c.1009A > G [p.R337G], c.1178A > T [p.D393V], c.1114C > T [p.L372F], c.1354G > A [p.A452T], c.1483G > A [p.G495R], c.2159G > A [p.R720Q], and c.2335C > T [p.R779C], are associated with Aicardi–Goutières syndrome (AGS) (Box 1) 36, 37. Intriguingly, the c.2336G > A [p.R779H] GOF mutation was found not only in patients with phenotypes indicative of AGS but also in systemic lupus erythematosus (SLE) (Box 1) 37, 38. Similarly, a IFIH1 c.2836G > A [p.A946T] variant was identified as a risk factor for type 1 diabetes, SLE, and Graves’ disease (Box 1) 39, 40, 41. All these missense mutations affect the helicase domain and were associated with an increased IFN-I signature 36, 37, 38, 42, 43, 44. Direct evidence linking RLR GOF-associated IFN-I upregulation to inflammatory and autoimmune phenotypes is currently limited. However, the lupus-like symptoms observed in the mouse model harboring Ifih1 c.2475G > A [p.G821S] are associated with spontaneous IFN-I induction in multiple organs and were abolished on MAVS- or IFN-I receptor-deficient backgrounds, thus providing the first direct link between MDA5 GOF mutation and an autoimmune phenotype [45].
Box 1. Autoimmune and Inflammatory Diseases Associated with RIG-I and MDA5 SNPs.
Singleton–Merten Syndrome (SMS)
A rare genetic multisystem disorder. The most common symptoms include, but are not limited to, tooth abnormalities, calcification of major arteries and aorta, reduced bone density affecting limbs, atrophy, and abnormal development in general. SMS is classified as either classical or atypical. The large majority of patients exhibit typical clinical presentation. However, patients exhibiting SMS symptoms but with normal dentition have been reported. These cases of SMS have been classified as atypical SMS 44, 86.
Aicardi–Goutières Syndrome (AGS)
A rare neurological and developmental genetic disorder causing severe mental and physical handicap. It is described as an early-onset disorder of the nervous system with rapid progression toward a vegetative state and death. AGS patients exhibit, among other phenotypes, signs of encephalopathy including radiological abnormalities in the white matter and an increased number of lymphocytes in the cerebrospinal fluid (CSF). Other relevant signs and symptoms include brain atrophy and increased levels of type I interferon (IFN-I) in the CSF, as well as abnormal enlargement of the liver and skin lesions [87].
Systemic Lupus Erythematosus (SLE)
A systemic multiorgan autoimmune disorder with heterogeneous disease manifestations and organs affected, including skin rashes, chronic fatigue, arthritis, severe glomerulonephritis, and neurological impairments. The loss of immune tolerance to self-antigens in SLE autoimmunity results from dysregulation of multiple cellular components, including several proinflammatory cytokines (notably type I interferons), disruption of the clearance of apoptotic waste and immunocomplexes, the presence of autoantibodies, abnormalities of the complement pathways, and disrupted lymphocyte immune responses 88, 89.
Type-1 Diabetes
Also called insulin-dependent diabetes, type 1 diabetes is a chronic condition in which the pancreas does not produce sufficient insulin, the hormone necessary for cellular uptake and metabolism of glucose. This results in accumulation of glucose in the bloodstream, hence causing symptoms such as extreme fatigue, weight loss, frequent urination, blurred vision, etc. Complications resulting from type 1 diabetes can result to death. Diabetic patients need to receive recombinant insulin for the rest of their life [90].
Graves’Disease
Also called toxic diffuse goiter, Graves’ disease is the most common cause of hypothyroidism which significantly affects the quality of life. Clinical manifestations, which depend on the age of the patient at the time of hypothyroidism onset, include, but are not limited to, weight loss, weak muscle, irritability, anxiety, sleeping problems, and eye problems such as swelling, pain, redness, and double vision [91].
Alt-text: Box 1
Structural analyses of RIG-I led to the proposed model where atypical SMS-causing mutations affecting the helicase 1 domain inhibit RIG-I ATPase function, thereby relieving the inhibitory mechanisms that prevent RIG-I from being activated by endogenous self-RNA [46]. Whether this mechanism explains the constitutive activation of disease-causing IFIH1 mutations that affect the ATPase domain remains to be determined. An alternative model proposes that AGS-causing IFIH1 mutations result in increased dsRNA avidity and constitutive activation of MDA5 by endogenous RNA recognition [37]. However, this view has been challenged because some distinct MDA5 AGS-causing mutations were found to lose ligand responsiveness and ATPase activity 36, 45. That single RLR gene mutations associated with a constitutive or increased IFN-I signature lead to distinct inflammatory and autoimmune diseases is still puzzling. Future studies using novel mouse models should aim to further our understanding of the mechanisms underlying specific disease-causing mutations of RIG-I and MDA5 to demonstrate the causal effects of the various polymorphisms.
The structural similarities between RIG-I and MDA5, as well as the well-documented signaling through the common adaptor MAVS, initially led to the consensus view that RIG-I and MDA5 provide multiple redundant immune mechanisms to offer effective protection of the host. This concept was further supported by the observation that several viruses, including the Paramyxoviridae Sendai virus (SeV) and measles virus (MeV), the Pneumoviridae respiratory syncytial virus (RSV), the Flaviviridae West Nile virus (WNV) and dengue virus (DENV), and the Reoviridae rotavirus, engage both RIG-I and MDA5 to trigger an IFN response 31, 47, 48, 49, 50, 51, 52, 53. However, accumulating evidence reveals distinct pathways regulated by RIG-I or MDA5 (Figure 2). Engagement of both RLRs is well documented to activate IRF3 and NF-κB transcription factors that cooperate to initiate the expression of antiviral and proinflammatory cytokines that are essential to limit virus replication and spreading 54, 55.
Recent work yielded data that that support temporal involvement of RIG-I and MDA5 in the initiation and duration of the cytokine response. In a mouse model of SeV infection, MDA5 is required for IFN-I and -III, tumor necrosis factor (TNF)-α, and IFN-γ expression at late (day 5), but not earlier (day 2), times of infection [49]. In vitro, RIG-I is involved in the initiation, while MDA5 is essential for the persistence, of antiviral IFIT1 expression induced by RSV [50]. This temporal involvement might either reflect the ability of RLRs to activate distinct, yet to be identified, signaling pathways downstream of MAVS or the generation of RIG-I and MDA5-specific PAMPs at different times of viral replication. In support of the latter, sensing of WNV infection by RIG-I and MDA5 was demonstrated to be dependent on distinct viral RNA species accumulating with different kinetics [53]. On the other hand, in the context of SeV and RSV infections, IRF3 activation is dependent on RIG-I, whereas MDA5 is only essential to dampen ubiquitination and subsequent proteasome-mediated degradation of IRF3 (Figure 2) [50].
The proinflammatory arm of the RLR-dependent response is also subject to specific sensor regulation. Both RIG-I and MDA5 are required for RSV-induced NF-κB activation; however, only RIG-I acts upstream of the classical IκBα-dependent NF-κB pathway that is typically responsible for rapid and transient NF-κB activity (Figure 2) [56]. This points to a distinct role of MDA5 in the regulation of the NF-κB response through a non-canonical pathway. In addition, RIG-I and MDA5 both induce the activation of NF-κB-dependent transcription of proinflammatory interleukin (IL)-6 and pro-IL-1β through a MAVS-dependent pathway involving the interaction of CARD9 with the CARD-containing adaptor BCL10, a well-known activator of apoptosis and NF-κB signaling 57, 58. Interestingly, RIG-I, but not MDA5, triggers inflammasome assembly to activate caspase-1-dependent cleavage of pro-IL-1β ultimately resulting in the extracellular production of mature bioactive IL-1β 52, 53, 54 (Figure 2). Thus, RIG-I and MDA5 play distinct roles in the induction and duration of the antiviral and inflammatory responses. Overall, our understanding of the redundancy and divergence of RIG-I and MDA5 functions is still sparse; much remains to be learned to fully comprehend the consequences of therapeutic activation of RIG-I or MDA5.
Targeting RLRs for Therapeutic and Prophylactic Strategies
The available data hold great promise for the use of specific RIG-I and MDA5 ligands as novel pan-antivirals, vaccine adjuvants, or potentiators of anticancer immunotherapies 5, 59, but they also highlight a knowledge gap and challenges that remain to be addressed. Many studies have reported the use of RIG-I ligands in preclinical studies, but specificity for RIG-I over MDA5 and Toll-like receptors (TLRs) has only been demonstrated for the agonists discussed hereafter. Intracellular delivery of an ivt 5′ppp RNA derived from the 5′- and 3′-UTRs of vesicular stomatitis virus (VSV) – VSV-UTR 5′ppp-RNA – induced an antiviral and inflammatory transcriptional program that significantly protected cells against influenza virus (IAV), VSV, DENV, HCV, human immunodeficiency virus (HIV), and chikungunya virus (CHIKV) in vitro and ex vivo 60, 61. In vivo, VSV-UTR 5′ppp-RNA moderately protected mice against IAV H1N1 and the H5N1-reassortant (H5N1-RE), but modification of the internal RNA sequence yielded an optimized agonist (M8) with increased antiviral activity against H5N1-RE IAV and CHIKV 30, 60 (Table 1 ). An alternative synthetic RIG-I-specific agonist composed of bent RNA formed by G–U wobble base pairs mimicking the structure of the IAV panhandle promoter (CBS-13-BPS) exhibited potent antiviral activity against IAV H1N1 and IAV resistant to the antiviral drug oseltamivir in vitro, as well as strong adjuvant potential in vivo in mice, when incorporated into the A/H1N1 PR8 inactivated IAV vaccine [62]. In addition, CBS-13-BPS triggered significant tumor regression in a murine pancreatic tumor model, indicating potential as a cancer immunotherapeutic [62]. A third type of RIG-I agonist, in the form of 5′ppp RNA harboring a stem-loop (SLR10 and SLR14), was recently found to induce greater IFN-I response compared to poly(I:C) in vivo; however, it remains to be tested in prophylactic or therapeutic contexts [9].
Table 1.
Synthetic and Natural RLR-Specific dsRNA Ligands (Agonists) Evaluated In Vivoa
| RLR ligand | Salient features | RLR specificity | In vivo pharmacological evaluation | Observations | Administration | Refs |
|---|---|---|---|---|---|---|
| M8 | Synthetic Ivt 5′-ppp Optimized from VSV 5′- and 3′-UTRs |
RIG-I In vivo JetPEI used to avoid TLR3 |
Antiviral (mice) | ↑ Anti-H5N1-RE protection | Intravenous | [30] |
| ↑ Anti-CHIKV protection | Intraperitoneal | [30] | ||||
| CBS-13 BPS | Synthetic Ivt 5′-OH Bent structure |
RIG-I In vivo JetPEI used to avoid TLR3 |
Vaccine adjuvant (mice) | ↑ Efficiency of an inactivated H1N1 vaccine | Intranasal | [62] |
| Cancer immunotherapeutic (mice) | ↓ Pancreatic tumor | Intratumor injection | [62] | |||
| SLR10 and SLR14 | Synthetic Ivt 5′ppp Stem-loop structure |
RIG-I In vivo JetPEI used to avoid TLR3 |
IFN-inducer and immunostimulant (mice) | ↑ Serum IFN-I and TNF-α | Intravenous | [9] |
| ↑ IFN-I and IFN-III mRNA in spleen | Intravenous | [9] | ||||
| Poly(IC:LC) | Synthetic Modification of poly(I:C) |
MDA5 TLR3 |
Immunostimulant (mice) | ↑ IFN-I in BALF, lungs, and serum | Intranasal and intramuscular | [63] |
| Antiviral (mice) | ↑ Anti-SARS-CoV protection | Intranasal | [64] | |||
| ↑ Anti-H1N1 and -H3N2 protection | [66] | |||||
| ↓ RSV replication ↓ Morbidity |
[63] | |||||
| Cancer immunotherapeutic (human) | ↑ Immunogenicity of an ovarian cancer vaccine | Subcutaneous | [69] | |||
| ↑ Immunogenicity of a glioma vaccine | Intramuscular | [68] | ||||
| Safely incorporated into a pancreatic cancer vaccine | Intramuscular | [67] | ||||
| rb-dsRNA | Natural Endornavirus genome From edible plants |
MDA5 TLR3 |
Immunostimulant (mice) | ↑ IFN-I and IL-1β in BALF |
Intranasal | [70] |
| MDA5 TLR3 |
Antiviral (mice) | ↑ Anti-H1N1 and -SeV protection | Intranasal | [70] | ||
| NAB2 | Natural Genome of Saccharomyces cerevisiae virus of the Totoviridiae family |
MDA5 TLR3 |
Cancer immunotherapeutic (mice) | ↑ Efficacy of a MUC1-positive lymphoma vaccine | Subcutaneous | [71] |
| MDA5 TLR3 |
IFN-inducer and immunostimulant (mice) | ↑ Serum IFN-I | Intraperitoneal | [72] |
Abbreviations and symbols: ↑, increased; ↓, decreased; BALF, bronchoalveolar lavage fluid; MUC1, mucin 1.
The difficulty associated with template-dependent synthesis of HMW RNA, added to the poorly defined specific features of MDA5 ligands, have limited the large majority of MDA5-related studies to the use of HMW poly(I:C) analogs. In the late 1960s, HMW poly(I:C) was evaluated as an immunostimulant in vivo with little to no success [4]. A more stable derivative, poly(IC:LC), obtained by modification with poly-L-lysine and carboxymethyl cellulose, has been successfully used in mice for prophylactic and therapeutic protection against a broad spectrum of human viruses including lethal severe acute respiratory syndrome coronavirus (SARS-CoV), lethal H5N1 IAV, and RSV (Table 1) 63, 64, 65, 66. Incorporating poly(IC:LC) as adjuvant also enhanced the clinical efficiency of experimental vaccines in patients with recurrent malignant glioma, ovarian cancer, or pancreatic cancer 67, 68, 69. The undefined structure, heterogeneity, and polydispersity of HMW poly(I:C) and analogs together with unpredictable pharmacokinetics and toxicity, present a hurdle for therapeutic use [4]. Identifying novel, structurally defined MDA5 ligands is an attractive avenue for developing new therapeutics.
Towards this, two natural MDA5-activating HMW dsRNA have recently been reported (Table 1). A dsRNA (rb-dsRNA) corresponding to the genome of endornavirus extracted from rice bran, that is found in several plants, significantly reduces the mortality and morbidity caused by H1N1 IAV and SeV, respectively, in mice [70]. The nucleic acid band 2 (NAB2), a dsRNA corresponding to a yeast virus of the Totoviridiae family, induces a stronger IFN-I response compared to poly(I:C) ex vivo, and strongly increases the efficacy of an attenuated poxvirus modified vaccinia virus Ankara (MVA)-based vaccine targeting cancer cells expressing the vaccine antigen mucin-1 (MUC-1) in mice 71, 72. However, data on MDA5 agonists should be interpreted with caution in relation to the impact on specific MDA5 stimulation. The poly(I:C) analogs, rb-dsRNA and NAB2, concomitantly activate TLR3, which is associated with a strong inflammatory response [73]. Complex formulations, such as the JetPEI transfection reagent, have been successfully used in vivo in mice for intracellular delivery of RIG-I ligands to bypass TLR3 activation 9, 30, 62 (Table 1). This approach would be worth testing with MDA5 ligands in preclinical studies. However, the application of these strategies to specifically engage RLRs in human trials remains challenging [74].
Although the above studies have started to build a case for the beneficial therapeutic usage of specific RIG-I and MDA5 agonists, evidence is missing to make an informed choice on the best ligand to trigger an appropriate response for particular applications. Comparative analyses of the antiviral or immunomodulatory activities of RIG-I and MDA5-specific agonists against various tumors and viruses in animals and humans should be instructive. An efficient antiviral response against several viruses involves both RLR sensors 31, 33, 47, 48, 49, 50, 51, 52, 56. Furthermore, ectopic expression of MDA5 in vitro prolongs RIG-I-dependent IRF-3 activation, suggesting that dual engagement of RIG-I and MDA5 has the potential to trigger a sustained antiviral response [50]. Therefore, one could speculate that strategies targeting both RIG-I and MDA5 could provide therapeutic advantages. The complete assessment of this hypothesis is limited by the availability of ligands proven to be capable of equally activating both RLRs. A strategy of mixing the RIG-I and MDA5 ligands would most likely be biased by different rates of stability and uptake of each ligand by the host cells. Moreover, the design of a universal RLR ligand is in turn hampered by incomplete knowledge of a common PAMP signature.
The advantages of using RIG-I or MDA5 ligands over IFNs as pan-antivirals is a question worth discussing. Recent studies have highlighted differences in the contributions of IFN-I and IFN-III to the antiviral response 75, 76. Extensive clinical use of IFN-I has clearly documented dose-related adverse events, highlighting the deleterious effect of an increased or prolonged IFN-I response [77]. However, a clinical trial using IFN-III in patients with chronic HCV infection reported an antiviral activity similar to that of IFN-I but with fewer systemic adverse effects, likely owing to the restricted distribution of the IFN-III receptor [78]. Therefore a type III response seems to be more favorable than a type I response for antiviral applications. Both IFN-I and -III transcriptional signatures are induced in vitro by VSV-derived 5′ppp-RNA RIG-I ligands (Table 1), but the balance between the two responses remains to be fully evaluated in vivo 30, 60. SLR10 and 14 ligands (Table 1) selectively induce high IFN-I gene expression associated with a weak type IFN-III response, a balance that is opposite to that triggered by poly(I:C) [9]. Although data using poly(I:C), which is recognized by MDA5, suggest a more favorable IFN balance for antiviral applications, such evidence using MDA5-specific ligands is not yet available. Further studies clarifying the relevance of the balance between IFN-I and -III in the quality of the antiviral immunity will be necessary to determine the best ratio for therapeutic indications and whether this can be achieved using RLR ligands.
The idea of inducing physiologically relevant levels of IFNs through the activation of signaling pathways leading to the transcriptional control of endogenous IFN genes might also limit the deleterious effects observed with exogenous IFNs. With this in mind, an alternative approach to RLR ligands that is currently at early stage of investigation is the use of IRF3 agonists to induce the IFN response 79, 80, 81. The use of RLR ligands, however, presents the particularity of inducing a proinflammatory response in addition to triggering the induction of IFNs. Inflammation, although necessary to resolve virus infection, is also responsible for disease pathogenesis when the strength and duration of the response are not strictly controlled [82]. A major gap in knowledge that needs to be addressed for appropriate pharmacological use of specific RLR RNA agonists concerns the beneficial versus the undesirable effects of the induced inflammatory response. Identification of SNPs in human DDX58, and to a greater extent in IFIH1, teaches us that uncontrolled activation of RLRs is associated with the onset of chronic inflammatory or autoimmune diseases (Figure 1) 36, 37, 38, 42, 43, 44. Further studies will be necessary to thoroughly evaluate if RIG-I- and/or MDA5-specific agonists provide immunostimulatory effects in vivo at the expense of a systemic elevated cytokine response that is responsible for long-lasting adverse effects [82]. Moreover, the inflammasome response should also be analyzed carefully: although it can be beneficial, it can also promote hyperinflammation and cell death [83]. Whether RIG-I ligand-induced inflammasome activation would provide an advantage over, or induce more adverse effects than, a non-inflammasome activating MDA5-specific strategy is a question worth addressing 57, 70, 84.
Concluding Remarks and Future Perspectives
Less than two decades after the first reports characterizing the role of RLRs in immunity, we are closer than ever to having RLR ligands in clinical applications. Pilot trial and ongoing clinical trials in humans are yielding promising results, notably for cancer immunotherapy 67, 68, 69, 85. Although we have come a long way and key milestones have been reached, there are still significant knowledge gaps that need to be filled by use of molecular, cellular, and structural biology approaches to not only be able to design exclusive and potent RIG-I or MDA5 ligands but also to decide on the most appropriate target for use in antiviral, vaccine adjuvant, or cancer immunotherapeutic applications (see Outstanding Questions). As discussed here, accumulating evidence demonstrates that distinct functions are induced following RIG-I or MDA5 engagement. We opine that these functional divergences need to be considered in the choice of agonists for therapeutic purposes. For instance, although ligand-induced cell death might be associated with adverse effects in antiviral applications, it might be beneficial in cancer therapeutics – where this could be harnessed to induce tumor cell death in conjunction with stimulating antitumor immunity. To achieve this, we need a more systematic and thorough characterization of the specificity of ligands used in preclinical studies. This will also allow determining the pros and cons of targeting RIG-I, MDA5, or the two simultaneously. Failing to do so might yield an incomplete picture of the clinical relevance and efficiency of ligand-based strategies to be considered for large-scale clinical trials and commercialization. Finally, in the era of personalized medicine that aims at using tailor-made drugs for a unique genetic profile, instead of one-size-fits-all medicine, one would think that having specific ligands for RIG-I or MDA5 would provide a range of options for patients with a receptor genetic polymorphism that could affect ligand binding.
Outstanding Questions.
What are the features of RNA ligands that will allow the synthesis of RIG-I- and MDA5-specific agonists: is there a maximum ligand length required for RIG-I activation and a minimum length for MDA5 binding?
Can the nucleotide sequence or 5′-terminal and internal nucleotide modifications alter the affinity of RIG-I and MDA5 to ligands?
What are the functions, and the underlying mechanisms, specifically mediated by RIG-I or MDA5? Could this knowledge be harnessed to develop the most appropriate antiviral, vaccine adjuvant, or cancer immunotherapeutics?
Is it necessary to avoid TLR3 engagement in every RLR-targeting strategy? If necessary, what formulation will allow intracellular delivery of RLR agonists for clinical applications in humans?
Would strategies specifically targeting either RIG-I or MDA5 have an advantage over the simultaneous engagement of both RIG-I and MDA5 in the context of particular prophylactic or therapeutic applications?
To what extent would RLR ligands provide efficient antiviral therapeutic effects when given after the infection? What would be the window of therapeutic opportunity?
Are RLR ligands appropriate as a counter-measure in the farming industry to limit the spread of viruses such as influenza during outbreaks in swine or poultry?
Why do distinct RIG-I and MDA5 polymorphisms associated with an increased IFN response result in different autoimmune and inflammatory diseases? Does this predict adverse effects that could result from clinical use of ligand-mediated activation of RIG-I and MDA5?
How do polymorphisms impact on the response of patients to RLR ligands?
Acknowledgments
Research in our laboratory is funded by grants from the Canadian Institutes of Health Research (CIHR) (MOP-130527, MOP-137099, III-134054 and PJT-159570) and by a Research Chair in Signaling in Virus Infection and Oncogenesis from the Université de Montréal to N.G. D.K. and N.G. are members of the Réseau en Santé Respiratoire du Fonds de la Recherche en Santé du Québec (RSR-FRQS).
Glossary
- Adjuvant
a molecule that potentiates the host immune response to an antigen.
- Agonist
a molecule that triggers a physiological response when bound to its receptor.
- Autoimmune disease
a condition in which the immune system produces antibodies that attack the healthy tissues of the host.
- Caspase-recruitment domain (CARD)
a motif found in many proteins involved in inflammation and apoptosis that mediates homotypic interactions between two proteins.
- Cytokines
small proteins released by cells to communicate with other cells.
- Gain-of-function (GOF) mutation
a type of mutation in a gene that results in an altered protein exhibiting a new function or an increased activity.
- Helicase
a protein capable of unwinding nucleic acids through the hydrolysis of ATP.
- Homotypic interaction
interaction through similar domains found in interacting protein partners.
- Immunostimulant
a molecule capable of boosting the immune response.
- Inflammasome
an intracellular multiprotein complex that triggers inflammatory responses through the activation of the thiol protease, caspase 1. It notably results in the production of mature interleukin (IL)-1β and IL-18, and induces cell death by pyroptosis.
- Interferon (IFN)
a subgroup of cytokines produced by host cells in response to infections that act on the infected cell or surrounding cells to trigger the induction of genes encoding proteins with antiviral activities.
- Melanoma differentiation-associated protein 5 (MDA5)
a cytoplasmic PRR that senses viral RNA or viral RNA intermediates, and whose activation leads to cytokine production.
- Panhandle
a form of secondary structure found in RNA molecules arising from pairing of complementary sequences in the 3′ and 5′ ends.
- Pathogen-associated molecular pattern (PAMP)
a molecular signature unique to a pathogen that is recognized as a danger signal by PRRs of host cells to induce an immune response.
- Pattern-recognition receptors (PRRs)
a group of receptors that recognize molecular patterns unique to pathogens to induce an immune response.
- Polydispersity
a measure of the molecular weight distribution. A polydisperse poly(I:C) solution means that the chain length varies, leading to a mixture of particles of different sizes.
- Retinoic acid-inducible gene I (RIG-I)
a cytoplasmic PRR that senses viral RNA or viral RNA intermediates; activation leads to cytokine production.
- RNA capping
the addition of a cap structure to mRNA in eukaryotic cells to protect them from degradation by host exonucleases and to facilitate recruitment of translation/protein factors. The cap-0 structure consists of N-7-methylguanylate connected to the first nucleotide. A cap-1 structure is formed by the addition of a methyl group to the second hydroxyl group of the first ribose sugar.
- Single-nucleotide polymorphism (SNP)
a variation at a single position in the genome sequence within a population.
- Toll-like receptors (TLRs)
a family of membrane-bound PRRs that are activated by different PAMPs. This activation results in the induction of immune events, such as the production of cytokines. Among other members, TLR3 is activated by double-stranded (ds) RNA.
References
- 1.Chow K.T., et al. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M., et al. Viral RNA. detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Vazquez C., Horner S.M. MAVS coordination of antiviral innate immunity. J. Virol. 2015;89:6974–6977. doi: 10.1128/JVI.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy H.B. Historical overview of the use of polynucleotides in cancer. J. Biol. Response Mod. 1985;4:475–480. [PubMed] [Google Scholar]
- 5.Yong H.Y., Luo D. RIG-I-like teceptors as novel targets for pan-antivirals and vaccine adjuvants against emerging and re-emerging viral infections. Front. Immunol. 2018;9:1379. doi: 10.3389/fimmu.2018.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devarkar S.C., et al. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U. S. A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohlway A., et al. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013;14:772–779. doi: 10.1038/embor.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linehan M.M., et al. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018;4 doi: 10.1126/sciadv.1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q., et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berke I.C., Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berke I.C., et al. MDA5 assembles into a polar helical filament on dsRNA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J., et al. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015;43:1216–1230. doi: 10.1093/nar/gku1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peisley A., et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peisley A., et al. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Wu B., et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 17.Yu Q., et al. Cryo-EM structures of MDA5–dsRNA filaments at different stages of ATP hydrolysis. Mol. Cell. 2018 doi: 10.1016/j.molcel.2018.10.012. Published online November 7, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui S., et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Schlee M., et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D.H., et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 22.Pichlmair A., et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuberth-Wagner C., et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zust R., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin A.F., et al. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016;7 doi: 10.1128/mBio.00833-16. e00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ThÇvenin C., et al. Induction of nuclear factor-κB and the human immunodeficiency virus long terminal repeat by okadaic acid, a specific inhibitor of phosphatases. New Biol. 1991;2:793–800. [PubMed] [Google Scholar]
- 27.Saito T., et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell G., et al. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzri D., Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang C., et al. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J. Virol. 2015;89:8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Runge S., et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H., et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Kato H., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., et al. Rig-I-/- mice develop colitis associated with downregulation of G alpha i2. Cell Res. 2007;17:858–868. doi: 10.1038/cr.2007.81. [DOI] [PubMed] [Google Scholar]
- 35.Lu C., MacDougall M. RIG-I-like receptor signaling in Singleton–Merten syndrome. Front. Genet. 2017;8:118. doi: 10.3389/fgene.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oda H., et al. Aicardi–Goutières syndrome is caused by IFIH1 mutations. Am. J. Hum. Genet. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice G.I., et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Eyck L., et al. Brief report: IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis Rheumatol. 2015;67:1592–1597. doi: 10.1002/art.39110. [DOI] [PubMed] [Google Scholar]
- 39.Gateva V., et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth D.J., et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 41.Sutherland A., et al. Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves’ disease susceptibility. J. Clin. Endocrinol. Metab. 2007;92:3338–3341. doi: 10.1210/jc.2007-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bursztejn A.C., et al. Unusual cutaneous features associated with a heterozygous gain-of-function mutation in IFIH1: overlap between Aicardi–Goutières and Singleton–Merten syndromes. Br. J. Dermatol. 2015;173:1505–1513. doi: 10.1111/bjd.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Carvalho L.M., et al. Musculoskeletal disease in MDA5-related type I interferonopathy: a Mendelian mimic of Jaccoud’s arthropathy. Arthritis Rheumatol. 2017;69:2081–2091. doi: 10.1002/art.40179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang M.A., et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton–Merten syndrome. Am. J. Hum. Genet. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funabiki M., et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Lassig C., et al. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. eLife. 2015;4 doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broquet A.H., et al. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 48.Fredericksen B.L., et al. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gitlin L., et al. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grandvaux N., et al. Sustained activation of interferon regulatory factor 3 during infection by paramyxoviruses requires MDA5. J. Innate Immun. 2014;6:650–662. doi: 10.1159/000360764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim W.K., et al. Deficiency of melanoma differentiation-associated protein 5 results in exacerbated chronic postviral lung inflammation. Am. J. Respir. Crit. Care Med. 2014;189:437–448. doi: 10.1164/rccm.201307-1338OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loo Y.M., et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Errett J.S., et al. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J. Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seth R.B., et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Yoboua F., et al. Respiratory syncytial virus-mediated NF-kappa B. p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J. Virol. 2010;84:7267–7277. doi: 10.1128/JVI.00142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poeck H., et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 58.Bertin J., et al. CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 2000;275:41082–41086. doi: 10.1074/jbc.C000726200. [DOI] [PubMed] [Google Scholar]
- 59.Wu Y., et al. The anticancer functions of RIG-I-like receptors RIG-I and MDA5, and their applications in cancer therapy. Transl. Res. 2017;190:51–60. doi: 10.1016/j.trsl.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 60.Goulet M.L., et al. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olagnier D., et al. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 2014;88:4180–4194. doi: 10.1128/JVI.03114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee J., et al. Systematic editing of synthetic RIG-I ligands to produce effective antiviral and anti-tumor RNA immunotherapies. Nucleic Acids Res. 2018;46:1635–1647. doi: 10.1093/nar/gky039. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Guerrero-Plata A., et al. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J. Virol. 2005;79:10190–10199. doi: 10.1128/JVI.79.16.10190-10199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumaki Y., et al. Prophylactic and therapeutic intranasal administration with an immunomodulator Hiltonol® (Poly IC:LC), in a lethal SARS-CoV-infected BALB/c mouse model. Antiviral Res. 2017;139:1–12. doi: 10.1016/j.antiviral.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy H.B., et al. A modified polyriboinosinic–polyribocytidylic acid complex that induces interferon in primates. J. Infect. Dis. 1975;132:434–439. doi: 10.1093/infdis/132.4.434. [DOI] [PubMed] [Google Scholar]
- 66.Wong J.P., et al. Prophylactic and therapeutic efficacies of poly(IC.LC) against respiratory influenza A virus infection in mice. Antimicrob. Agents Chemother. 1995;39:2574–2576. doi: 10.1128/aac.39.11.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehrotra S., et al. Vaccination with poly(IC:LC) and peptide-pulsed autologous dendritic cells in patients with pancreatic cancer. J. Hematol. Oncol. 2017;10:82. doi: 10.1186/s13045-017-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okada H., et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with α-type 1 polarized dendritic cells and polyinosinic–polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J. Clin. Oncol. 2011;29:330–336. doi: 10.1200/JCO.2010.30.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabbatini P., et al. Phase I trial of overlapping long peptides from a tumor self-antigen and poly-ICLC shows rapid induction of integrated immune response in ovarian cancer patients. Clin. Cancer Res. 2012;18:6497–6508. doi: 10.1158/1078-0432.CCR-12-2189. [DOI] [PubMed] [Google Scholar]
- 70.Kasumba D.M., et al. A plant-derived nucleic acid reconciles type I IFN and a pyroptotic-like event in immunity against respiratory viruses. J. Immunol. 2017;199:2460–2474. doi: 10.4049/jimmunol.1700523. [DOI] [PubMed] [Google Scholar]
- 71.Claudepierre M.C., et al. Yeast virus-derived stimulator of the innate immune system augments the efficacy of virus vector-based immunotherapy. J. Virol. 2014;88:5242–5255. doi: 10.1128/JVI.03819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberson A., et al. NAB2 is a novel immune stimulator of MDA-5 that promotes a strong type I interferon response. Oncotarget. 2018;9:5641–5651. doi: 10.18632/oncotarget.23725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen G.Y., Nunez G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu T.Y. Strategies for designing synthetic immune agonists. Immunology. 2016;148:315–325. doi: 10.1111/imm.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kotenko S.V., Durbin J.E. Contribution of type III interferons to antiviral immunity: location, location, location. J. Biol. Chem. 2017;292:7295–7303. doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galani I.E., et al. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 77.Sleijfer S., et al. Side effects of interferon-alpha therapy. Pharm. World Sci. 2005;27:423–431. doi: 10.1007/s11096-005-1319-7. [DOI] [PubMed] [Google Scholar]
- 78.Muir A.J., et al. A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 2014;61:1238–1246. doi: 10.1016/j.jhep.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 79.Bedard K.M., et al. Isoflavone agonists of IRF-3 dependent signaling have antiviral activity against RNA viruses. J. Virol. 2012;86:7334–7344. doi: 10.1128/JVI.06867-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pattabhi S., et al. Targeting innate immunity for antiviral therapy through small molecule agonists of the RLR pathway. J. Virol. 2015;90:2372–2387. doi: 10.1128/JVI.02202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Probst P., et al. A small-molecule IRF3 agonist functions as an influenza vaccine adjuvant by modulating the antiviral immune response. Vaccine. 2017;35:1964–1971. doi: 10.1016/j.vaccine.2017.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tisoncik J.R., et al. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michelle D.T., Ashley M. An update on the NLRP3 inflammasome and influenza: the road to redemption or perdition? Curr. Opin. Immunol. 2018;54:80–85. doi: 10.1016/j.coi.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Pothlichet J., et al. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kyi C., et al. Therapeutic immune modulation against solid cancers with intratumoral poly-ICLC: a pilot trial. Clin. Cancer Res. 2018;24:4937–4948. doi: 10.1158/1078-0432.CCR-17-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feigenbaum A., et al. Singleton–Merten syndrome: an autosomal dominant disorder with variable expression. Am. J. Med. Genet. A. 2013;161A:360–370. doi: 10.1002/ajmg.a.35732. [DOI] [PubMed] [Google Scholar]
- 87.Crow Y.J., Manel N. Aicardi–Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 88.Lisnevskaia L., et al. Systemic lupus erythematosus. Lancet. 2014;384:1878–1888. doi: 10.1016/S0140-6736(14)60128-8. [DOI] [PubMed] [Google Scholar]
- 89.Fava A., Petri M. Systemic lupus erythematosus: diagnosis and clinical management. J. Autoimmun. 2018 doi: 10.1016/j.jaut.2018.11.001. Published online November 15, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Atkinson M.A., et al. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith T.J., Hegedus L. Graves’ disease. N. Engl. J. Med. 2016;375:1552–1565. doi: 10.1056/NEJMra1510030. [DOI] [PubMed] [Google Scholar]