Abstract
A reverse transcriptase polymerase chain reaction (RT-PCR) for the detection of feline coronavirus (FCoV) messenger RNA in peripheral blood mononuclear cells (PBMCs) is described. The assay is evaluated as a diagnostic test for feline infectious peritonitis (FIP). It is based on a well-documented key event in the development of FIP: the replication of virulent FCoV mutants in monocytes/macrophages. To detect most feline coronavirus field strains, the test was designed to amplify subgenomic mRNA of the highly conserved M gene. The test was applied to 1075 feline blood samples (424 from healthy, 651 from sick cats suspected of FIP) and returned 46% of the diseased cats as positive for feline coronavirus mRNA in their peripheral blood cells; of the healthy cats, 5% tested positive. Of a group of 81 animals in which FIP had been confirmed by post-mortem examination, 75 (93%) tested positive, whereas 17 cats with different pathologies (non-FIP cases) all tested negative. In view of the low rate of false-positive results (high specificity) the mRNA RT-PCR may be a valuable addition to the diagnostic arsenal for FIP.
Keywords: Feline infectious peritonitis, Coronavirus, mRNA, RT-PCR
1. Introduction
Coronaviruses are enveloped, positive-stranded ssRNA viruses, a genus in the family Coronaviridae, order Nidovirales. They are ubiquitous in cat populations, with particularly high prevalence in catteries and multiple-cat households. Feline coronaviruses (FCoVs) show a bimodal pathogenicity distribution, with subclinical or mild enteric infections in young kittens at one extreme and the deadly feline infectious peritonitis (FIP) at the other. The low virulence strains are referred to as feline enteric coronaviruses (FECV), the highly virulent ones as FIP viruses (FIPV). Though occurring only sporadically (i.e. not causing epidemics), FIP is an important disease: it is mostly fatal, its biology is still poorly understood and prevention is difficult, to say the least. FIP is an immune mediated, progressive polyserositis and pyogranulomatosis. It occurs worldwide, affecting both domestic and wild felids (Holzworth, 1973, Horzinek and Osterhaus, 1979).
Antibodies against FCoVs are found in 80–90% of the animals living in catteries or multiple-cat households and in up to 50% of solitary cats; however, only some 1–5% of the seropositive cats eventually come down with FIP. The reason for this discrepancy became clear when the biological and genetic properties of FECV and FIPV isolates had been studied (Addie and Jarrett, 1992, Hohdatsu et al., 1992, Horzinek and Osterhaus, 1979): the avirulent FCoV strains causing inconspicuous infections are responsible for the high seroprevalence; in cats experiencing some immunosuppressive event, expansion of the quasispecies cloud and mutations in the FECV genome lead to virulent variants that induce FIP (Vennema et al., 1998).
At present, there are no routine serological and virological assays available for an aetiological diagnosis of FIP, and to distinguish avirulent from virulent FCoVs. Although serology is still used in the diagnosis of FIP, it is of very limited value. Results can only be interpreted in correlation with clinical symptoms. Currently the presumptive diagnosis of FIP is based on clinical data and characteristic changes in some blood parameters (Cammarata Parodi et al., 1993, Gouffeux et al., 1975). A definite diagnosis can only be made on the basis of histological examination of biopsy material or post-mortem (Sparkes et al., 1991, Sparkes et al., 1992).
Our PCR technique using primers targeted to conserved regions of the viral genome, the 3′-UTR (Lai and Cavanagh, 1997), and its modifications (using the S gene (Gamble et al., 1997)) became a valuable tool for the detection of FCoV in body fluids and tissue samples. Unfortunately, the technique detects also avirulent FCoVs in healthy cats. Although the percentage of PCR-positive healthy animals is much lower when compared to FIP cats, a positive PCR result alone does not allow a definite diagnosis (Egberink et al., 1995, Gunn-Moore et al., 1998).
An important event in FIP pathogenesis is the infection of monocytes and macrophages (Stoddart and Scott, 1989). Originally, it was thought that the avirulent FECVs would remain confined to the digestive tract and not spread beyond the intestinal epithelium and regional lymph nodes. Virulent FIPVs, on the other hand, would leave the gut, enter the bloodstream, generalize and reach different organ parenchymas via infected monocytes. Not unexpectedly, however, FCoV were detected in blood samples of healthy cats that never developed FIP, and also after experimental FCoV infection (Gamble et al., 1997, Gunn-Moore et al., 1998, Herrewegh et al., 1995, Kipar et al., 1999).
There may be a difference between the sheer presence of FCoV in peripheral blood mononuclear cells (PBMCs) and their replication in PBMCs, and we hypothesized that the latter may be a correlate of virulence. A RT-PCR that detects messenger RNA of the highly conserved M gene of the FCoV genome in peripheral blood cell samples (Lai and Cavanagh, 1997, Zhang et al., 1994) would detect the macrophage-tropic variants and bypass non-virulent FCoV strains in blood. The present study presents the results of this approach.
2. Materials and methods
2.1. Virus and clinical specimens
The FCoV reference strains and their sources are listed in Table 1 . Strains FIPV 79-1146, FECV 79-1683, and FIPV UCD1 were grown in felis catus whole fetus (fcwf) cells. FIPV UCD3 was obtained from tissue cell culture from infected fcwf cells (Pedersen et al., 1981b). FECV UCD was acquired from feline faeces as described by Pedersen (1987) and grown to low titers in fcwf cells. FIPV Dahlberg was obtained from brain of a mouse inoculated with homogenate as described by Osterhaus et al. (1978). FIPV Wellcome was derived from feline embryonic lung (FEL) culture cells as described by O’Reilly et al. (1979).
Table 1.
Feline, canine and porcine coronavirus reference strains
| Strain | Serotypea | Source | Reference |
|---|---|---|---|
| FECV UCD | I | Feline faeces | Pedersen (1987) |
| FECV RM | I | Feline faeces | Hickman et al. (1995) |
| FIPV Dahlberg | I | Mouse brain homogenate | Osterhaus et al. (1978) |
| FIPV UCD1 | I | Fcwf cells | Pedersen et al. (1981b) |
| FIPV UCD3 | Ib | Fcwf cells | Pedersen and Floyd (1985) |
| FECV 79-1683 | II | Fcwf cells | Evermann et al. (1981) |
| FIPV 79-1146 | II | Fcwf cells | McKeirnan et al. (1981) |
| FIPV Wellcome | II | Feline embryonic lung cells | O’Reilly et al. (1979) |
| CCV K378 | Vennema et al. (1992) | ||
| TGEV Purdue | Kapke and Brian (1986) |
Assignment according to Pedersen et al., 1981a.
Tentative assignment (Hohdatsu et al., 1992).
Blood samples were collected from diseased cats suspected of having FIP based on clinical symptoms (n = 651) as well as from healthy cats (n = 424). The healthy cats were mainly animals living in the same household or cattery as the cats suspected of having FIP. These samples were obtained from different veterinary clinics in The Netherlands.
2.2. Sample preparation for RT-PCR
Blood: A maximum of 1 ml of non-coagulated EDTA blood was centrifuged for 10 min at 2500 rpm. Plasma was separated from the cell pellet and stored at −20 °C. One volume of PBS was added to the blood cells and total RNA was isolated following the Total Quick RNA Blood Kit Protocol (Talent).
2.3. Primer selection
The oligonucleotide primers were chosen from the highly conserved M gene sequence (primer 212) of the FCoV genome combined with a primer aiming at the leader sequence of the FCoV-genome (primer 1179). Primer sequences are shown in Table 2 .
Table 2.
Oligonucleotide primers used in the mRNA RT-PCR
| Oligo | Sequence (5′–3′) | Size (nucleotides) | Positiona | Orientation |
|---|---|---|---|---|
| 212a | TAATGCCATACACGAACCAGCT | 22 | 26440–26461 | Antisense |
| 1179a | GTGCTAGATTTGTCTTCGGACACC | 25 | 60–83 | Sense |
| 1181b | CAAAGTTGTCATGGATGACC | 20 | – | Antisense |
| 1180b | CCATGGAGAAGGCTGGGG | 18 | – | Sense |
Numerical position on the genome of FIPV 79-1179 as determined from the 5′ ATG start codon; M-gene (de Groot et al., 1988).
Feline GAPDH gene.
As a control to check the efficiency of the RNA isolation from all the blood samples and the subsequent reverse transcriptase reaction, a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RT-PCR was performed for every clinical sample (primers 1180 and 1181).
2.4. Reverse transcription
For the reverse transcriptase (RT) reactions, 10 μl of the RNA solution and 2 μl of reverse primer 212 or 1181 (each 5 mM) were mixed and incubated for 2 min at 95 °C and immediately cooled on ice. Subsequently, a mix consisting of 4 μl RT-Buffer (10×; GibcoBRL Life Technologies), 2 μl dithiothreitol (DTT) (100 mM; GibcoBRL Life Technologies), 1 μl deoxyribonucleotide triphosphates (dNTPs) mix (25 mM each dNTP; GibcoBRL Life Technologies), 0.5 μl RNAguard/RNase inhibitor (40 U/μl; Pharmacia Biochemicals) and 0.5 μl Moloney Murine leukaemia virus (MMLV) reverse transcriptase (200 U/μl; GibcoBRL Life Technologies) was added. The reaction mixture was spun down and incubated for 60 min at 37 °C. The enzyme was inactivated by incubation at 95 °C for 5 min. Samples were stored at −20 °C before using it in the mRNA RT-PCR assay.
2.5. Polymerase chain reaction
Following reverse transcription, 3 μl of the RT reaction mixture was added to 27 μl of the PCR reaction mixture. The PCR mix consisted of 3 μl PCR Buffer 2 (10×; Perkin Elmer USA, 1 × 10 mM Tris–HCl, pH 8.3, 50 mM KCl), 2.5 μl magnesium chloride (25 mM; GibcoBRL Life Technologies), 1 μl dNTPs (25 mM each dNTP; GibcoBRL Life Technologies), 1 μl primer 212 (5 mM), and 1 μl primer 1179 (5 mM) (both Invitrogen), 0.25 μl Taq Polymerase (5 U/ μl; GibcoBRL Life Technologies). For the GAPDH RT-PCR reaction the same PCR mix was used but with different primers: 1 μl primers 1180 and 1181 (each 5 mM) (both Invitrogen).
The reaction mixture was placed in a thermal cycler (Biozym). The temperature cycling protocol consisted of 10 min incubation at 95 °C followed by 30 cycles of 1 min denaturation at 95 °C, 1 min primer annealing at 62 °C and 1 min primer extension at 72 °C. The 30 cycles were followed by 10 min at 72 °C and finally the reaction mixture was cooled to 4 °C.
2.6. Analysis of amplified products
Twenty microliters of each PCR sample was analysed by electrophoresis using a 1.5% TAE agarose gel (GibcoBRL Life Technologies) for 45 min at 100 V. A 100 bp molecular weight marker (Invitrogen) was used to control the size of the amplified PCR product. Amplification products were visualised using ethidium bromide staining and UV radiation. Samples revealing a 295 bp fragment for the primers 212 and 1179 and another fragment of 195 bp for the primers 1180 and 1181 were considered positive for coronavirus. Amplification products were photographed using the BioRad GelDoc 1000.
Twenty-three of the obtained PCR products were sequenced to confirm the RT-PCR product.
In order to avoid contamination due to carry-over of amplification products several precautions were taken including physical separation of the pre- and post-PCR procedures, the use of aerosol-resistant filter tips (Biozym), and during each step from RNA isolation to reverse transcriptase and amplification, negative controls of RNase free water were included to try to rule out any false positives.
2.7. Necropsy
If possible, necropsy of the cat was performed to confirm or rule out a clinical diagnosis of FIP. A total of 98 cats were subjected to post-mortem examination. When macroscopic observations were inconclusive, sections of different organs like liver, kidney, spleen etc. were prepared and examined histopathologically.
3. Results
3.1. Primer sensitivity
To determine if a RT-PCR for M gene mRNA detects different coronavirus isolates, several laboratory isolates were subjected to this assay (Table 1). RNA from FIPV serotype I (strains UCD1, UCD3), and serotype II (strains 79-1146, NOR15, Wellcome), FECV serotype I (UCD, RM), FECV serotype II (79-1683), FIPV Wellcome, CCV-K378 and TGEV Purdue could all be detected in cell culture and faeces material or tissue homogenates. After amplification, fragments of the expected size of 295 bp were obtained with all isolates, as shown in Fig. 1 .
Fig. 1.
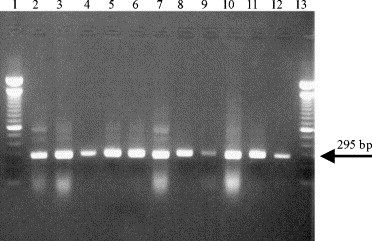
Amplification mRNA (RT-PCR products) from several coronavirus strains. Lane 1: 100 bp Molecular Weight Marker (MWM; Invitrogen); lane 2: FECV UCD; lane 3: FIPV Dahlberg; lane 4: FIPV UCD1; lane 5: FIPV UCD3; lane 6: FIPV NOR15; lane 7: FECV RM; lane 8: FIPV 79-1146; lane 9: FIPV Wellcome; lane 10: FECV 79-1683; lane 11: CCV K378; lane 12: TGEV Purdue; lane 13: 100 bp MWM.
In all samples tested, GAPDH amplicons were demonstrated. The GAPDH gene, which is constitutively expressed at high levels in most tissues, was used for reference as a positive result in the GAPDH RT-PCR will rules out any failure of sample RNA isolation or reverse transcription. An example of a positive mRNA RT-PCR assay and GAPDH control is shown in Fig. 2 .
Fig. 2.
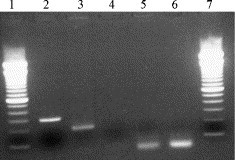
Specificity controls in FCoV mRNA RT-PCR assay. Lane 1: 100 bp MWM; lane 2: FCoV mRNA positive; lane 3: GAPDH mRNA positive; lane 4: RT-negative control p212; lane 5: mRNA negative PCR control; lane 6: GAPDH negative PCR control; lane 7: 100 bp MWM.
3.2. Detection of FCoV in the blood of healthy cats
Blood samples from 424 healthy cats were assessed for the presence of FCoV in peripheral blood cells. These animals had been living in catteries or multiple cat households, where other cats with FIP-related clinical signs were living. Twenty-three cats out of the 424 cats (5%) indeed tested positive for FCoV in the mRNA RT-PCR test. Two cats from the 23 PCR-positive animals became sick within 2 months, both showing different clinical symptoms, one of them indicative of FIP. Unfortunately, the cause of death could not be assessed by necropsy.
3.3. Detection of FCoV in blood of diseased cats
Veterinary practitioners had submitted 651 samples from cats they suspected to suffer from FIP. The animals had shown one or several of the following symptoms: fever, anorexia, weight loss, diarrhea, poor growth, enlarged abdomen, presence of ascitic or thoracic fluid, uveitis and neurological signs. Of these, 301 samples (46%) were positive for FCoV mRNA in blood cells. A summary of the PCR results is shown in Table 3 .
Table 3.
Results of FCoV mRNA RT-PCR in cats with clinical symptoms consistent with FIP and in healthy cats
| Cats (n = 1075) | mRNA positive | mRNA negative |
|---|---|---|
| Cats with clinical symptoms indicative of FIP (n = 651) | 301/651 (46%) | 350/651 (54%) |
| Cats without clinical symptoms (n = 424) | 23/424 (5%) | 401/424 (95%) |
3.4. Detection of FCoV in blood of cats with FIP confirmed by necropsy
Microscopy was performed on 98 cats tested for FCoV mRNA in the blood. In 81 cases FIP was confirmed. Of these, 75 cats (93%) were found to have FCoV mRNA in their peripheral blood cells (Table 4 ).
Table 4.
Results of FCoV mRNA RT-PCR of cats examined post-mortem
| Cats (n = 98) | mRNA positive | mRNA negative |
|---|---|---|
| Cats with proven FIP (n = 81) | 75/81 (93%) | 6/81 (7%) |
| Cats with other diseases (n = 17) | 0/17 (0%) | 17/17 (100%) |
In none of 17 animals that were shown to have suffered from other diseases than FIP, FCoV mRNA was detected in peripheral blood cells (e.g. heart failure, neoplastia, and bacterial infections. The obtained results were statistically significant when controlled by a chi-square (Table 5 ).
Table 5.
Chi-square statistical analysis of PCR results confirmed by necropsy
| PCR results confirmed by necropsy | |||
|---|---|---|---|
| mRNA positive | mRNA negative | Total | |
| FIP | 75 | 6 | 81 |
| Non-FIP | 0 | 17 | 17 |
| Total | 75 | 23 | 98 |
Degrees of freedom: 1; chi-square = 67.0692431561997; p is less than or equal to 0.001; the distribution is significant.
4. Discussion
This report describes an RT-PCR assay to detect FCoV mRNA in blood samples of cats. Our approach was based on the assumption that during the pathogenesis of FIP, the mutant virus would replicate in monocytes and macrophages. We postulated that detection of FCoV mRNA in blood samples would correlate with the development of FIP. There are several observations that led to this assumption.
Infection of monocytes and macrophages is considered as the most important pathogenetic event in FIP. The mutant virus has acquired a new tropism and replicates to high titers in monocytes and macrophages (Stoddart and Scott, 1989, Kipar et al., 1999). In vitro, the virulence of FCoV strains correlated with their ability to infect macrophages: avirulent FCoVs infected fewer cells and produced lower titers than virulent FCoVs. The avirulent FCoVs were also inferior in sustaining viral replication and spreading to other macrophages (Haijema, personal communication).
In coronavirus-infected cells a nested set of subgenomic mRNAs is synthesized, each molecule possessing a “leader sequence”. This stretch of 60–98 nucleotides (coronavirus species dependent) has been derived from the 5′-end of the genome through discontinuous transcription and is not translated. Making use of primers specific for the M-gene mRNA leader sequence (Lai and Cavanagh, 1997) and a conserved part of the M-gene, a molecule of 295 bp will be amplified. Using the mRNA RT-PCR assay, type II FCoV genomes like FIPV 79-1146, FIPV Wellcome, FIPV NOR15 and FECV 79-1783 could be detected in cell culture. Type I FCoVs like FIPV UCD1, FIPV UCD3, FECV UCD, and FIPV Dahlberg were detected as well. In view of the fact that also canine (CCV K378) and porcine (TGEV Purdue) coronaviruses tested positive the assay should detect most, if not all, FCoV variants. The high detection rate of FCoV from cats suspected of suffering from FIP in the field supports this assumption. From its design, the mRNA assay would appear to be more specific (only replicating virus detected) and more sensitive (only nucleated blood cells employed) for the diagnosis of FIP than previous RT-PCR assays focused on genomic RNA in body fluids, feces, and tissues (Gamble et al., 1997, Gunn-Moore et al., 1998, Herrewegh et al., 1995).
Using this assay, we detected mRNA in about 93% of EDTA blood samples from confirmed FIP cases. In the genomic RNA PCR, 78–92% of FIP cats were found to test positive (Gamble et al., 1997, Gunn-Moore et al., 1998, Herrewegh et al., 1995). More importantly, of the healthy cats living in catteries or multiple cat households with a notoriously large virus burden, only 5% tested positive for FCoV. The presence of FCoV RNA in blood monocytes in healthy cats infected with FCoV is an indication that the development of FIP is not associated with the capability of an FCoV to cause viraemia and systemic infection (Meli et al., 2004)
Previous studies quote figures between 20 and 40% (Gamble et al., 1997, Gunn-Moore et al., 1998, Herrewegh et al., 1995), which can be expected in view of the high sensitivity of the PCR. The specificity of our test format would therefore appear as a significant improvement over previously published methods. The question remains if the mRNA positive, healthy cats harbour virulent mutants in an early stage of FIP pathogenesis. Quantitative analyses of FCoV mRNA levels would be needed to identify potential differences between healthy and diseased cats.
Acknowledgements
The authors would like to thank the veterinary practitioners and referring cat owners; without their help this study could not have been completed. This study was supported by a research grant from ID-Lelystad, The Netherlands.
References
- Addie D.D., Jarrett J.O. A study of naturally occurring feline coronavirus infections in kittens. Vet. Rec. 1992;130:133–137. doi: 10.1136/vr.130.7.133. [DOI] [PubMed] [Google Scholar]
- Cammarata Parodi M., Cammarata G., Paltrinieri S., Lavazza A., Ape F. Using direct immunofluorescence to detect coronavirus in peritoneal and pleural effusions. J. Small Animal Prac. 1993;34:609–613. [Google Scholar]
- de Groot R.J., Andeweg A.C., Horzinek M.C., Spaan W.J. Sequence analysis of the 3′-end of the feline coronavirus FIPV 79-1146 genome: comparison with the genome of porcine coronavirus TGEV reveals large insertions. Virology. 1988;167:370–376. doi: 10.1016/0042-6822(88)90097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egberink H.F., Herrewegh A.P., Schuurman N.M., van der Linde-Sipman J.S., Horzinek M.C., de Groot R.J. FIP, easy to diagnose? Vet. Q. 1995;17:24–265. [PubMed] [Google Scholar]
- Evermann J.F., Baumgartener L., Ott R.L., Davis E.V., McKeirnan A.J. Characterization of a feline infectious peritonitis virus isolate. Vet. Pathol. 1981;18:256–265. doi: 10.1177/030098588101800214. [DOI] [PubMed] [Google Scholar]
- Gamble D.A., Lobbiani A., Gramegna M., Moore L.E., Colucci G. Development of a nested PCR assay for detection of feline infectious peritonitis virus in clinical specimens. J. Clin. Microbiol. 1997;35:673–675. doi: 10.1128/jcm.35.3.673-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouffeux M., Pastoret P.P., Henroteaux M., Massip A. Feline infectious peritonitis. Proteins of plasma and ascitic fluid. Vet. Pathol. 1975;12:335–348. doi: 10.1177/0300985875012005-00601. [DOI] [PubMed] [Google Scholar]
- Gunn-Moore D.A., Gruffydd-Jones T.J., Harbour D.A. Detection of feline coronaviruses by culture and reverse transcriptase-polymerase chain reaction of blood samples from healthy cats and cats with clinical feline infectious peritonitis. Vet. Microbiol. 1998;62:193–205. doi: 10.1016/S0378-1135(98)00210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A., de Groot R.J., Cepica A., Egberink H.F., Horzinek M.C., Rottier P.J. Detection of feline coronavirus RNA in feces, tissues, and body fluids of naturally infected cats by reverse transcriptase PCR. J. Clin. Microbiol. 1995;33:784–789. doi: 10.1128/jcm.33.3.684-689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.A., Morris J.G., Rogers Q.R., Pedersen N.C. Elimination of feline coronavirus infection from a large experimental specific pathogen-free catbreeding colony by serologic testing and isolation. Feline Pract. 1995;23:96–102. [Google Scholar]
- Hohdatsu T., Okada S., Ishizuka Y., Yamada H., Koyama H. The prevalence of type I and II feline coronavirus infections in cats. J. Vet. Med. Sci. 1992;54:557–562. doi: 10.1292/jvms.54.557. [DOI] [PubMed] [Google Scholar]
- Holzworth J. Some important disorders of cats. Cornell. Vet. 1973;53:157–160. [PubMed] [Google Scholar]
- Horzinek M.C., Osterhaus A.D. The virology and pathogenesis of felineinfectious peritonitis. Arch. Virol. 1979;59:1–15. doi: 10.1007/BF01317889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapke P.A., Brian D.A. Sequence analysis of the porcine transmissible gastroenteritis coronavirus nucleocapsid protein gene. Virology. 1986;151(1):41–49. doi: 10.1016/0042-6822(86)90102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A., Belman S., Gunn-Moore D.A., Leukert W., Menger S., Reinacher M. Histopathological alterations of lymphatic tissues in cats without feline infectious peritonitis after long-term exposure to FIP virus. Vet. Microbiol. 1999;69:131–137. doi: 10.1016/S0378-1135(99)00101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus. Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeirnan A.J., Evermann J.F., Hargis A., Ott. R.L. Isolation of Feline Coronaviruses from two cats with divers disease manifestations. Feline Pract. 1981;11:16–20. [Google Scholar]
- Meli M., Kipar A., Muller C., Jenal K., Gonczi E., Borel N., Gunn-Moore D., Chalmers S., Lin F., Reinacher M., Lutz H. High viral loads despite absence of clinical and pathological findings in cats experimentally infected with feline coronavirus (FCoV) type I and in naturally FCoV-infected cats. J Feline Med Surg. 2004;6(2):69–81. doi: 10.1016/j.jfms.2003.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly K.J.B., Fishman, Hitchcock L.M. Feline infectious peritonitis: Isolation of a coronavirus. Vet. Rec. 1979;104:348. doi: 10.1136/vr.104.15.348. [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D., Horzinek M.C., Wirahadiredja R.M.S., Kroon A. Feline Infectious Peritonitis (FIP) virus; propagation in suckling rat and hamster brain. Zbl. Vet. Med. 1978;25:816–825. [PubMed] [Google Scholar]
- Pedersen N.C. Virologic and immunologic aspects of feline infectious peritonitis virus infection. Adv. Exp. Med. biol. 1987;218:529–550. doi: 10.1007/978-1-4684-1280-2_69. [DOI] [PubMed] [Google Scholar]
- Pedersen N.C., Boyle J.F., Floyd K. Infection studies in kittens, using feline infectious peritonitis virus propagated in cell culture. Am. J. Vet. Res. 1981;42:367–373. [PubMed] [Google Scholar]
- Pedersen N.C., Boyle J.F., Floyd K., Fudge A., Barker J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 1981;42:377–378. [PubMed] [Google Scholar]
- Pedersen N.C., Floyd K. Experimental studies with three new strains of feline infectious peritonitis virus: FIPV-UCD2, FIPV-UCD3, and FIPV-UCD4. 34th Annual Symposium of Viral Diseases of Small Animals. 1985;7(12):1001–1011. [Google Scholar]
- Sparkes A.H., Gruffydd-Jones T.J., Harbour D.A. Feline infectious peritonitis: a review of clinicopathological changes in 65 cases, and a critical assessment of their diagnostic value. Vet. Rec. 1991;129:209–212. doi: 10.1136/vr.129.10.209. [DOI] [PubMed] [Google Scholar]
- Sparkes A.H., Gruffydd-Jones T.J., Howard P.E., Harbour D.A. Coronavirus serology in healthy pedigree cats. Vet. Rec. 1992;131:35–36. doi: 10.1136/vr.131.2.35. [DOI] [PubMed] [Google Scholar]
- Stoddart C.A., Scott F.W. Intrinsic resistance of feline peritoneal macrophages to coronavirus infection correlates with in vivo virulence. J. Virol. 1989;73(1):436–440. doi: 10.1128/jvi.63.1.436-440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Rossen J.W., Wesseling J., Horzinek M.C., Rottier P.J. Genomic organization and expression of the 3′ end of the canine and feline enteric coronaviruses. Virology. 1992;191(1):134–140. doi: 10.1016/0042-6822(92)90174-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liao C.L., Lai. M.M. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and cis. J. Virol. 1994;78:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]


