Abstract
The nucleocapsid (N) gene of human coronavirus strain OC43 (HCoV-OC43) was amplified by reverse transcriptase-polymerase chain reaction, and cloned in pENTR™/D-TOPO® plasmid. This plasmid containing the N gene was recombined with in a BaculoDirect™ baculovirus DNA designed in order to express N protein in fusion with a C-terminal polyhistidine tag containing V5 epitope. Sf21 cells were transfected with recombinant baculovirus DNA. Recombinant N protein was extracted from infected cells, analysed by SDS-PAGE and Western blot, and purified by Ni2+ affinity procedure. Sera from 100 healthcare workers and five 2–3-year-old children were tested in a Western blot assay using the purified recombinant N protein. All of the sera from adults and two of the sera from children have a positive result.
Keywords: Coronavirus, OC43 strain, Nucleocapsid protein, Baculovirus, Insect cells, Western blot
1. Introduction
Coronaviruses (family Coronaviridae, order Nidovirales) are enveloped viruses with a linear, non-segmented, positive-sense, single-stranded RNA. The RNA genomes are the largest genomic RNA molecules known to date (27–31 kb). These viruses are divided into three distinct groups named 1, 2, and 3. Five types of human coronaviruses (HCoVs) have been described: HCoVs-229E and OC43 have been recognized since the mid-1960s and belong to groups 1 and 2, respectively. Recently, three other human coronaviruses were discovered. The SARS-associated coronavirus (SARS-CoV) was identified in 2003 during a worldwide epidemic starting from the Guangdong province (Ksiazek et al., 2003, Kuiken et al., 2003). In 2004, another group 1 coronavirus, HCoV-NL63 was reported in the Netherlands (Fouchier et al., 2004, Van der Hoek et al., 2004). In January 2005, a new group 2 coronavirus, HCoV-HKU1, was found in two patients suffering from pneumonia in Hong-Kong (Woo et al., 2005). Most of the seroepidemiological studies on human classical coronaviruses were conducted in 1970s using neutralization on cell cultures, hemagglutination inhibition or complement fixation methods, which are labor intensive and time consuming. As there is no available validated test as yet, it is necessary to develop a serological test for the detection of HCoV-OC43 antibodies to obtain recent data about the seroprevalence of this virus in the population. Development of new serodiagnostic tests requires the use of viral proteins. But the cultivation of HCoV-OC43 is difficult and does not allow for the production of large quantities of viral antigen. The objective here is to describe the expression of a recombinant nucleocapsid protein of HCoV-OC43 in a baculovirus/insect cell system and the development of a Western blot immunoassay for the detection of human antibodies against HCoV-OC43.
2. Materials and methods
2.1. Virus and cells
The cell-line adapted strain of prototype human coronavirus OC43 was obtained from American Type Culture Collection (ATCC), Rockville, MD. HCoV-OC43 was propagated by inoculation into a 1-day cultivated human rectal tumour cells (HRT18) and incubated for 48 h or 72 h at 35 °C in a humidified atmosphere containing 5% CO2 in RPMI 1640 medium (Gibco™) supplemented with 2% foetal bovine serum, HEPES 20 mM (EUROBIO™), NaHCO3 0.1% and antibiotics. Propagation of recombinant baculovirus and expression of N recombinant protein were made in Spodoptera frugiperda derived Sf21 cell line. Sf21 was maintained in suspension or adherent cultures in Grace's insect cell culture medium (Gibco™) supplemented with 10% fetal bovine serum. BaculoDirect™ C-Term Linear DNA (Invitrogen) derived from Autographa californica multiple nuclear polyhedrosis virus (AcMNPV) DNA was used to build the recombinant baculovirus. It allows transferring gene of interest from the entry plasmid to the baculovirus DNA directly in vitro without the need of recombination in bacterial cells, using specific recombination sites from bacteriophage lambda. The presence of Herpes simplex virus thymidine kinase gene (HSV1tk) and lacZ gene located between the two recombination sites allows to inhibit replication of non-recombinant baculovirus in presence of ganciclovir, and to determinate viral purity using β-galactosidase staining (Godeau et al., 1992, Lalumiere and Richardson, 1995). The expressed protein is placed in fusion with a tag containing hexahistidine and V5 epitope sequence (a 14 amino-acid peptide derived from the proteins P and V of the simian paramyxovirus SV5) allowing to detect and purify the recombinant fusion protein Southern et al. (1991).
2.2. Construction of baculovirus entry vector
HCoV-OC43 RNA was extracted from 200 μL of the supernatant of infected HRT18 culture by matrix affinity chromatography with QIAamp® Viral RNA Mini Kit (Qiagen). HCoV-OC43 N gene was amplified by reverse transcriptase (RT)-polymerase chain reaction (PCR) with two custom primers designed from previously reported nucleotide sequence of HCoV-OC43 N gene (GenBank accession no. AY391777) and following industrial guideline instructions: TMNT: 5′-CACCATGTCTTTTACTCCTGGTAAG-3′ and NTHO3: 5′-TATTTCTGAGGTGTCTTCAGT-3′. Expand High Fidelity PCR System (Roche) was used to amplify the N gene. The amplification product was cloned into pENTR™/D-TOPO® (Invitrogen) and transformed in electrocompetent Escherichia coli strain TOP10F’ cells. Clones containing the N gene were amplified in LB broth and plasmids were purified with QIAprep® Spin Miniprep Kit (Qiagen). To confirm the insertion in frame and the absence of mutation in the N gene, plasmids were sequenced by BIOFIDAL Society (170 avenue Gabriel Péri, 69120 Vaulx en Velin, France) using M13 forward and reverse primers, and TMNT/NTHO3 primers.
2.3. Construction of the recombinant baculovirus
Recombination reaction was performed 18 h at room temperature in a microcentrifuge tube containing 100 ng (2 μL) of the purified entry vector, 300 ng (10 μL) of the BaculoDirect™ Linear DNA, 4 μL of 5× LR Clonase™ Reaction Buffer and 4 μL of LR Clonase™ Enzyme mix. After incubation time, 2 μL of Proteinase K solution (Invitrogen) was added to the reaction, and incubated 10 min at 37 °C. Lipid mediated transfection of the Sf21 cells was performed with Cellfectin® Reagent (Invitrogen) in six well plates. Each well was seeded with 1.5 × 106 Sf21 cells. Cells were allowed to attach for 1 h at room temperature. Transfection mixture was prepared with 10 μL of LR recombination reaction, 6 μL of Cellfectin® Reagent and 200 μL of unsupplemented Grace's Insect Medium, and incubated at room temperature for 45 min. Medium was removed from each wells and carefully rinsed with unsupplemented Grace's insect medium. Eight hundred microliters of unsupplemented Grace's Insect Medium was added to the transfection mixture and drop onto the cells. Plate was incubated at 26 °C for 5 h. After incubation time, transfection mixture was removed and 2 mL of complete growth media with 10% FBS, antibiotics and 100 μM ganciclovir, was added to each well. Plate was incubated at 27 °C for 72 h in a moisturized box. When the first signs of infection appeared, cell culture medium containing virus was harvested. It was designated as P1 viral stock. To prepare a high-titer viral stock, 500 μL of the P1 viral stock was used to infect 1.5 × 106 Sf21 cells in 1.5 mL of complete growth media with antibiotics and 100 μM ganciclovir. Plate was incubated at 27 °C for 72 h in a moisturized box. When the first signs of infection appeared, cell culture medium containing virus was harvested. It was designated as P2 viral stock. To ensure that non-recombinant virus were eliminated by ganciclovir selection, β-galactosidase staining of three wells containing, respectively, non-infected cells, cells used to produce P1 viral stock, and cells used to produce P2 viral stock, was proceeded using β-Gal Staining Kit (Invitrogen). PCR was used to confirm the presence and orientation of HCoV-OC43 N gene in the recombinant baculovirus. Extraction of total DNA was operated on infected cells using QIAmp® DNA Mini Kit (Qiagen). PCR used for detection of N gene was applied to extracted DNA. Another PCR assay using a combination of a forward primer PHED-F (5′-AAATGATAACCATCTCGC-3′) located in the polyhedrin gene (recommended by Invitrogen) and the reverse primer NTHO3 of the insert was applied to extracted DNA in the same conditions as below.
2.4. Production and analysis of recombinant protein
Sf21 cells in 25 cm2 flasques were infected with high titer recombinant baculovirus suspension (P2 viral stock) with or whithout 2% FBS. Insect cells and culture medium were harvested 48, 72, 96 and 168 h post-infection. Harvested cells were suspended in 50 mM NaPO4, 500 mM NaCl, pH 8.0 solution, and broken by 10 freeze–thaw cycles using liquid nitrogen and a 37 °C water bath. After centrifugation of cell lysate at 4000 rpm for 15 min, supernatant was stored for SDS-PAGE analysis. Proteins from culture medium and cell lysates were analysed on a 4.8% stacking, 10% resolving polyacrylamide gel by a discontinuous SDS-PAGE system. Precision Plus Protein™ Standards (BioRad) was used as a molecular weight standard. After SDS-PAGE, proteins were transferred onto Trans-Blot® Transfer Medium (Biorad) with Criterion™ Blotter (BioRad). Membrane was air-dried and coloured with Ponceau red. Membrane was then washed twice with Tris-buffered saline 10 mM (TBS) to remove Ponceau red. Membrane was incubated 1 h at room temperature in block solution consisting of 5% non-fat dried milk in TBS-Tween 0.05% solution. Membrane was then incubated for 1 h at room temperature with 1:5000 dilution of anti V5 antibody (Invitrogen) in block solution. Membrane was washed for 15 min with 2 changes of TBS-Tween 0.05% and incubated for 1 h at room temperature with a 1:2000 dilution of peroxydase-labelled goat anti mouse IgG + IgM antibody (Argene) in block solution. Membrane was washed as above and reacted with an Opti-4CN™ Substrate (BioRad) or Super Signal® West Pico Chemiluminescent Substrate (Pierce) solution for 5 min. Chemiluminescent signal was acquired with Fluor-S™ Multimager (BioRad).
2.5. Purification of recombinant protein
Supernatant of infected cell lysate was applied to Ni2+-NTA resin (Invitrogen) equilibrated with a pH 8.0 binding buffer containing 10 mM imidazole, 50 mM NaPO4 and 500 mM NaCl. The column was incubated in agitation 1 h at room temperature. The column was then washed four times with 6 mL pH 8.0 wash buffer containing 20 mM imidazole, 50 mM NaPO4 and 500 mM NaCl. Recombinant polyhistidine-tagged protein was finally eluted with 8 × 1 mL pH 8.0 elution buffer containing 250 mM imidazole, 50 mM NaPO4 and 500 mM NaCl. Each 1 mL aliquot containing purified recombinant protein was analysed by SDS-PAGE as described above.
2.6. Selection of the human serum specimens
Serum specimens collected from a hundred healthcare workers (16 male/84 female; mean age: 34.9 ± 11.2) were tested for OC43 serology in an indirect immunofluorescence assay. Five serum specimens from 2 to 3 years old children admitted in surgical departments were also tested. Two days HCoV-OC43 infected HRT18 cells were trypsined and washed with PBS. After the cells were suspended in PBS, they were put upon slides, dried and fixed with acetone at −20 °C for 10 min. Ten microliters of each 1/10 diluted serum was put upon slides and incubated at 37 °C for 30 min. After 5 min wash in 1/10 diluted PBS, 10 μL of 1/80 diluted sheep FITC labelled conjugate F(ab’)2 anti-IgG (Bio-Rad) prepared with Evans blue were put upon slides and incubated at 37 °C for 30 min. This secondary antibody was used on infected cells without human antibodies to check the absence of false positive result due to the conjugate itself. After a 5 min wash, slides were observed with a fluorescent microscope by two experienced technicians. A 1/2500 diluted polyclonal antibody directed against nucleocapsid of BCoV (BVN-80B24 kindly provided by M Vautherot J.F., INRA, Tours) was used as a positive control.
2.7. Western blot immunoassay using human sera
The same protocol as described above (Section 2.4) was used to test human sera by Western blotting. Each well contained 50 ng of recombinant N protein. Human sera were diluted 10-fold in blocking solution. HRP-labelled goat anti human antibody (BioRad) at a dilution of 1:3000 was used as a secondary antibody.
3. Results
3.1. Construction of the baculovirus entry vector
RT-PCR amplification assay using TMNT/NTHO3 primers generate a unique fragment having the expected molecular level (1348 bp) and visible on agarose gel under ultraviolet light. After cloning of the PCR product in pENTR and transformation of TOP10F’ cells, 32 clones were isolated and screened with specific PCR using TMNT/NTHO3 primers. Only two clones were transformed with plasmid containing N gene of HCoV-OC43 in the correct orientation. The two plasmids were designated p26 and p28. The two entry vectors p26 and p28 were sequenced. Both constructs contained the HCoV-OC43 N gene in frame. Sequencing showed a substitution in position 242 of the N gene (T242C) in both plasmids, resulting in amino-acid change from Valine to Alanine in position 81 of the protein. This mutation was already described and might be a polymorphism of the HCoV-OC43 nucleoprotein (GenBank accession no. AAQ93016). In p28 two other mutations were observed. Entry vector p26 was chosen to set up the recombination reaction.
3.2. Construction of the recombinant baculovirus
Recombinant baculovirus was obtained after recombination of p26 with linearized baculovirus DNA and transfection of Sf21 cells with the bacmid. Three days after transfection, cessation of cell growth, increase in cell diameter and detachment of cells from the monolayer were observed, and signed cell infection. After re-infection of new Sf21 cells and staining with X-Gal, only cells used to produce P1 viral stock were blue, showing that there were no more non-recombinant baculovirus in P2 viral stock. Recombinant baculovirus was designated as boc26. PCR controls of the presence and orientation of N gene in baculovirus DNA were all positive.
3.3. Expression of the recombinant N protein
After infection of Sf21 cells with boc26, recombinant N protein with a molecular weight of about 55 kDa was detected by SDS-PAGE. Analysis by Western blot shows the presence of two proteins with molecular weights of 54 and 60 kDa. The size of the recombinant protein approximates to the predicted molecular weight of the N fusion protein containing the C-terminal tag. Analysis of cell lysate and cell culture medium shows that the recombinant protein is majorly expressed in infected cells (Fig. 1 ). Varying incubation time of infected cells shows that the maximal expression was achieved 48 h after infection. Presence of 2% FBS in the culture medium increases the production of the recombinant N protein.
Fig. 1.
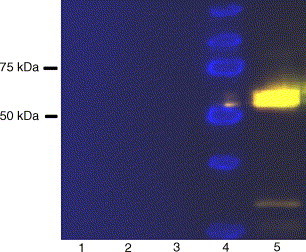
Western blot analysis of recombinant N protein of HCoV-OC43. Proteins were separated by SDS-PAGE and analysed by immunoblotting using HRP-labelled anti V5 antibody. Chemiluminescent signal was acquired for 60 s. Lane 1: non-infected Sf21 cell culture medium; lane 2: boc26-infected Sf21 cell culture medium; lane 3: non-infected Sf21 cell lysate; lane 4: molecular weight marker; lane 5: boc26-infected Sf21 cell lysate.
3.4. Purification of the N protein
Recombinant N protein was purified to near homogeneity by a simple one-step Ni2+ affinity purification procedure. Protein purity was analysed and confirmed by SDS-PAGE and Western immunoblot (Fig. 2, Fig. 3 ). The concentration of the protein was determined with a Micro BC Assay (Uptima). A total of 0.5 mg of purified recombinant protein was obtained from 5 × 107 Sf21 cells. After purification, 6.25 ng of purified protein was needed to give a positive signal with anti V5 antibody, and 12.5 ng with anti BCV antibody (Fig. 4, Fig. 5 ).
Fig. 2.
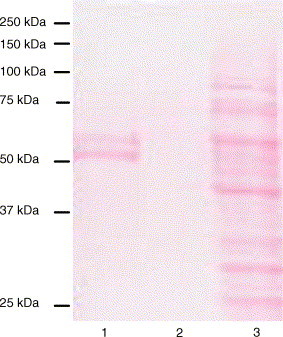
Ten percent SDS-PAGE analysis of the purified recombinant N protein of HCoV-OC43. Lane 1: purified recombinant protein by Ni2+ affinity procedure; lane 2: washing residue of the Ni2+ affinity purification; lane 3: non-purified recombinant N protein extracted from boc26-infected Sf21 cells.
Fig. 3.
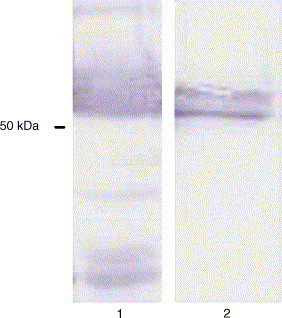
Western blot analysis of the purified recombinant N protein of HCoV-OC43 using anti V5 antibody. Lane 1: non-purified recombinant N protein extracted from boc26-infected Sf21 cells. Lane 2: purified recombinant protein from Ni2+ affinity procedure.
Fig. 4.
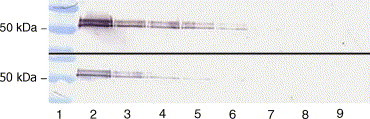
Verification of antigenic integrity of the purified recombinant N protein of HCoV-OC43 by Western blot analysis. The analysis was done with anti V5 antibody (up) and anti N BCoV antibody (down). To determine the sensitivity of the test with each antibody, decreasing quantity of the protein was used. Lane 1: molecular weight marker. Lanes 2–8: 100, 50, 25, 12.5, 7.25, 3.125, 1 ng. Lane 9: negative control.
Fig. 5.
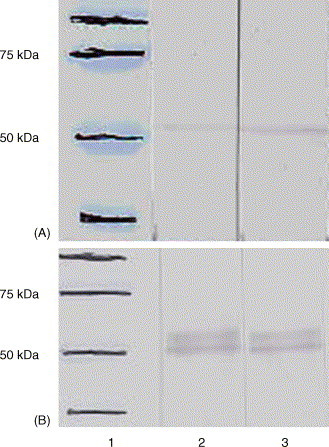
Detection of human antibodies against nucleocapsid protein of HCoV-OC43. Proteins were separated by 10% SDS-PAGE and analysed by immunoblotting. Human sera positive for the detection of OC43 antibody by Western blot analysis using recombinant N protein and equivocal in an immunofluorescent assay were used as primary antibody (lanes 2 and 3). HRP-labelled goat anti human IgG antibody was used as a secondary antibody. Both N protein extracted from HRT18 cells infected with HCoV-OC43 (A) and recombinant N protein extracted from boc26-infected Sf21 cells (B) were tested and give positive results.
3.5. Serological assay
Among the 100 serum specimens taken from healthcare workers, 88 showed positive signals using the indirect immunofluorescence assay (IFI), and 12 showed equivocal signal. All the results of the IFI were verified by two experienced people. Among these 100 serum specimens, 100 were positive (100%) according to Western blotting analysis. Among the five serum specimens collected from 2 to 3-year-old children, three were negative and two were positive for the detection of HCoV-OC43 antibodies. To avoid non-specific cross-reaction with V5 epitope, the twelve positive sera by Western blotting analysis using recombinant N protein and equivocal by immunofluorescence, were tested in a Western blot immunoassay using HCoV-OC43 HRT18 infected cell lysate. All of the 12 sera give a weak positive signal with the natural N protein of HCoV-OC43.
4. Discussion
The emergence of SARS-CoV during the winter of 2002–2003 has greatly increased the interest of scientists in coronaviruses (Ksiazek et al., 2003, Kuiken et al., 2003). For the detection of SARS-CoV, several commercial kits are available. “Classical” coronaviruses are more often responsible of upper and mild respiratory tract illnesses, but can also be responsible for severe infections (Pene et al., 2003, Vabret et al., 2003). Current diagnosis of acute coronaviral infection is based on molecular methods (Vabret et al., 2001). Results from epidemiological surveys conducted in the 1970s have led to the conclusion that these viruses are worldwide in distribution, that seroprevalence increases rapidly during childhood reaching up to 90% in adults (McIntosh et al., 1970, Hasony and Macnaughton, 1982). Nevertheless, these studies were carried out more than 25 years ago, and it would be useful to collect recent data. To this end, we decided to develop new tools for epidemiological studies. In order to produce monoclonal and polyclonal antibodies and also to make immunoassays, we have chosen to express recombinant nucleocapsid protein of HCoV-OC43. Nucleocapsid protein was chosen because it is the most abundant structural protein in infected cells and its production is important during the whole infection cycle. Furthermore, the immunogenicity of this protein induces the production of high titer antibodies (Laude and Masters, 1995). Baculovirus expression systems have often been used to express large quantities of recombinant proteins. Expressed proteins are antigenically close to native proteins because of the post-translational modifications, similar to those observed in mammalian cells. Furthermore, baculovirus-expressed nucleocapsid proteins have been successfully used as antigens in ELISA-based assays for detection of antibodies specific of many virus including SARS-CoV and several animal coronaviruses (Carattoli et al., 2005, Guy et al., 2002). The recombinant baculovirus containing HCoV-OC43 N gene, boc26, produced two proteins (54 and 60 kD). The predicted molecular weight of the recombinant HCoV-OC43 N protein was determined to be 54 kD, including C-terminal tag. The size of the recombinant protein determined by SDS-PAGE and Western immunoblot analysis is approximately the same as the predicted size. The presence of these two proteins has already been described (Laude and Masters, 1995, Liu et al., 2001). Studies on the biosynthesis of coronavirus nucleocapsid proteins in infected cells have revealed the presence of one major intracellular form, and of several related polypeptides. The presence of these two proteins on the Western blot can result from proteolytic cleavage at preferential sites, premature termination or translation of deleted transcripts (Laude and Masters, 1995). The fact that anti-BCoV directed against nucleocapsid specifically recognizes both recombinant proteins assured that these proteins were antigenically similar.
Nevertheless, two points need to be considered. First, the cross reactivity with other group 2 human coronaviruses cannot be excluded with our test. Today, there are two identified group 2 human coronaviruses: HCoVs OC43 and HKU1. The latter virus has been identified recently, precisely in January 2005. No adapted cell-cultured strain could be obtained, and no antibody against HCoVs HKU1 is available. Thus, even if the cross-reactivity could not be excluded, it cannot be verified today. Then, it is important that the assay is specific for OC43 and is not influenced by binding of antibodies directed to the V5 epitope. Neither a control-protein nor V5 epitope is available. In order to verify that there is no reactivity with V5 epitope in our test, we looked for human sera negative in this test. We tested five sera sampled from 2 to 3-year-old children. At this age, 100% of children are known to have already been infected by paramyxoviruses such as respiratory syncytial virus and thus carry antibodies against these viruses. Three of these five sera tested negative for the serological test (WB). These results suggest that our assay is specific for OC43 and is not influenced by the paramyxovirus V5 epitope. As expected, our study confirms that the seroprevalence against HCoV-OC43 in adults is really high. However, it would be interesting to test more serum samples from children to determine the mean age of seroconversion.
The production of this recombinant protein will be used to produce mono- and polyclonal antibodies in order to develop a rapid, easy and direct diagnosis method of HCoV-OC43 infections by immunofluorescence, and could be used for the development of an ELISA test. The Western blot immunoassay described in this publication will help to confirm coronavirus infections by seroconversion detection, and could be used in further epidemiologic studies.
Acknowledgements
We are very grateful to J.F. Vautherot (INRA, Tours) who supplied us with anti BCV antibodies, to J. Gauduchon (University of Caen) and F. Mouthon (CEA, Fontenay aux roses) for their advice in analysing proteins, and to N. Schnepf and A. Garbarg-Chenon (Hôpital Trousseau, Paris) who supplied us with insect cells.
This project was supported by the European Commission EPISARS contract (No. SP22-CT-2004-511063) and the Programme de Recherche en Réseaux Franco-Chinois (Epidémie de SRAS: de l’émergence au contrôle).
References
- Carattoli A., Bonito P.D., Grasso F., Giorgi C., Blasi F., Niedrig M., Cassone A. Recombinant protein-based ELISA and immuno-cytochemical assay for the diagnosis of SARS. J. Med. Virol. 2005;76(2):137–142. doi: 10.1002/jmv.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H., Osterhaus A.D. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U.S.A. 2004;101(16):6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeau F., Saucier C., Kourilsky P. Replication inhibition by nucleoside analogues of a recombinant Autographa californica multicapsid nuclear polyhedrosis virus harboring the herpes thymidine kinase gene driven by the IE-1(0) promoter: a new way to select recombinant baculoviruses. Nucleic Acids Res. 1992;20(23):6239–6246. doi: 10.1093/nar/20.23.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.S., Smith L.G., Breslin J.J., Pakpinyo S. Development of a competitive enzyme-linked immunosorbent assay for detection of turkey coronavirus antibodies. Avian Dis. 2002;46(2):334–341. doi: 10.1637/0005-2086(2002)046[0334:DOACEL]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hasony H.J., Macnaughton M.R. Prevalence of human coronavirus antibody in the population of southern Iraq. J. Med. Virol. 1982;9(3):209–216. doi: 10.1002/jmv.1890090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalumiere M., Richardson C.D. Production of recombinant baculoviruses using rapid screening vectors that contain the gene for beta-galactosidase. Methods Mol. Biol. 1995;39:161–177. doi: 10.1385/0-89603-272-8:161. [DOI] [PubMed] [Google Scholar]
- Laude H., Masters P.S. The coronavirus nucleocapsid protein. In: Sidell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 141–164. [Google Scholar]
- Liu C., Kokuho T., Kubota T., Watanabe S., Inumaru S., Yokomizo Y., Onodera T. A serodiagnostic ELISA using recombinant antigen of swine transmissible gastroenteritis virus nucleoprotein. J. Vet. Med. Sci. 2001;63(11):1253–1256. doi: 10.1292/jvms.63.1253. [DOI] [PubMed] [Google Scholar]
- McIntosh K., Kapikian A.Z., Turner H.C., Hartley J.W., Parrott R.H., Chanock R.M. Seroepidemiologic studies of coronavirus infection in adults and children. Am. J. Epidemiol. 1970;91(6):585–592. doi: 10.1093/oxfordjournals.aje.a121171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene F., Merlat A., Vabret A., Rozenberg F., Buzyn A., Dreyfus F., Cariou A., Freymuth F., Lebon P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37(7):929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern J.A., Young D.F., Heaney F., Baumgartner W.K., Randall R.E. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991;72(7):1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- Vabret A., Mouthon F., Mourez T., Gouarin S., Petitjean J., Freymuth F. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J. Virol. Methods. 2001;97(1–2):59–66. doi: 10.1016/S0166-0934(01)00343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mourez T., Gouarin S., Petitjean J., Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 2003;36(8):985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


