Abstract
The live equine infectious anemia virus (EIAV) vaccine strain EIAVDLV121 was developed by in vitro attenuation of a virulent strain, EIAVLN40, in the 1970s, and it has been demonstrated to induce protective immunity under laboratory and natural EIAV infection conditions. The detailed biological features of this attenuated virus remain to be further investigated. Experimental inoculation with EIAVDLV121 did not result in clinical symptoms even with immunosuppressive treatment in our previous studies. Here, we further investigated whether the replication of the vaccine strain EIAVDLV121 in experimentally infected horses causes histopathological lesions to develop in the targeted organs. Both the lungs and the spleen have been demonstrated to support EIAV replication. By evaluating the gross macroscopic and histological changes, we found that EIAVDLV121 did not cause detectable histopathological lesions and that it replicated several hundred times more slowly than its parental virulent strain, EIAVLN40, in tissues. Immunochemical assays of these tissues indicated that the primary target cells of EIAVDLV121 were monocytes/macrophages, but that EIAVLN40 also infected alveolar epithelial cells and vascular endothelial cells. In addition, both of these viral strains promoted the up- and down-regulation of the expression of various cytokines and chemokines, implicating the potential involvement of these cellular factors in the pathological outcomes of EIAV infection and host immune responses. Taken together, these results demonstrate that the EIAV vaccine strain does not cause obvious histopathological lesions or clinical symptoms and that it induces a unique cytokine response profile. These features are considered essential for EIAVDLV121 to function as an effective live vaccine.
Keywords: Horses, Equine infectious anemia virus, Pathological lesions, Macrophages, Cytokine expression
1. Introduction
Equine infectious anemia (EIA), which is caused by equine infectious anemia virus (EIAV), is a worldwide disease of equids that is often endemic (Cook et al., 2013, Leroux et al., 2004). Persistent infection can occur in horses, characterized by recurring cycles of viremia and clinical symptoms, including fever, anemia, ventral edema, thrombocytopenia and general wasting (Leroux et al., 2004). Most EIAV-infected horses typically experience a classical disease process, which is divided into three stages, including acute, chronic and asymptomatic stages (Cook et al., 2013, Leroux et al., 2004). Published studies have consistently confirmed that during the acute phase, persistent replicative sites of EIAV are macrophage-rich tissues, including the spleen, bone marrow, liver, kidney and lymph nodes. Peripheral blood mononuclear cells (PBMCs) and other tissues contain fewer viruses in horses infected with the wild-type virulent strain EIAVWyo (Kono et al., 1971, Sellon et al., 1992). The primary target cells for EIAV replication are of the monocyte/macrophage lineages. Macrovascular endothelial cells can also be infected during the acute stage (Harrold et al., 2000, Maury et al., 1998, Oaks et al., 1999). However, during the asymptomatic stage, although the spleen and liver have low levels of viral replication, the virus is usually undetectable in PBMCs and other tissues of horses infected with the weakly virulent tissue culture-adapted strain EIAVPV (Harrold et al., 2000). In addition, viral mRNA has been found in tissue macrophages during the asymptomatic stage (Oaks et al., 1998). Therefore, there are differences in the tissue distribution and cellular tropism between the acute and asymptomatic phases of EIAV infection.
The live attenuated vaccine strain EIAVDLV121 was developed in the 1970s by successively culturing the pathogenic strain EIAVDV117, which was adapted from a horse-tropic virulent strain, EIAVLN40, and is pathogenic to both horses and donkeys, in donkey monocyte-derived macrophages (MDMs) for 121 passages. EIAVDLV121 was extensively applied in China for the vaccination of horses, mules and donkeys and controlled the pandemic of EIA(Shen, 1983). This vaccine strain was further adapted in donkey dermal cells (FDD) to improve the induction of immune protection and facilitate manufacturing (Shen et al., 2001).
Whole-blood transfer from horses in the asymptomatic state to naïve animals has been shown to result in EIAV infection and disease in recipient horses (Issel et al., 1982), and the treatment of horses in the asymptomatic state with immunosuppressive drugs such as dexamethasone has been reported to lead to the recrudescence of disease (Kono et al., 1976). These results demonstrate that this long-term unapparent or asymptomatic state is reversible. However, the same approach of immunosuppression did not induce either a significant boost in the plasma viral load or active EIA in horses infected with an attenuated EIAV vaccine strain (Ma et al., 2009, Shen and Wang, 1985). The above results suggests that the live EIAV vaccine strain has a unique biological features. Tissue or cell tropism is an important characteristic of viruses that affects their pathogenesis. It is therefore of interest to examine whether the cellular tropism and replication of this vaccine strain in tissues are different from those of other EIAV strains.
The abnormal expression of cytokines and chemokines has been demonstrated in infections with lentiviruses, including human immunodeficiency virus-1 (HIV-1), feline immunodeficiency virus (FIV) and simian immunodeficiency virus (SIV), and has been shown to contributes to systemic tissue damage (Biancotto et al., 2007, Qin et al., 2010, Scott et al., 2011). The expression levels of IL-2 and IL-15 have been reported to be increased in lymphoid tissues in chronic HIV-1 infection, resulting in lymphadenopathy (Biancotto et al., 2007). A study of FIV-infected cats has demonstrated that cytokine dysregulation in early- and late-term placentas dramatically impacts pregnancy outcome and that the viral load during late pregnancy is positively correlated with IL-6 expression (Scott et al., 2011). In addition, SIV infection alters the chemokine networks and local immune environments in the lungs, which are critical to the outcome of viral infection (Qin et al., 2013). The pulmonary symptoms associated with SIV infection are closely associated with the increased expression of MCP-1 and CXCL10 in the lungs of rhesus macaques (Qin et al., 2010). EIAV infection also results in interstitial pulmonary pathology similar to that observed in HIV-infected children (Doffman and Miller, 2013), SIV-infected macaques (Qin et al., 2010) and FIV-infected cats (Bolfa et al., 2013, Cadore et al., 1997). In vitro infection of primary monocytes/macrophages also induces the differential expression of several cytokines (e.g., IL-1α, IL-1β, IL-4, IL-10, IFN-α and TNF-α) and chemokines (e.g., IL-8, MCP-1, MCP-2, MIP-1α, MIP-1β and IP-10) between the EIAV virulent strain EIAVLN40 and the vaccine strain EIAVDLV121 (Lin et al., 2011). However, little is known about the cytokine responses in tissues from EIAVLN40- or EIAVDLV121-infected horses.
Comparative virology and immunology studies between Chinese pathogenic strains and partial or completely attenuated strains have been extensively performed in our laboratory for 10 years (Lin et al., 2011, Liu et al., 2015, Ma et al., 2014, Wang et al., 2008). To better understand the differences in tissue tropism and pathogenesis between Chinese pathogenic strains and completely attenuated strains, six horses (three in each group) were inoculated with either the pathogenic EIAVLN40 strain or completely attenuated EIAVDLV121 vaccine strain. An additional three horses were placed into a negative control group. The pathological damage, replication status, and cellular tropism, as well as host responses in the lungs and spleens of infected animals, were examined.
2. Materials and methods
2.1. Animals
Nine two-year-old male horses were used in this study. All horses were negative for equine arteritis virus (EAV), equine influenza virus (EIV) and equine herpes virus (EHV) at the initiation of the study. EIAV infection was screened three months prior to the experiment using an agar gel immunodiffusion test (AGID) (VMRD, USA) three times with an interval of one month. All animal experiments were carried out under approved animal handling protocols and according to the guidelines of the Animal Management Committee of Chinese Harbin Veterinary Research Institute.
2.2. Viruses and infections
The EIAV strains used in this study were stored at the Harbin Veterinary Research Institute. The horses were randomly separated into three groups. The three horses in Group A (A1–A3) were subcutaneously inoculated with 1 × 105 TCID50 (50% tissue culture infective doses) of the vaccine strain EIAVDLV121. The horses inoculated with this dose of EIAVDLV121 developed protective immunity to challenge with EIAVLN40 (Shen and Wang, 1985, Shen, 1983). Group B contained three (B1–B3) horses that were inoculated with the same titer of the pathogenic strain EIAVLN40 following a previously described protocol (Shen and Wang, 1985; Shen, 1983). The last three horses (Group C, including C1, C2 and C3) were injected with the same volume of saline as the negative controls. The horses in each group were separately fed, and the breeding conditions were controlled to ensure for animal welfare.
2.3. Sample collection
The body temperature, platelets counts and clinical symptoms of the horses inoculated with EIAV were monitored for 30 days. Blood samples were obtained, and tissues were collected for histopathological, virological and cytokine assays on the day of euthanization. PBMCs were separated from the blood as previously described (Lin et al., 2011). Three different regions of the lungs and spleen were sampled. All tissue samples were obtained in duplicate, one of which was fixed in 10% neutralized phosphate-buffered (PB) formalin for histopathology assays, and the other of which was stored separately at −80 °C for viral load and cytokine analyses.
2.4. Histopathology and immunohistochemistry
After fixation for 24 h, all tissues were processed for routine histology. Tissue blocks were sliced into 5 μm sections and then stained with standard hematoxylin and eosin (H&E). Immunohistochemical staining was performed to detect the viral p26 antigen in the tissues according to a published protocol (Bolfa et al., 2013). In the present study, the monoclonal anti-p26 antibody CA26, prepared in this laboratory, was used as the primary antibody and a goat anti-mouse IgG labeled with horseradish peroxidase was used the secondary antibody (Beyotime, China). DAB (Diaminobenzidine) (Beyotime, China) was used as a chromogen, and hematoxylin (Beyotime, China) was used as a counterstain. The controls for specificity included uninfected and infected tissue sections that were not incubated with the primary antibody. All tissue sections were examined by light microscopy (Leica, Germany).
2.5. RNA extraction and reverse transcription
Tissue homogenates were prepared by grinding tissue samples in liquid nitrogen in a mortar; then, the total RNA was extracted from of the tissue samples using Trizol regent (Invitrogen, USA), according to the manufacturer's instructions. The total RNA was treated to remove genomic DNA using a PrimeScript RT reagent Kit with gDNA Eraser from a Perfect Real Time Kit (Takara, Japan) prior to undergoing reverse transcription. Briefly, to remove genomic DNA, a reaction mixture containing 2 μl of total RNA (1 μg), 2 μl 5 × gDNA buffer, 1 μl gDNA Eraser and 5 μl DEPC-treated water was prepared. The reaction conditions were as follows: 42 °C for 2 min, followed by storage at 4 °C. Reverse transcription was performed in a total volume of 20 μl; 10 μl of reverse-transcription mix was added to the reaction solution from the above step, which included 4 μl of 5 × PrimeScript buffer 2 (for real-time PCR), 1 μl PrimeScript RT Enzyme Mix I, 1 μl RT Primer Mix and 4 μl RNase-Free dH2O. The reaction conditions were as follows: 37 °C for 15 min; 85 °C for 5 s; and storage at 4 °C. Three independent RNA extractions were performed for each tissue site, and the resulting cDNA was used as the corresponding template for viral transcripts amplification and tissue viral load and cytokine analyses. These samples were stored at −80 °C until use.
2.6. Nested PCR
Viral transcripts were amplified by performing reverse transcription of the cDNA templates. The nested PCR primers used, including the outer and inner primers, and the annealing temperature for each primer set are indicated in Supplementary Table 2. PCR was performed in a 50 μl reaction volume using a Takara LA PCR Amplification Kit Version 2.1 (Takara, Japan) following the manufacturer's instructions. Sterile distilled water and cDNA templates prepared from healthy tissues were used as the blank and negative controls, respectively. All PCR products were analyzed by 1% agarose gel electrophoresis.
2.7. Viral loads in tissues
EIAV RNA in the lungs and spleens was measured using quantitative real-time PCR (qRT-PCR) and presented as viral copy numbers per μg of total tissue RNA. Standards were prepared as previously described (Wei et al., 2012). The reaction was performed in a 96-well optical plate (Roche, Switzerland) in a 20 μl volume, which included 1 μl cDNA template, 10 μl SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara, Japan), 1 μl PCR forward primer (10 μM), 1 μl PCR reverse primer (10 μM) and 7 μl RNase-free water. The reaction conditions were as follows: 95 °C for 30 s and 40 cycles of 95 °C for 5 s (the annealing temperature used for each gene is indicated in Supplementary Table 2) and a final extension step at 72 °C for 20 s. The samples were tested in quadruplicate. Data were collected and analyzed with an MxPro3005p qPCR system (Stratagene, USA).
2.8. Quantitative analysis of cytokines
The expression levels of IFN-α, IFN-β, IFN-γ, IL-1α, IL-1β, IL-6, IL-8, IL-10, CXCL10, MX1, CXCL12 and CCL21 in the lungs and spleens were determined by RT-PCR. Briefly, RNA was extracted as described above. The samples were assessed in quadruplicate, and β-actin was used as the reference mRNA. The target mRNA samples were run in parallel on the same plate. The reaction conditions were the same as those described for the qRT-PCR of EIAV genomic RNA. The relative cytokine expression levels were calculated from the normalized ΔC T values. To determine the differentially expressed target genes in the horses infected with either EIAVDLV121 or EIAVLN40, the fold changes in cytokine expression levels from the tissues of the EIAV-infected horses were compared to those of uninfected horses, which served as the calibrator, as calculated using the 2−ΔΔCT method (Abel et al., 2002).
2.9. Statistical analysis
All statistical analyses of the real-time RT-PCR data, including the viral loads and cytokine mRNA levels in the PBMCs and tissues from the EIAV-infected horses, were performed to compare the experimental groups using two-tailed Student's t test. The results were considered statistically significant or very significant when P values were less than 0.05 or 0.01, respectively.
Spearman's correlation coefficients (r) were determined between the viral loads and the fold changes of specific cytokines in the samples obtained from the same tissue site. All analyses were performed using SPSS 16.0 software (IBM, USA), and all analytic graphs were constructed using GraphPad Prism (GraphPad Software, USA).
3. Results
3.1. Obvious pathological changes occurred in tissues from horses infected with EIAVLN40 but not with EIAVDLV121
To examine the pathogenicity of the vaccine strain EIAVDLV121 in horses, we experimentally infected three horses (Group A) with 1 × 105 TCID50 of this strain. Another two groups of horses (Group B and Group C, three horses each) were inoculated either with the same volume of the pathogenic strain EIAVLN40 or the saline as the positive and negative controls, respectively. All three of the horses infected with the EIAVLN40 (B1–B3) developed active EIA (body temperature >39 °C, platelet count < 100,000/μl, and plasma viral load > 1 × 106/ml) within 1 month post-infection and were euthanized at 30 days post-infection. No detectable EIA-associated symptoms were observed in the horses from the other two groups (Groups A and C; Supplementary Table 1).
Clear alveolar outlines and normal bronchial structures in the lungs (Fig. 1 A and E) and a normal profile of the white pulp and moderate cell density in the red pulp in the spleens were observed in the horses in Groups A and C (Fig. 1B and F). In contrast, alveolar wall thickening (arrowheads), severe histological inflammation, the emergence of macrophages, cell degeneration and necrosis of the alveolar wall (arrows) were identified in the lungs of the horses infected with EIAVLN40 (Fig. 1C). Furthermore, serious hyperemia was observed in the red pulp (arrowheads), and necrotic zones of varable sizes appeared in the white pulp, within which either the structures of blurred cytoplasm or ruptured nuclei due to chromatin dissolution in the lymphocytes were observed in the spleens from EIAVLN40-infected horses (arrows) (Fig. 1D).
Fig. 1.
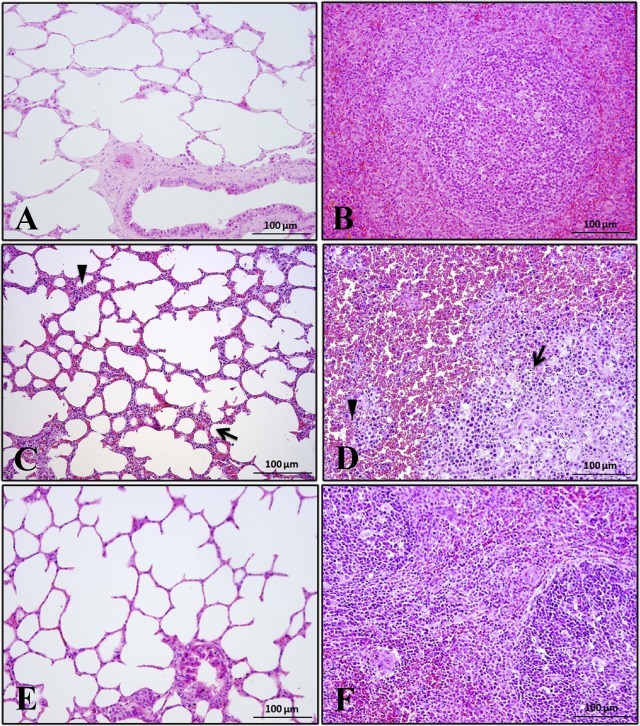
Histopathological observation of lungs and spleens. Tissue sections from horses infected with EIAVDLV121 (A and B) or EIAVLN40 (C and D) and uninfected negative controls (E and F) were stained with hematoxylin and eosin (H&E). Arrowheads indicate alveolar wall thickening; and arrows indicate severe histological inflammation and the emergence of macrophages, cell degeneration and necrosis of the alveolar walls (C). Arrowheads indicate serious hyperemia in the red pulp; and arrows indicate necrotic zones of variable sizes that appeared in the white pulp (D). The magnification of the lung is 100× and that of the spleen is 100×.
3.2. EIAVDLV121 infected a different panel of target cells at a much lower replication level compared with EIAVLN40
The capsid protein (p26), which contains most antigenic epitopes for T cell-mediated immune responses, is the most abundant protein of EIAV (Alvarez et al., 2007, Hu et al., 2014). Immunohistology for p26 revealed that in addition to being detected in the alveolar walls in the lungs (Fig. 2 A and C) and the white pulp in the spleens (Fig. 2B and D) from the EIAVDLV121- and EIAVLN40-infected horses, this viral protein was found in the vessels of both tissues from the EIAVLN40-infected horses (Fig. 2C and D). According to cell morphology and location, the p26 antigen was mainly distributed in alveolar epithelial cells and vascular endothelial cells in the lungs (Fig. 2C) and in vascular endothelial cells in the spleens of the horses infected with EIAVLN40 (Fig. 2D), as well as in macrophages in the alveolar walls of the lungs (Fig. 2A and C) and in the macrophages in the spleens (Fig. 2B and D) of both the EIAVDLV121-infected and EIAVLN40-infected horses. No signals were detected in the lungs or spleens of the negative control group C (Fig. 2E and F).
Fig. 2.
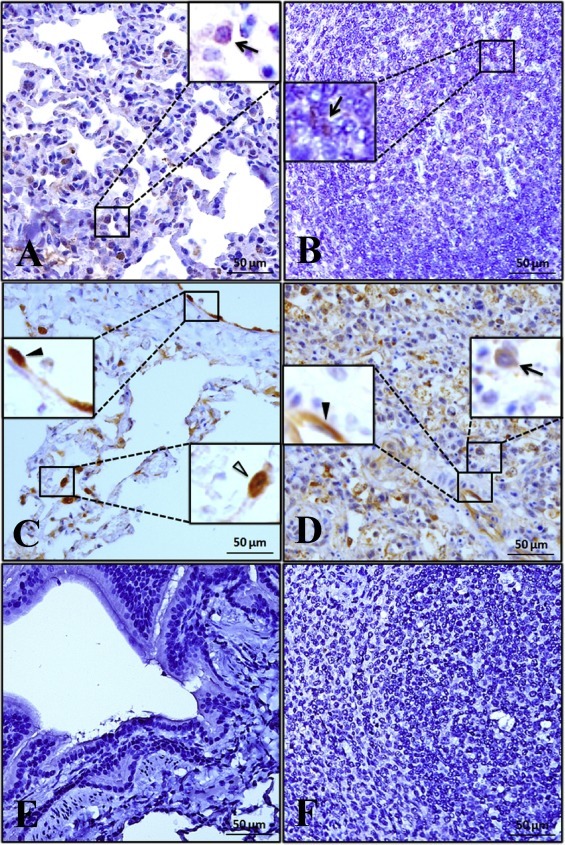
Immunohistochemistry of lungs and spleens. Immunohistochemical assays of the EIAV p26 capsid protein in these tissues from horses infected with either EIAVDLV121 (A and B) or EIAVLN40 (C and D) or from uninfected horses (E and F) were performed using a p26 monoclonal antibody and a horseradish peroxidase-conjugated secondary antibody. The arrows show the positively stained macrophages. The solid and hollow arrowheads indicate positively stained vascular endothelial cells and alveolar epithelial cells, respectively. The magnification is 50×.
The level of viral load in the host are considered as thresholds for the type of host responses characterized as thresholds of virus detection, seroconversion, vaccine and disease (Whitney and Ruprecht, 2004). Horses infected with EIAV exhibit typical clinical symptoms when the circulating viral load reaches 107 copies/ml or higher (Cook et al., 2003). Therefore, we detected the EIAV RNA copy numbers in the indicated tissues to evaluate their correlations with the pathological changes in these tissues. To compare the cell-associated viral RNA levels in the tissues between the EIAVDLV121-infected and EIAVLN40-infected horses, viral RNA was quantified by qRT-PCR. Viral RNA was detected in the lungs and spleens of all three EIAVDLV121-inoculated horses in Group A, with mean levels of 1.1 × 103 and 2.8 × 103 viral copies/μg total RNA, respectively (Fig. 3 A). The mean viral RNA level in the PBMCs was significantly higher (1.2 × 104 viral copies/μg of total RNA) (Fig. 3A). Significant differences were observed in the viral loads between the PBMCs and the lungs for Group A (P < 0.01). However, in the EIAVLN40-infected horses (Group B), the mean viral RNA levels were 4.4 × 105 and 2.0 × 106 copies/μg total RNA in the lungs and spleens, respectively, and a higher level (6.5 × 106 viral copies/μg of total RNA) was observed in the PBMCs (Fig. 3B). Furthermore, there were no significant differences in the viral loads among the tissues of Group B. However, the viral RNA levels in the lungs, spleens and PBMCs of the EIAVLN40-infected horses were significantly higher than those of the EIAVDLV121-infected horses (P < 0.01 for all three).
Fig. 3.
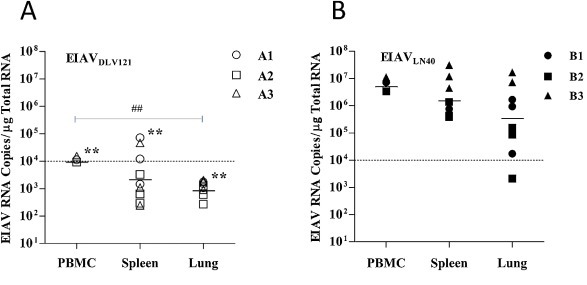
Detection of EIAV RNA in peripheral blood mononuclear cells (PBMCs), lungs and spleens. Total RNA was extracted from these tissues. Levels of cell-associated EIAV RNA were quantitated by amplifying the gag gene fragment using real-time (RT)-PCR in tissue samples from animals infected with either EIAVDLV121 (A) or EIAVLN40 (B). Three samples from each lung and spleen were measured. *, P < 0.05 and **, P < 0.01 compared with the corresponding sample between groups. #, P < 0.05 and ##, P < 0.01 compared with the different samples within the same group. The triangle, square and circle represent different animals, respectively. The results for PBMCs from each horse are expressed as the mean values and those for each tissue site are expressed as the means ± SD (n = 4).
To confirm the virus replication in the lungs and spleens of the EIAVDLV121-inoculated horses, nested PCR was performed to amplify the viral transcripts, including gag, pol and env. The three transcripts were clearly detected in the lung and spleen tissues from all EIAV-infected horses (Fig. S1A and B). These results indicated that viral RNAs of both the vaccine and virulent strains were most likely present in their full-length forms in the tissues, suggesting the synthesis of viral genomes. None of the negative control animals tested positive for viral RNA.
3.3. Infection with EIAVDLV121 and EIAVLN40 differentially regulated the expression of multiple cytokines and chemokines
EIAVLN40 promotes the up- and down-regulation of the expression of different cytokines and chemokines that are critically involved in host responses, including immunity and pathogenesis. Our previous study has revealed that the expression levels of cytokines and chemokines differ in cultivated MDMs in the presence of pathogenic versus attenuated EIAV strains (Lin et al., 2011).
To investigate whether the EIAV vaccine strain EIAVDLV121 also induced different cytokine responses from those induced by the pathogenic strain EIAVLN40 in vivo, the relative mRNA expression levels of eight cytokines (IL-1α, IL-1β, IL-6, IL-10, IFN-α, IFN-β, IFN-γ, and MX1) and four chemokines (IL-8, CCL21, CXCL10 and CXCL12) were examined in the lungs and spleens of horses inoculated with these viruses, and these levels were compared with those of the uninfected control group. According to the amplification curve, all PCR efficiencies were >95%, demonstrating the similar amplification efficiencies among the target (cytokines) and reference (β-actin) mRNAs.
In both the EIAVDLV121- and EIAVLN40-infected lungs and spleens, the mean fold changes of the IL-1α, IL-6, IL-8 and CXCL10 mRNA levels were increased (by more than two-fold) relative to the uninfected group, but the fold changes in the EIAVDLV121-inoculated horses were much lower than those in the EIAVLN40-infected horses, especially those of IL-6 and IL-8, which differed by close to or over a two-fold. In addition, the expression levels of IL-1β, IL10, IFN-α, IFN-β and CXCL12 in EIAVLN40-infected lungs and spleens, and only those of IL-1β, IFN-α, IFN-β in the EIAVDLV121-infected spleens, were enhanced following the EIAV infection (Table 1 ).
Table 1.
Measurement of cytokine and chemokine mRNA expression levels in tissues by quantitative real-time PCR.
| mRNA | Fold changea |
Pd |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen (n = 9) |
Lung (n = 9) |
Intra-group (lung versus spleen) |
Inter-group (EIAVLN40 versus EIAVDLV121) |
||||||||
| EIAVLN40 versus UI |
EIAVDLV121 versus UI |
EIAVLN40 versus UI |
EIAVDLV121 versus UI |
EIAVLN40 | EIAVDLV121 | Lung versus lung |
Lung versus spleen |
Spleen versus lung |
Spleen versus spleen |
||
| Cytokines (increased)b |
IL-1α | 16.9 ± 9.8 | 4.3 ± 2.2 | 7.5 ± 2.3 | 3.5 ± 0.9 | NS | NS | NS | NS | NS | NS |
| IL-1β | 14.8 ± 3.9 | 2.9 ± 0.6 | 33.2 ± 17.3 | 1.0c ± 0.1 | NS | 0.008 | NS | NS | 0.003 | 0.009 | |
| IL-6 | 269.9 ± 90.5 | 2.9 ± 0.4 | 149.4 ± 64.7 | 2.9 ± 1.2 | NS | NS | 0.04 | 0.04 | 0.009 | 0.009 | |
| IL-10 | 25.7 ± 12.8 | 1.4c ± 0.4 | 4.2 ± 1.0 | 1.3c ± 0.3 | NS | NS | 0.014 | 0.019 | NS | NS | |
| IFN-α | 323.8 ± 228.0 | 2.6 ± 1.4 | 9.3 ± 2.9 | 2.0 ± 1.1 | NS | NS | 0.009 | NS | NS | NS | |
| IFN-β | 28.1 ± 13.2 | 4.0 ± 2.0 | 1325.0 ± 872.8 | 1.5c ± 0.3 | NS | NS | NS | NS | NS | NS | |
| IFN-γ | 1.2c ± 0.3 | 33.4 ± 7.7 | 14.2 ± 4.0 | 14.8 ± 4.0 | 0.002 | 0.04 | NS | 0.04 | 0.004 | <0.001 | |
| MX1 | 5.1 ± 1.0 | 4.8 ± 1.3 | 1.4c ± 0.3 | 3.8 ± 0.9 | 0.003 | NS | 0.02 | 0.019 | NS | NS | |
| Chemokines (increased)b |
IL-8 | 535.6 ± 99.0 | 4.5 ± 1.3 | 318.7 ± 111.9 | 4.3 ± 2.2 | NS | NS | <0.001 | <0.001 | 0.01 | 0.01 |
| CXCL10 | 147.7 ± 95.2 | 4.3 ± 0.5 | 31.8 ± 13.0 | 3.3 ± 0.5 | NS | NS | 0.034 | 0.04 | NS | NS | |
| Anti-apoptotic chemokinesb | CXCL12 | 48.2 ± 37.8 | 1.4c ± 0.3 | 2.3 ± 0.8 | 17.2 ± 2.8 | NS | <0.001 | 0.013 | NS | NS | NS |
| CCL21 | 17.0 ± 9.7 | 1.2c ± 0.6 | 1.5c ± 0.6 | 4.7 ± 0.9 | NS | NS | NS | NS | NS | NS | |
UI: uninfected.
NS: not significant.
The arithmetic mean of the fold change of the expression level target gene relative to that of uninfected animals, and data are the mean ± SEM.
Increased expression level of cytokines and chemokines with a fold change >2.
Unchanged expression level was defined as fold change <2. Underlined data: decreased expression level with a fold change >2.
Student's t-tests were used for all comparisons.
EIAVDLV121 makedly induced the expression of some cytokines, which differed from those induced by EIAVLN40. The CCL21 and CXCL12 mRNA levels were decreased by 4.7- and 17.2-fold, respectively, in the EIAVDLV121-infected lungs, but they were up-regulated by 2.3- and 1.5-fold by EIAVLN40. Notably, the vaccine strain enhanced IFN-γ mRNA expression by a mean of 33.4-fold in the spleens, whereas the pathogenic EIAVLN40 strain had almost no effect on IFN-γ mRNA expression (1.2-fold). Similarly, the expression of MX1 in the lungs was significantly higher than that in the EIAVLN40-infected group, and the difference in the mean fold change was 2.4 (Table 1).
Host immunity and inflammation are considered to be associated with the proliferation status of an infecting virus. We next examined the possible correlation between the levels of EIAV replication and cytokine expression at corresponding sites in the same tissues infected with either EIAVDLV121 or EIAVLN40. In the EIAVDLV121-inoculated horses, the viral RNA copy numbers were positively correlated with the mRNA expression levels of IFN-α (r = 0.663, P = 0.003) and IL-10 (r = 0.508, P = 0.031) (Fig. 4 and Table 2 ). In the EIAVLN40-infected group, IL-6 was positively correlated with the viral RNA copy numbers (r = 0.660, P = 0.003), but two cytokines, IFN-β (r = −0.689, P = 0.002) and IL-1β (r = −0.687, P = 0.002), were negatively correlated (Table 2). In addition, chemokines mediate cell migration and the recruitment of leukocytes, which process and secrete cytokines and chemokines, which regulate distinct intracellular signaling events (Baggiolini, 1998; Rollins, 1997). Thus, we examined the possible correlations among the expression of cytokines/chemokines at corresponding sites. In the EIAVDLV121-inoculated horses, the expression of IL-1α was positively correlated with that of IL-10 (r = 0.530, P = 0.024) and MX1 (r = 0.623, P = 0.006), and the expression of CXCL10 and IFN-γ was positively correlated with that of CCL21 (r = 0.485, P = 0.041 and r = 0.484, P = 0.042, respectively). In addition, the expression of CXCL12 was positively correlated with that of IFN-β (r = 0.495, P = 0.037) and IL-1β (r = 0.765, P < 0.001). In the EIAVLN40-inoculated horses, the expression of IFN-β was positively correlated with that of IL-1β (r = 0.993, P < 0.001), and the expression of IL-1α was positively correlated with that of CCL21 (r = 0.881, P < 0.001). Additionally, the expression of MX1 was positively correlated with that of IFN-α (r = 0.501, P = 0.034) but negatively correlated with that of IFN-γ (r = −0.486, P = 0.041) (Table 2).
Fig. 4.
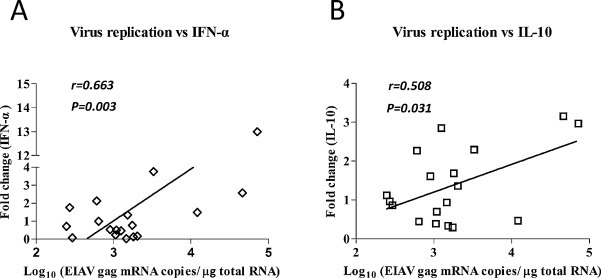
An example of correlation analyses between cytokine mRNA expression levels and viral RNA copy numbers in tissues from EIAVDLV121-infected horses. EIAV genomic RNA and IFN-α (A) and IL-10 (B) mRNA were quantified by RT-PCR, as described in Fig. 3. Spearman's correlation coefficients (statistically significant: 0.5 < r < 0.8) and P values (significant: P < 0.05, very significant: P < 0.01) are provided in each graph.
Table 2.
Correlations between relative mRNA expression levels of the cytokines in tissues during EIAV infection.
| Group | Gene-specific mRNA |
Comparison gene-specific mRNA |
Pearson's correlation coefficient (P value)a |
|---|---|---|---|
| EIAVDLV121 | EIAV | IFN-α | 0.663 (0.003) |
| IL-10 | 0.508 (0.031) | ||
| IFN-α | EIAV | 0.663 (0.003) | |
| IL-10 | 0.521 (0.026) | ||
| IFN-β | CXCL12 | 0.495 (0.037) | |
| IFN-γ | CCL21 | 0.484 (0.042) | |
| IL-1α | IL-10 | 0.53 (0.024) | |
| MX1 | 0.623 (0.006) | ||
| IL-1β | CXCL12 | 0.765 (<0.001) | |
| IL10 | EIAV | 0.508 (0.031) | |
| IFN-α | 0.521 (0.026) | ||
| IL-1α | 0.53 (0.024) | ||
| MX1 | IL-1α | 0.623 (0.006) | |
| CXCL10 | CCL21 | 0.485 (0.041) | |
| CXCL12 | IFN-β | 0.495 (0.037) | |
| IL-1β | 0.765 (<0.001) | ||
| CCL21 | IFN-γ | 0.484 (0.042) | |
| CXCL10 | 0.485 (0.041) | ||
| EIAVLN40 | EIAV | IFN-β | −0.689(0.002) |
| IL-1β | −0.687(0.002) | ||
| IL6 | 0.66 (0.003) | ||
| IFN-α | MX1 | 0.501 (0.034) | |
| IFN-β | EIAV | −0.689(0.002) | |
| IL-1β | 0.993 (<0.001) | ||
| IFN-γ | MX1 | −0.486 (0.041) | |
| IL-1α | CCL21 | 0.881 (<0.001) | |
| IL-1β | EIAV | −0.687(0.002) | |
| IL6 | EIAV | 0.66 (0.003) | |
| MX1 | IFN-α | 0.501 (0.034) | |
| IFN-γ | −0.486 (0.041) | ||
| CCL21 | IL-1α | 0.881 (<0.001) | |
Only Pearson's correlations for which P < 0.05 are shown.
4. Discussion
In this study, we observed that the excessive responses of several cytokines were associated with a high level of proliferation of the pathogenic strain EIAVLN40 and multiple severe organ lesions. These cellular factors were mainly proinflammatory cytokines, antiviral factors and chemokines, primarily including IL-6, IL-8, IFN-α, IFN-β, CXCL10 and CXCL12. IL-6 and IL-8 (also called CXCL8) are both pro-inflammatory cytokines that attract and activate immunocytes, including T cells, B cells and phagocytes (Baggiolini and Clark-Lewis, 1992, Wong et al., 1988). Although they have important roles in inducing and regulating immune protection to invade microbes, these two cytokines, particularly IL-6, are largely involved in the development of lesions caused by overactive immune responses, such as those observed in infections of influenza virus (Cillóniz et al., 2009, de Jong et al., 2006), HIV-1, SIV and severe acute respiratory syndrome (SARS) coronavirus (Imai et al., 2008, Mukura et al., 2012, Roberts et al., 2004). IFN-α and IFN-β, which are both type I interferons that trigger potent innate immune responses in host defense systems (Isaacs and Lindenmann, 1957), play important roles in antiviral, antibacterial and anticancer responses. IFN-α/β activate immature dendritic cells and upregulate the expression of chemokine and chemokine receptors. IFN-α/β have been found to regulate adaptive immunity by promoting the generation of neutralizing antibodies and enhancing T cell-dependent antibody responses. The knockout of functional type I IFN receptor in mouse models has been shown to deactivate its associated signaling pathways (Trinchieri, 2010) and to dramatically increase the severity of symptoms in virus-infected mammalian hosts (Muller et al., 1994). In contrast, the vaccine strain EIAVDLV121 had a significantly lower replication level and induced a moderate elevation in the expression of cytokines tested in parallel compared with the pathogenic strain EIAVLN40, except for IFN-γ in the spleen and CXCL12 in the lungs. Therefore, the present results indicate that differences in EIAV pathogenicity are largely determined by differences in viral replication and the host responses in multiple organs, such as the spleen and lungs.
CXCL10, also termed IFNγ-induced protein 10 (IP-10), is produced in response to IFN-γ in monocytes, endothelial cells and fibroblasts to attract different types of immunocytes (Neville et al., 1997). CXCL10 expression has been reported to be increased in an encephalitic brain and lungs affected directly and indirectly by infection of simian/human immunodeficiency virus (SHIV) or SIV (Sasseville et al., 1996). CXCL10 has also been shown to be associated with the development of pneumonia and to recruit additional inflammatory cells to the lungs during viral infection (Schaefer et al., 2006, Sui et al., 2005). Interstitial pneumonia has been reported in EIAV-infected horses, similar to SIV-infected macaques and HIV-1-infected humans (Bolfa et al., 2013). Up-regulation of CXCL10 expression coupled with pathological damage implicate the involvement of this chemokine in the development of pneumonia associated with EIAV infection. CXCL12, also known as stromal cell-derived factor 1 (SDF1), is an anti-apoptotic chemokine, as are CCL21 and CCL25. All of these chemokines are involved in enhancing cell survival by inactivation of members of the apoptotic pathways at the post-translational level (Banas et al., 2002, Guo et al., 2005, Qiuping et al., 2004). Down-regulation of the mRNA expression of these chemokines in the lungs and lymphoid tissues during SIV infection has been reported to contribute to the increased apoptosis of host cells (Qin et al., 2010, Qin et al., 2008). The 40- and 17-fold upregulation of CXCL12 and CCL21 mRNA expression, respectively, in the EIAVLN40-infected spleens and the 17-fold increase in CXCL12 mRNA expression in the EIAVDLV121-infected lungs observed in this study suggest that the host response is triggered by EIAV infection to reduce cellular apoptosis.
The present study revealed that both the pathogenic and vaccine strains replicated in some of the host tissues, however, they caused different clinical and histopathological outcomes. It has been reported that viral tropism, which refers to the specificity of a certain virus for a particular cell (cellular tropism) or tissue type (tissue tropism), is an important factor in determining the outcome of viral infection (McFadden et al., 2009). Consistent with other reports (Harrold et al., 2000, Maury et al., 1998, Oaks et al., 1998, Oaks et al., 1999), our data demonstrated that macrophages were the primary target cells of EIAV infection and replication in the EIAVDLV121-inoculated horses and that alveolar epithelial cells and vascular endothelial cells in the lungs and vascular endothelial cells in the spleen were also EIAV positive in EIAVLN40-infected horses. Previous studies have observed that multiple consensus mutations in macrophage- and fibroblast-adapted EIAV strains during the process of in vitro attenuation, especially in the LTR (Payne et al., 1999) and env regions(Wang et al., 2008). The LTR region contains the transcription initiation site and thus largely determines the cellular tropism (Payne et al., 1999). The discontinuous area in the C-terminus of the EIAV gp90 glycoprotein was identified as the binding domain of EIAV receptor (Sun et al., 2008, Zhang et al., 2008). Thus, it is reasonable to speculate that cellular tropism can be affected by the sequence variability among strains.
Notably, in contrast with the other investigated cytokines and chemokines, which were more strongly up-regulated by EIAVLN40, the mRNA expression of IFN-γ, an important regulator of immune responses, was effectively stimulated (an approximately 30-fold increase) in the spleen by EIAVDLV121 but fluctuated around the baseline of the uninfected control group in the spleens infected with EIAVLN40. These results are consistent with those of a study showing that the production of IFN-γ in the serum, together with that of IL-2 and IL-12, are markedly increased and are maintained at a high concentration at approximately 50 dpi in horses inoculated with the vaccine strain EIAVDLV121 but that they exhibit no or a low level of up-regulation in horses infected with EIAVLN40(Zhang et al., 2007). It is well accepted that the expression of IFN-γ is an important index of cell-mediated immunity, especially cytotoxic T lymphocyte (CTL) responses to lentiviral infections, including HIV-1 and SIV, and that it is correlated with vaccine-induced immunity (Martins et al., 2010, Roff et al., 2014). Furthermore, IFN-γ stimulates functional polarization of monocyte-derived macrophages into the classical M1 cells, which express high levels of pro-inflammatory cytokines and provide protection from viral and microbial infections (Mantovani et al., 2004, Mantovani et al., 2005). In addition, IFN-γ is a powerful inducer of programmed death-1 (PDL-1), which strongly attenuates immune reactions (Flies et al., 2011, Keir et al., 2008). We speculate that the enhancement of IFN-γ expression by the EIAV vaccine strain is involved in the protective immune response against the pathogenic strains and in the remarkable decrease in the number of inflammatory-related tissue lesions caused by EIAV infection.
Correlation analysis between the EIAV vRNA copy numbers and cytokine expression levels revealed that the vRNA copy number of attenuated EIAVDLV121 was positively correlated with IFN-α and IL-10 and that the pathogenic EIAVLN40 proliferation level was positively correlated with IL-6 and negatively correlated with IFN-β and IL-1β. These data indicate that although the several hundred-fold higher proliferation level of EIAVLN40 largely determined the significantly higher expression levels of a panel of cytokines/chemokines, as shown in Table 1, the differences in the sequences of double-stranded (ds)RNAs between the vaccine and pathogenic viral genomes may have had different effects on inducing the expression of some cytokines that play important roles in immune responses. In addition to the type of viral protein, the features of viral genomic materials also greatly influence intracellular signaling networks. We have previously demonstrated that compared with the similar proliferation level of the pathogenic strain, the EIAV vaccine strain induced the activation of Toll-like receptor 3 (TLR3) by approximately 10-fold, which is an intracellular pattern recognition receptor (PPR) that is specifically activated by dsRNA (Ma et al., 2014). It is assumed that the vaccine vRNA copy number-dependent induction of the expression of anti-viral IFN-α and anti-inflammatory IL-10 is partly responsible for the attenuated virulence of EIAVDLV121. In addition, the negatively correlation of the EIAVLN40 vRNA copy number with IL-1β expression suggests that the EIAV pathogenic strain has an inhibitory effect on this potent antiviral protein. This effect was demonstrated in the EIAVLN40 inoculated horses by the markedly increased upregulation of IL-1β expression in the lungs compared with the spleen, which is a major peripheral immune organ that harbors much more macrophages, which are the principal target cells of EIAV, and a ten-fold greater number of EIAV virions.
In summary, there were no visible pathological lesions resulting from infection observed in the horses experimentally infected with the attenuated vaccine strain, EIAVDLV121 in contrast with those infected with its parental pathogenic strain EIAVLN40, which causes severe hyperemia, hemorrhage and necrosis in multiple organs of infected animals. The following biological characteristics of the attenuated virus may have caused the differences in pathology: (i) the relatively low copy number and stable replication status in tissues; (ii) the different cellular tropism; and (iii) differences in the induction of cytokines and chemokines, both in the expression levels and spectra. Both the attenuated virulence and the appropriate induction of cytokine responses of the EIAV vaccine strain may provide the host with sufficient time (immune maturation) to develop protective immunity elicited by the correct antigenic stimulation by the vaccine.
Conflicts of interest
All authors report that there were no conflicts of interest in the preparation of this manuscript.
Acknowledgements
This study was supported by grants from the Chinese National Key Programs for Infectious Diseases (2012ZX10001-008) and the National Natural Science Foundation of China (31070809 to J-H Z and 31302066 to X-F W).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2016.01.006.
Contributor Information
Xi-Jun He, Email: hexijun@caas.cn.
Xiaojun Wang, Email: xjw@hvri.ac.cn.
Jian-Hua Zhou, Email: jianhua_uc@126.com.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Abel K., Alegria-Hartman M.J., Rothaeusler K., Marthas M., Miller C.J. The relationship between simian immunodeficiency virus RNA levels and the mRNA levels of alpha/beta interferons (IFN-alpha/beta) and IFN-alpha/beta-inducible Mx in lymphoid tissues of rhesus macaques during acute and chronic infection. J. Virol. 2002;76:8433–8445. doi: 10.1128/JVI.76.16.8433-8445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez I., Gutierrez G., Vissani A., Rodriguez S., Barrandeguy M., Trono K. Standardization and validation of an agar gel immunodiffusion test for the diagnosis of equine infectious anemia using a recombinant p26 antigen. Vet. Microbiol. 2007;121:344–351. doi: 10.1016/j.vetmic.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- Baggiolini M., Clark-Lewis I. Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 1992;307:97–101. doi: 10.1016/0014-5793(92)80909-z. [DOI] [PubMed] [Google Scholar]
- Banas B., Wörnle M., Berger T., Nelson P.J., Cohen C.D., Kretzler M., Pfirstinger J., Mack M., Lipp M., Grone H.J., Schlöndorff D. Roles of SLC/CCL21 and CCR7 in human kidney for mesangial proliferation, migration, apoptosis, and tissue homeostasis. J. Immunol. 2002;168:4301–4307. doi: 10.4049/jimmunol.168.9.4301. [DOI] [PubMed] [Google Scholar]
- Biancotto A., Grivel J.C., Iglehart S.J., Vanpouille C., Lisco A., Sieg S.F., Debernardo R., Garate K., Rodriguez B., Margolis L.B., Lederman M.M. Abnormal activation and cytokine spectra in lymph nodes of people chronically infected with HIV-1. Blood. 2007;109:4272–4279. doi: 10.1182/blood-2006-11-055764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolfa P., Nolf M., Cadore J.L., Catoi C., Archer F., Dolmazon C., Mornex J.F., Leroux C. Interstitial lung disease associated with equine infectious anemia virus infection in horses. Vet. Res. 2013;44:113. doi: 10.1186/1297-9716-44-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadore J.L., Steiner-Laurent S., Greenland T., Mornex J.F., Loire R. Interstitial lung disease in feline immunodeficiency virus (FIV) infected cats. Res. Vet. Sci. 1997;62:287–288. doi: 10.1016/s0034-5288(97)90206-x. [DOI] [PubMed] [Google Scholar]
- Cillóniz C., Shinya K., Peng X., Korth M.J., Proll S.C., Aicher L.D., Carter V.S., Chang J.H., Kobasa D., Feldmann F., Strong J.E., Feldmann H., Kawaoka Y., Katze M.G. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 2009;5:e1000604. doi: 10.1371/journal.ppat.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R.F., Cook S.J., Berger S.L., Leroux C., Ghabrial N.N., Gantz M., Bolin P.S., Mousel M.R., Issel C.J. Enhancement of equine infectious anemia virus virulence by identification and removal of suboptimal nucleotides. Virology. 2003;313:588–603. doi: 10.1016/s0042-6822(03)00351-9. [DOI] [PubMed] [Google Scholar]
- Cook R.F., Leroux C., Issel C.J. Equine infectious anemia and equine infectious anemia virus in 2013: a review. Vet. Microbiol. 2013;167:181–204. doi: 10.1016/j.vetmic.2013.09.031. [DOI] [PubMed] [Google Scholar]
- Doffman S.R., Miller R.F. Interstitial lung disease in HIV. Clin. Chest Med. 2013;34:293–306. doi: 10.1016/j.ccm.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Flies D.B., Sandler B.J., Sznol M., Chen L. Blockade of the B7-H1/PD-1 pathway for cancer immunotherapy. Yale J. Biol. Med. 2011;84:409–421. [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Hangoc G., Bian H., Pelus L.M., Broxmeyer H.E. SDF-1/CXCL12 enhances survival and chemotaxis of murine embryonic stem cells and production of primitive and definitive hematopoietic progenitor cells. Stem Cells. 2005;23:1324–1332. doi: 10.1634/stemcells.2005-0085. [DOI] [PubMed] [Google Scholar]
- Harrold S.M., Cook S.J., Cook R.F., Rushlow K.E., Issel C.J., Montelaro R.C. Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J. Virol. 2000;74:3112–3121. doi: 10.1128/jvi.74.7.3112-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Chang H., Ge M., Lin Y., Wang X., Guo W. Development of antigen capture ELISA for the quantification of EIAV p26 protein. Appl. Microbiol. Biotechnol. 2014;98:9073–9081. doi: 10.1007/s00253-014-6078-8. [DOI] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H.C., Wang H.L., Liu H.L., Sun Y., Pasparakis M., Kopf M., Mech C., Bavari S., Peiris J.S.M., Slutsky A.S., Akira S., Hultqvist M., Holmdahl R., Nicholls J., Jiang C.Y., Binder C.J., Penninger J.M. Identification of oxidative stress and toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B: Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- Issel C.J., Adams W.V., Jr., Meek L., Ochoa R. Transmission of equine infectious anemia virus from horses without clinical signs of disease. J. Am. Vet. Med. Assoc. 1982;180:272–275. [PubMed] [Google Scholar]
- de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J., Chau T.N.B., Hoang D.M., Chau N.V.V., Khanh T.H., Dong V.C., Qui P.T., Cam B.V., Ha D.Q., Guan Y., Peiris J.S.M., Chinh N.T., Hien T.T., Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y., Hirasawa K., Fukunaga Y., Taniguchi T. Recrudescence of equine infectious anemia by treatment with immunosuppressive drugs. Natl. Inst. Anim. Health Q. (Tokyo) 1976;16:8–15. [PubMed] [Google Scholar]
- Kono Y., Kobayashi K., Fukunaga Y. Distribution of equine infectious anemia virus in horses infected with the virus. Natl. Inst. Anim. Health Q. (Tokyo) 1971;11:11–20. [PubMed] [Google Scholar]
- Leroux C., Cadore J.L., Montelaro R.C. Equine infectious anemia virus (EIAV): what has HIV's country cousin got to tell us? Vet. Res. 2004;35:485–512. doi: 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- Lin Y.Z., Cao X.Z., Li L., Jiang C.G., Wang X.F., Ma J., Zhou J.H. The pathogenic and vaccine strains of equine infectious anemia virus differentially induce cytokine and chemokine expression and apoptosis in macrophages. Virus Res. 2011;160:274–282. doi: 10.1016/j.virusres.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang X.F., Ma J., He X.J., Wang X.J., Zhou J.H. Characterization of equine infectious anemia virus integration in the horse genome. Viruses. 2015;7:3241–3260. doi: 10.3390/v7062769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Jiang C., Lin Y., Wang X., Zhao L., Xiang W., Shao Y., Shen R., Kong X., Zhou J. In vivo evolution of the gp90 gene and consistently low plasma viral load during transient immune suppression demonstrate the safety of an attenuated equine infectious anemia virus (EIAV) vaccine. Arch. Virol. 2009;154:867–873. doi: 10.1007/s00705-009-0378-9. [DOI] [PubMed] [Google Scholar]
- Ma J., Wang S.S., Lin Y.Z., Liu H.F., Liu Q., Wei H.M., Wang X.F., Wang Y.H., Du C., Kong X.G., Zhou J.H., Wang X. Infection of equine monocyte-derived macrophages with an attenuated equine infectious anemia virus (EIAV) strain induces a strong resistance to the infection by a virulent EIAV strain. Vet. Res. 2014;45:82. doi: 10.1186/s13567-014-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Mantovani A., Sica A., Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Martins M.A., Wilson N.A., Reed J.S., Ahn C.D., Klimentidis Y.C., Allison D.B., Watkins D.I. T-cell correlates of vaccine efficacy after a heterologous simian immunodeficiency virus challenge. J. Virol. 2010;84:4352–4365. doi: 10.1128/JVI.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury W., Oaks J.L., Bradley S. Equine endothelial cells support productive infection of equine infectious anemia virus. J. Virol. 1998;72:9291–9297. doi: 10.1128/jvi.72.11.9291-9297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G., Mohamed M.R., Rahman M.M., Bartee E. Cytokine determinants of viral tropism. Nat. Rev. Immunol. 2009;9:645–655. doi: 10.1038/nri2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukura L.R., Ghosh M., Fahey J.V., Cu-Uvin S., Wira C.R. Genital tract viral load in HIV type 1-positive women correlates with specific cytokine levels in cervical-vaginal secretions but is not a determinant of infectious virus or anti-HIV activity. Aids Res. Hum. Retroviruses. 2012;28:1533–1539. doi: 10.1089/aid.2011.0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Neville L.F., Mathiak G., Bagasra O. The immunobiology of interferon-gamma inducible protein 10 kD (IP-10): a novel: pleiotropic member of the CXC chemokine superfamily. Cytokine Growth Factor Rev. 1997;8:207–219. doi: 10.1016/s1359-6101(97)00015-4. [DOI] [PubMed] [Google Scholar]
- Oaks J.L., McGuire T.C., Ulibarri C., Crawford T.B. Equine infectious anemia virus is found in tissue macrophages during subclinical infection. J. Virol. 1998;72:7263–7269. doi: 10.1128/jvi.72.9.7263-7269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks J.L., Ulibarri C., Crawford T.B. Endothelial cell infection in vivo by equine infectious anaemia virus. J. Gen. Virol. 1999;80(Pt. 9):2393–2397. doi: 10.1099/0022-1317-80-9-2393. [DOI] [PubMed] [Google Scholar]
- Payne S.L., La Celle K., Pei X.F., Qi X.M., Shao H., Steagall W.K., Perry S., Fuller F. Long terminal repeat sequences of equine infectious anaemia virus are a major determinant of cell tropism. J. Gen. Virol. 1999;80(Pt. 3):755–759. doi: 10.1099/0022-1317-80-3-755. [DOI] [PubMed] [Google Scholar]
- Qin S., Fallert Junecko B.A., Trichel A.M., Tarwater P.M., Murphey-Corb M.A., Kirschner D.E., Reinhart T.A. Simian immunodeficiency virus infection alters chemokine networks in lung tissues of cynomolgus macaques: association with Pneumocystis carinii infection. Am. J. Pathol. 2010;177:1274–1285. doi: 10.2353/ajpath.2010.091288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Junecko B.A., Lucero C.M., Klamar C.R., Trichel A.M., Murphey-Corb M.A., Tarwater P.M., Kirschner D.E., Reinhart T.A. Simian immunodeficiency virus infection potently modulates chemokine networks and immune environments in hilar lymph nodes of cynomolgus macaques. J. Acquir. Immune Defic. Syndr. 2013;63:428–437. doi: 10.1097/QAI.0b013e31828ac85f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Sui Y., Soloff A.C., Junecko B.A.F., Kirschner D.E., Murphey-Corb M.A., Watkins S.C., Tarwater P.M., Pease J.E., Barratt-Boyes S.M., Reinhart T.A. Chemokine and cytokine mediated loss of regulatory T cells in lymph nodes during pathogenic simian immunodeficiency virus infection. J. Immunol. 2008;180:5530–5536. doi: 10.4049/jimmunol.180.8.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiuping Z., Jei X., Youxin J., Wei J., Chun L., Jin W., Qun W., Yan L., Chunsong H., Mingzhen Y., Qingping G., Kejian Z., Zhimin S., Qun L., Junyan L., Jinquan T. CC chemokine ligand 25 enhances resistance to apoptosis in CD4+ T cells from patients with T-cell lineage acute and chronic lymphocytic leukemia by means of livin activation. Cancer Res. 2004;64:7579–7587. doi: 10.1158/0008-5472.CAN-04-0641. [DOI] [PubMed] [Google Scholar]
- Roff S.R., Noon-Song E.N., Yamamoto J.K. The significance of interferon-gamma in HIV-1 pathogenesis, therapy, and prophylaxis. Front. Immunol. 2014;4:498. doi: 10.3389/fimmu.2013.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E.S., Burudi E.M.E., Flynn C., Madden L.J., Roinick K.L., Watry D.D., Taffe M.A., Fox H.S. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. J. Neuroimmunol. 2004;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Rollins B.J. Chemokines. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- Sasseville V.G., Smith M.M., Mackay C.R., Pauley D.R., Mansfield K.G., Ringler D.J., Lackner A.A. Chemokine expression in simian immunodeficiency virus-induced AIDS encephalitis. Am. J. Pathol. 1996;149:1459. [PMC free article] [PubMed] [Google Scholar]
- Schaefer T.M., Fuller C.L., Basu S., Fallert B.A., Poveda S.L., Sanghavi S.K., Choi Y.K., Kirschner D.E., Feingold E., Reinhart T.A. Increased expression of interferon-inducible genes in macaque lung tissues during simian immunodeficiency virus infection. Microbes Infect. 2006;8:1839–1850. doi: 10.1016/j.micinf.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Scott V.L., Boudreaux C.E., Lockett N.N., Clay B.T., Coats K.S. Cytokine dysregulation in early- and late-term placentas from feline immunodeficiency virus (FIV)-infected cats. Am. J. Reprod. Immunol. 2011;65:480–491. doi: 10.1111/j.1600-0897.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon D.C., Perry S.T., Coggins L., Fuller F.J. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J. Virol. 1992;66:5906–5913. doi: 10.1128/jvi.66.10.5906-5913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Wang Z. EIAV: a National Review of Policies, Programs, and Future Objectives. American Quarter Horse Association; Amarillo: 1985. Development and use of an equine infectious anemia donkey leucocyte attenuated vaccine; pp. 135–148. [Google Scholar]
- Shen R.X. Development and use of an equine infectious anemia Donkey leucocyte attenuated vaccine. Proceedings of the International Symposium on Immunity to Equine Infectious anemia. 1983:21–53. [Google Scholar]
- Shen R.X., Wang Z.Y., Dong J.P., 2001. Fetal donkey dermal cells adapted attenuated vaccine of equine infectious anemia virus and its culture method. (Chinese patent No.01123620). In China (ed.).
- Sui Y., Li S., Pinson D., Adany I., Li Z., Villinger F., Narayan O., Buch S. Simian human immunodeficiency virus-associated pneumonia correlates with increased expression of MCP-1, CXCL10, and viral RNA in the lungs of rhesus macaques. Am. J. Pathol. 2005;166:355–365. doi: 10.1016/S0002-9440(10)62259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhang B., Jin J., Montelaro R.C. Binding of equine infectious anemia virus to the equine lentivirus receptor-1 is mediated by complex discontinuous sequences in the viral envelope gp90 protein. J. Gen. Virol. 2008;89:2011–2019. doi: 10.1099/vir.0.83646-0. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Type I interferon: friend or foe? J. Exp. Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.F., Jiang C.G., Guo W., Xiang W., Lv X.L., Zhao L.P., Wang F.L., Kong X.G., Zhang X.Y., Shao Y.M., Zhou J.H. Comparison of proviral genomes between the Chinese EIAV donkey leukocyte-attenuated vaccine and its parental virulent strain. Chin. J. Virol. 2008;24:443–450. [PubMed] [Google Scholar]
- Wei H.M., Wang X.F., Wang S.S., Du C., Liu H.F., Liu Q., Zhou J.H. The application of single-genome amplification and sequencing in genomic analysis of an attenuated EIAV vaccine. Chin. J. Virol. 2012;28:431–438. [PubMed] [Google Scholar]
- Whitney J.B., Ruprecht R.M. Live attenuated HIV vaccines: pitfalls and prospects. Curr. Opin. Infect. Dis. 2004;17:17–26. doi: 10.1097/00001432-200402000-00004. [DOI] [PubMed] [Google Scholar]
- Wong G.G., Witek-Giannotti J., Hewick R.M., Clark S.C., Ogawa M. Interleukin 6: identification as a hematopoietic colony-stimulating factor. Behring Inst. Mitt. 1988;83:40–47. [PubMed] [Google Scholar]
- Zhang B., Sun C., Jin S., Cascio M., Montelaro R.C. Mapping of equine lentivirus receptor 1 residues critical for equine infectious anemia virus envelope binding. J. Virol. 2008;82:1204–1213. doi: 10.1128/JVI.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang Y., Liang H., Wei L., Xiang W., Shen R., Shao Y. Correlation between the induction of Th1 cytokines by an attenuated equine infectious anemia virus vaccine and protection against disease progression. J. Gen. Virol. 2007;88:998–1004. doi: 10.1099/vir.0.82416-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


