Abstract
Coronaviruses (CoVs) can cause trivial or fatal disease in humans and in animals. Detection methods for a wide range of CoVs are needed, to understand viral evolution, host range, transmission and maintenance in reservoirs. A new concept, “Multiprobe QPCR”, which uses a balanced mixture of competing discrete non- or moderately degenerated nuclease degradable (TaqMan®) probes was employed. It provides a broadly targeted and rational single tube real-time reverse transcription PCR (“NQPCR”) for the generic detection and discovery of CoV. Degenerate primers, previously published, and the new probes, were from a conserved stretch of open reading frame 1b, encoding the replicase. This multiprobe design reduced the degree of probe degeneration, which otherwise decreases the sensitivity, and allowed a preliminary classification of the amplified sequence directly from the QPCR trace. The split probe strategy allowed detection of down to 10 viral nucleic acid equivalents of CoV from all known CoV groups. Evaluation was with reference CoV strains, synthetic targets, human respiratory samples and avian fecal samples. Infectious-Bronchitis-Virus (IBV)-related variants were found in 7 of 35 sample pools, from 100 wild mallards (Anas platyrhynchos). Ducks may spread and harbour CoVs. NQPCR can detect a wide range of CoVs, as illustrated for humans and ducks.
Abbreviations: Ct, threshold cycle; FRET, fluorescence resonance energy transfer; LNA, locked nucleic acid; NQPCR, nuclease-based real-time reverse transcription PCR; ORF, open reading frame; PCR, polymerase chain reaction; QPCR, quantitative real-time reverse transcription PCR; RNA, ribonucleic acid; RT-PCR, reverse transcriptase polymerase chain reaction; TET, tetrachloro-6-carboxyfluorescein; TD, touch down temperature protocol
Keywords: Coronaviruses, Real-time PCR, Nuclease-based probes, TaqMan, Avian coronavirus, Split probe strategy
1. Introduction
Coronaviruses (CoVs) are positive single-stranded RNA viruses. They belong to the family Coronaviridae in the order Nidovirales (Cavanagh, 1997). CoVs have the largest known nonsegmented viral RNA genome, 27–32 kb in length and is composed in its 5′-proximal two-thirds of two large open reading frames, ORF1a and ORF1b, encoding the replicase complex (Brian and Baric, 2005). The CoV family traditionally is subdivided into three groups based on serological and genetic properties (Cavanagh et al., 1993, Siddell et al., 1983). Groups 1 and 2 infect a large range of mammalian species whereas group 3 is restricted to birds (Saif, 2004). Classification of the severe acute respiratory syndrome coronavirus (SARS-CoV) in group 2 or as the prototype of a new group 4 is subject to controversy and is complicated by the seeming recombinant origin of its genome (Gorbalenya et al., 2004, Rest and Mindell, 2003, Snijder et al., 2003, Stavrinides and Guttman, 2004). The situation recently became more complicated because bats were found to harbour a wide range of mammalian CoVs (Li et al., 2005, Poon et al., 2005). It is thus likely that bats serve as reservoirs for CoVs in a similar manner as anseriform birds do for influenza A (Webster et al., 1992). CoVs are responsible for a broad spectrum of diseases, including respiratory and enteric pathologies, both in humans and in animals (Saif, 2004). Human CoVs (HCoVs) cause respiratory tract illnesses. However, there is also direct evidence for involvement in enteric (e.g. strain HECoV 4408) (Zhang et al., 1994, Zhu et al., 2006) and suggestive evidence for involvement in neurological disease (Clarke et al., 1979, Flewett et al., 1987, Foley and Leutenegger, 2001, Gerna et al., 1985, Gerna et al., 1984, Han et al., 2006, Jacomy et al., 2006, Luby et al., 1999, Resta et al., 1985, Saif, 2004, Schnagl et al., 1990, Schnagl et al., 1986, Sitbon, 1985, Vabret et al., 2006, Wang et al., 2007, Zhu et al., 2006). HCoV are frequent causes of the common cold; usually a self-limiting upper respiratory tract (URT) illness (Heikkinen and Jarvinen, 2003). The prototypic HCoVs, HCoV-229E, and HCoV-OC43, were identified in the 1960s (Hamre and Procknow, 1966, Vabret et al., 2001). They are members of groups 1 and 2, respectively. Infection with these viruses is thought to be responsible for ≈30% of common cold cases (Fields et al., 2001). Until the discovery of the SARS-CoV, CoVs received relatively little attention as human pathogens beyond their causal role in the common cold. The most recently discovered HCoVs, the SARS-CoV (Peiris et al., 2003), HCoV-NL63 (van der Hoek et al., 2006) and CoV HKU1 (Woo et al., 2005) causes severe respiratory tract infection. Additionally, the knowledge of the pathogenesis of CoVs in wildlife species is also limited. Among the many and highly diverse CoVs found in bats (see above), SARS-like CoVs were identified in horseshoe bats of the genus rhinolophus. Virological and serological studies indicated that masked palm civets (Paguma larvata) were infected with SARS-CoV or a closely related virus (Wang and Eaton, 2007). SARS-CoV thus may have spilled into civets from bats, and then emerged into humans. However, the lack of genomic sequences for many animal CoVs impeded clarification of the origin of SARS-CoV. Sequencing analysis of SARS-CoV revealed that the 5′ end, containing the polymerase gene, is of mammalian origin, and the structural genes (excluding spike glycoprotein) at the 3′ end are of avian origin (Jackwood, 2006). With some exceptions (Pantin-Jackwood et al., 2007, Spackman and Cameron, 1983, Spackman et al., 2005, Swayne et al., 2004, Woo et al., 2009), the spread of CoVs among birds is not well known (Cavanagh, 2005, Jonassen et al., 2005). Even though the number of known CoVs in birds is relatively small in comparison to the number in bats, like in bats, birds contain a wide diversity of CoVs. While this report was being prepared, three novel CoVs, classified as CoV group 3c, were identified in three different bird families, bulbul CoV HKU11, thrush CoV HKU11 and munia CoV HKU13 (BuCoV HKU11, ThCoV HKU12, MuCoV HKU13) (Woo et al., 2009). New diagnostic methods, which are able to detect a wide range of CoVs in various species and which can support the better understanding of CoV evolution, host range and reservoir conditions, are needed.
Currently, diagnostic tests are mostly limited to sequence amplification techniques as the CoVs have a restricted host range and are fastidious to propagate in cell culture. The clinical relevance of serological methods, mainly based on enzyme-linked immunosorbent assays, is limited, since they are generally time-consuming. A variety of assays for detection of CoV have been developed to obtain more sensitive and rapid diagnostic results, such as reverse transcriptase PCR (RT-PCR) (Vabret et al., 2001), nested RT-PCR (Myint et al., 1994) and, recently, also real-time RT-PCR (van Elden et al., 2004). For simplicity, real-time RT-PCR is in the following referred to as “QPCR”. QPCR is less time-consuming than nested or unnested RT-PCR methods, because the amplification and analysis are completed in a closed system, reducing contamination risks.
Although molecular techniques enable the detection of species or groups of CoVs, there are few reports of pan-CoV RT-PCRs (Ksiazek et al., 2003, Moes et al., 2005, Stephensen et al., 1999), especially those that have been adapted to a real-time format (Escutenaire et al., 2007). All methods are based on amplification of fragments from the replicase gene, which has a highly conserved structure and function. This region is suitable for design of primers allowing a broad-spectrum detection of genetically distant CoVs. The SYBRgreen method (Escutenaire et al., 2007) is both sensitive and specific according to our experience. The specificity is ascertained by a melting curve analysis, which in many cases can discriminate a true positive from a false signal (Escutenaire et al., 2007). However, a weakly positive true signal may occasionally be obscured by a stronger false one. Thus, SYBRgreen techniques to some degree lack the increased specificity provided by TaqMan® nuclease-based systems (Bustin, 2004). We therefore set out to develop a broadly targeted TaqMan® detection method system for Coronaviridae using a set of degenerate primers previously developed and reported by us (Escutenaire et al., 2007) for the generic detection of CoVs. The availability of an alternative hybridization-based assay is useful for verification of results based on the previous method. The greater specificity may also facilitate the interpretation of weak hybridization signals, which might be overlooked in the SYBRgreen technique. An optimised mixture of three probes, giving a signal arising from the best binding probes, was necessary to cover most of the known variation within the target stretch of Coronaviridae. They target discrete, short, conserved domains in coronaviral subsets. Their use avoided the extensive degeneration necessary with only one probe. LNA (locked nucleic acid) was used to increase the melting temperature of the relatively short probes. This enhances their affinity to the consensus target sequences. A somewhat similar approach has been used by others (Yip et al., 2005), but without the broad, virus family-specific, targeting and the increased specificity provided by the nuclease-based probe detection of the present method.
The goal of this study was to design a broadly targeted technique for detection of all known groups of CoVs in animals and humans for use in a single tube QPCR. It is intended to complement or replace the previous SYBRgreen pan-CoV QPCR (Escutenaire et al., 2007) published by our group. The new, broadly targeted, PCR is in the following referred to as the nuclease-based QPCR (NQPCR). It provides a probe-based positive identification that increases the specificity of a positive result and a provisional classification when the three probes are tagged with different fluorophores.
2. Materials and methods
2.1. Computer program for identification of conserved portions of variable viral genomes
The program, “ConSort”© (J. Blomberg et al., unpublished), uses alignments of 2-10000 viral sequences made using BLAST, CLUSTALW, MegAlign or MultAlin. Areas of sequence conservation were identified, and the degree of representativeness for all sequences in the alignment computed. The program shows first the most common variant for each position, then less common ones in decreasing order. Insertions and deletions are marked especially. A graphical overview of the degree of conservation is also given.
2.2. Primer and probe design and synthesis
A set of previously reported primers were used (Escutenaire et al., 2007). Oligonucleotides were designed with the aim of (i) primers having an approximately equal T m, (ii) the probe having a higher T m than the primers, (iii) minimizing primer–primer and primer–probe interactions, while (iv) still operating within a conserved region. All oligonucleotides were checked for homo- and heterospecific annealing as well as self-annealing loops, using a cutoff of −5 kcal/mol for the predicted Gibbs free energy [ΔG] of oligonucleotide interactions. The primer–primer interactions were studied using Oligo Analyzer 3.0 (http://207.32.43.70/biotools/oligocalc/oligocalc.asp), and http://www.idtdna.com/analyzer/Applications/OligoAnalyzer. Primers were purchased from Thermo Hybaid, Interactiva Division (Ulm, Germany). The probes with LNA modified nucleotides were purchased from Eurogentec (Seraing, Belgium). The primer sequences and their positions in the genome of SARS-TOR2 (AY274119) are: 11-FW: 5′-TGATGATGSNGTTGTNTGYTAYAA-3′ (nt 15467–15670) and 13-RV: 5′-GCATWGTRTGYTGNGARCARAATTC-3′ (nt 15801–15825). The probe sequences and their position in the genome of HCoV-229E (GenBank accession No. NC002645, nt 14836–14862), SARS-TOR2 (GenBank accession No. AY274119, nt 15743–15770) and NL63 (GenBank accession No. NC005831, nt 14788–14804) are probe_I: 5′-[FAM]TTGTATTATCAGAATGGYGTSTTYATG[EDQ]-3′, probe_II: 5′-[FAM]TGTGTTCATGTCWGARGCWAAATGTTGG[EDQ]-3′, probe_III: 5′-[FAM]TCTAARTGTTGGGTDGA[EDQ]-3′. In later experiments, the same probe sequences were labeled with separate fluorophores. Then, probe_I was labeled with ROX and probe_III with JOE instead of FAM. The black hole quencher EDQ was retained on these modified probes. Underlined positions denote LNA nucleotides. Introducing LNA residues at the most conserved positions was intended to increase the expected T m of the probes, to achieve approximately the same T m for all three probes.
The aim was to make the probes more resistant to viral variation, allowing hybridization in spite of a few mismatches. However, probe-based techniques inherently have a greater specificity than non-probe-based (SYBRgreen) QPCRs. Thus, there is normally a trade-off between specificity and broadness of detection. In epidemiological studies, with samples containing many different kinds of CoVs, several pan-CoV detection systems should be used to ensure a more complete coverage of the virus family. Alignment of the triple-probes with sequences from reference strains of CoV was generated using CLUSTALW version 1.83 (Fig. 1a).
Fig. 1.

(a) Alignment of the triple-probes with sequences from reference strains of CoV. Conserved regions are black. The most frequent variants are shown in gray. Variable positions have no background. The three probe sequences with IUPAC ambiguity codes are shown at the bottom, LNA positions are underlined. (b) Predicted hybridization of amplimer stretches from a broad range of coronaviruses to the three probes. An amplimer sequence from the Ottenby duck samples, with an IBV-like sequence (Ottenby 67433, obtained from a mallard. It was not mentioned in Section 2. It was however identical to the Ottenby spring 3 sample in 120 of 130 positions) was used as query in a BLAST search for similar nucleotide sequences in GenBank. The figure lists the BLAST hits in order of decreasing similarity to the query. The stretches corresponding to the query found by the BLAST search were subjected to analysis with the Visual OMP (VOMP) software, which can predict the degree of interaction (shown as an absolute ΔG value) of an oligonucleotide with another. VOMP automatically removes all identical sequences, which meant that some reference sequences were removed from the hit list. For example, the OC43 sequence was removed. It was identical to the human enteric CoV sequence. Remaining reference sequences are shown in bold. All variants of each of the three probes were tested against all BLAST hit sequences. For each probe, the average absolute ΔG of the three variants predicted to hybridize best to the respective target sequences is shown. The three lines depict the predicted binding of probe_I (229E-derived; dark blue), probe_II (SARS-derived; red) and probe_III (NL63-derived, yellow), respectively. The columns show the average of the predicted probe absolute ΔG values, to indicate whether the triple-probe system would be able to detect that target sequence. (c) Differential fluorescence signal obtained with the three separately labeled probes. CoV, either RNA from reference strains, or synthetic target oligonucleotides from published CoV strains, were run in NQPCR. Results were colour coded as in (b). For further explanation of strain names, see Table 1. TCoV: Turkey CoV; CoV-cons-synth: synthetic oligonucleotide made from a consensus sequence of the PCR amplimer stretch (see Section 2). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
The predicted Gibbs free energy [ΔG] of oligonucleotide and target interactions was analysed utilizing Visual OMP™ v6.6.0.0 (DNA Software; http://www.DNAsoftware.com). The software uses nearest-neighbour thermodynamic theory to simulate hybridization of nucleic acids in solution and provides structural and thermodynamic parameters in intra- or intermolecular situations (SantaLucia, 2007). Data entry included the sequences of the three probes and target (the sequence stretch of CoV strains that hybridizes with the probe). The predicted free energy for hybridization of probes with different CoVs as target was obtained for all variants of each probe (i.e. the probes have different degrees of degeneracy; probe_I: 8, probe_II: 8 and probe_III: 6). The following solution conditions were used in Visual OMP to simulate the experiments; assay T m 50 °C, Na+ 0.05, Mg2+ 1.5 mM, 100 nM of probe and 100 nM target. Under the above experimental conditions the average ΔG for each probe variant with its target was plotted (Fig. 1b) (cf. its experimental counterpart, Fig. 1c).
2.3. Collection of coronavirus strains
A collection of CoVs was used to assess the sensitivity of the real-time RT-PCR (Table 1 ). Twenty-two human and animal strains, representative of the four CoV groups, including SARS-CoV, were analysed. The following references describe the origin of the collection of CoV strains (Adzhar et al., 1995, Bohl et al., 1972, Cavanagh and Davis, 1993, Darbyshire et al., 1979, Davelaar et al., 1984, Drosten et al., 2003, Evermann et al., 1981, Fields, 1973, Hamre and Procknow, 1966, Kahn and McIntosh, 2005, Kourtesis et al., 2001, Lavi et al., 1984, Milane et al., 1997, Pratelli et al., 2003a, Pratelli et al., 2003b, Shaw et al., 1996).
Table 1.
Collection of viral strains used for the evaluation of the pan-CoV NQPCR.
| Coronavirus species | Strain | Source | Reference | Copies/reaction | Threshold cycle, Ct |
|---|---|---|---|---|---|
| Group 1 | |||||
| HCoV-NL63 | NL63 | L.van der Hoek | 2.31 × 106 | 12.96 | |
| HCoV-229E | 229E | ATCC | Hamre and Procknow (1966) | 8.65 × 103 | 22.14 |
| CCoV type I | Elmo/02 | C.Buonavoglia | Pratelli et al. (2003a) | 1.07 × 106 | 14.23 |
| CCoV type II | 144/01 | C.Buonavoglia | Pratelli et al. (2003b) | 8.95 × 106 | 10.74 |
| FCoV type I | FIPV DF2 | I.Kiss | Evermann et al. (1981) | 2.99 × 106 | 12.54 |
| PRCoV | PRCV | I.Kiss | 9.80 × 101 | 29.50 | |
| TGEV | Purdue | I.Kiss | Bohl et al. (1972) | 5.14 × 108 | 7.87 |
| PEDV | PEDV 7 | I.Kiss | 7.94 × 104 | 18.50 | |
| Group 2 | |||||
| HCoV-OC43 | OC43 | ATCC | Kahn and McIntosh (2005) | 3.03 × 106 | 12.52 |
| MHV | MHV-A59 | P.Rottier | Lavi et al. (1984) | 2.46 × 107 | 9.08 |
| BCoV | BCQ-4 | A.Kheyar | Milane et al. (1997) | 9.30 × 106 | 10.68 |
| BCoV | BCQ-3708 | A.Kheyar | Kourtesis et al. (2001) | 4.21 × 105 | 15.76 |
| Group 3 | |||||
| IBV | IBV 927 | I.Kiss | 4.42 × 106 | 15.75 | |
| IBV | 7/91 (793B) | D.Cavanagh | Adzhar et al. (1995) | 1.01 × 107 | 10.54 |
| IBV | HV-10 | D.Cavanagh | 1.14 × 107 | 10.35 | |
| IBV | Gray 390 | D.Cavanagh | Darbyshire et al. (1979) | 6.01 × 102 | 26.52 |
| IBV | D207 | D.Cavanagh | Davelaar et al. (1984) | 7.51 × 106 | 11.03 |
| IBV | Arkansas 99 | D.Cavanagh | Fields (1973) | 1.33 × 106 | 13.87 |
| IBV | UK/918/67 | D.Cavanagh | Cavanagh and Davis (1993) | 4.67 × 105 | 15.59 |
| IBV | AZRI 5508/95 | D.Cavanagh | 1.73 × 107 | 9.65 | |
| IBV | B1648 | D.Cavanagh | Shaw et al. (1996) | 1.96 × 105 | 17.02 |
| Group 4 | |||||
| SARS-CoV | FFM-ic | M.Niedrig | Drosten et al. (2003) | 4.60 × 101 | 30.75 |
HCoV: human coronavirus; CCoV: canine coronavirus; FCoV: feline coronavirus; PRCoV: porcine respiratory coronavirus; TGEV: transmissible gastroenteritis virus; PEDV: porcine epidemic diarrhea virus; MHV: murine hepatitis virus; BCoV: bovine coronavirus; IBV: infectious bronchitis virus.
2.4. Clinical samples and comparative methods
Nasopharyngeal aspirates were collected from 77 patients admitted to the Uppsala Academic Hospital with an episode of acute respiratory infection. The samples were taken routinely for respiratory virus diagnosis by indirect immunofluorescence (IFA) of influenza A and B, parainfluenza 1, 2 and 3 and human respiratory syncytial virus (the respiratory panel kit; cat. nr 3105, Chemicon International Inc., Temecula, CA, USA; or the Influenza A and B kits; cat. nr 5001 and 5002; Chemicon). For comparison, a nested RT-PCR for influenza A and B was used as previously described (Herrmann et al., 2001). Nasopharyngeal secretion samples were collected at the hospital by using a baby feeding tube and an aspiration trap. After suction the feeding tube was rinsed with approximately 2 mL of sterile saline and immediately transported to the lab. Anjuman Hyder, Kåre Bondeson and Björn Herrmann participated in the IFA diagnoses. Fecal samples of 100 mallard ducks (Anas platyrhynchos) were collected (Ottenby Bird Observatory, Sweden) and pooled in 11 pools of autumn 2003 and in 24 pools of spring 2004 and stored at −70 °C until RNA extraction.
2.5. Sample processing and RNA extraction
The samples were diluted 1:100 in nuclease-free water. RNA was extracted from 140 μl diluted sample using the QIAamp viral RNA kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. RNA was recovered in 60 μl of nuclease-free water and either used immediately or stored at −70 °C.
2.6. Real-time QPCR system (“NQPCR”)
A one-step real-time RT-PCR, nuclease-based QPCR (NQPCR), was developed based on the TaqMan® principle, using the iScript one-step RT-PCR kit for probes (Bio-Rad, Hercules, USA). The PCR was formed using a previously reported primer set (Escutenaire et al., 2007). All reactions were performed on the Corbett Research Rotor-Gene Real Time Amplification system (RG-3000, Corbett Research, Mortlake, NSW, Australia), with a total reaction volume of 50 μl containing 25 μl 2× RT-PCR reaction mix for probes, 800 nM of each primer and 200 nM of each probe, and 1 μl of iScript reverse transcriptase. The thermal conditions were as follows; reverse transcription at 50 °C for 30 min, followed by iScript reverse transcriptase inactivation and activation of the hot-start DNA polymerase at 95 °C for 10 min, followed by 5 touchdown steps, in which the annealing temperature was decreased from 56 °C, by 2 °C every second cycle down to 48 °C, followed by 50 cycles of 94 °C for 30 s and 46 °C 60 s. Fluorescence was measured during the latter period. The pan-CoV SYBRgreen QPCR (Escutenaire et al., 2007) was used as a control, both with the human respiratory, and the avian fecal, samples.
2.7. Sequences of PCR products and phylogenetic analysis
RT-PCR products from the SYBRgreen pan-CoV QPCR and NQPCR were gel purified (Qiaquick PCR purification kit; QIAGEN Inc.) and sequenced using the fluorescent dye terminator method, ABI PRISM® Big Dye™ Terminator Cycle Sequencing v3.1 Ready Reaction kit (PerkinElmer) and on an ABI PRISM® 310 genetic analyzer according to the manufacturer's recommendations (Applied Biosystems, Foster City, CA, USA). The 179 nucleotide long amplicon sequences from mallard ducks from Ottenby and human nasopharyngeal aspirates from Uppsala were aligned using CLUSTALW Version 1.83 (Suppl. Info, Fig. S1a and b). Neighbour joining bootstrap consensus (Fig. 2 ) and minimum evolution, bootstrap consensus and maximum parsimony, bootstrap consensus (Suppl. Info, Fig. S2a and b) trees were created from the alignments using Mega 4.1 (Tamura et al., 2007). The reference coronaviral sequences were obtained from GenBank, with the given accession numbers.
Fig. 2.
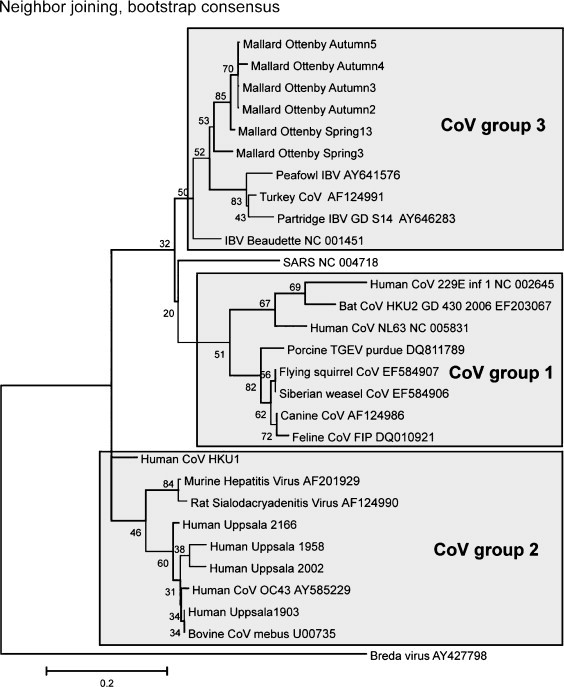
Neighbour-joining bootstrap consensus tree, based on the short (179 bp) amplicon stretch. The bootstrap percentage of 500 trees is shown for most branches. The number of informative sites is low in this conserved portion of the RNA polymerase gene. Therefore, the branching pattern cannot be expected to precisely reflect the phylogeny and classification of Coronaviridae. The tree is intended to approximately represent the observed CoV sequences in a context of most similar reference sequences. The major CoV groups, and reference sequences belonging to them, are shown as boxes. Seven CoV positive human samples were sequenced (“Human Uppsala” plus sample number), and aligned with reference sequences. Four of them, which clustered with CoV group 2 strains, are shown in the tree. Sequences from three CoV positive samples were incomplete. Six of seven sequenced amplicons from mallards (“Mallard Ottenby spring/autumn” plus pool number) are also shown (see Suppl. Info, alignment in Fig. S1a and b, and two trees, Fig. S2a and b, made with other techniques, for further information). A more exact classification of the observed sequences will require sequencing of a longer stretch, which was out of scope for this methodological paper.
2.8. Quantitation standards
Standard curves were generated for quantitation of assays with the help of three kinds of standard: (i) bovine CoV RNA, (ii) bovine CoV cDNA, and (iii) a synthetic CoV consensus oligonucleotide. The bovine CoV RNA is described in Table 1. The bovine cDNA was obtained by reverse transcription from this source. The RNA and DNA concentrations were determined spectrophotometrically using the Nanodrop ND_1000 instrument according to the manufacturer's instructions (Nanodrop, Wilmington, DE).
Standard curves were also generated using a 10-fold dilution series of a synthetic CoV consensus target DNA oligonucleotide (“CORONA_CONSENSUS”: TGATGATGGTGTTGTGTGTTATAACAATGTTTGTATTATCAGAATAA TGTGTTCATGTCTGAAGCTAAATGTTGG GTTGAACCAGACATAAATAAAGGACCTCATGAATTTTGTTCACAGCATACAATGC), where the first underlined sequence corresponds to probe_I, the italicised to probe_II and the second underlined sequence to probe_III. Standards for standard curves were diluted in RNAse free water including 20 ng/μl of yeast tRNA (Ambion, Huntingdon, United Kingdom).
The concentration interpolated from the standard curves was referred to as “equivalents” per PCR reaction.
3. Results
3.1. Identification of conserved portions in coronaviral genomes by using the ConSort© program
A ConSort© analysis of a CLUSTALW alignment of 32 full-length coronaviral sequences indicated that ORF1b was one of the most conserved portions. During the design of the NQPCR probes, representatives from three CoV groups were used as query in BLAST at the NCBI website to search and align the viral sequences in GenBank, i.e. probe_I: ORF1b of 229E; probe_II: ORF1b of SARS-TOR2; probe_III: ORF1b of NL63 were used as query. The ConSort output from the GenBank search was used for design of probes for QPCR.
3.2. Evaluation of the NQPCR on coronavirus strains
A major problem with nuclease-based TaqMan® QPCR probes is the sensitivity to mismatches. A in non-degenerated probe (5-[FAM]TATTATCAAAATAATGTCTTTATG[TAMRA]-3) was first synthesized. Not unexpectedly, it did not detect all CoV strains. A degenerated probe 5′-[FAM]TAYTAYCARAAYRRYGTBTTYATG[TAMRA]-3′, encompassing the whole variation, was then synthesized, with a degeneracy of 768. In our experience, probes with a high degeneracy give very low signals. In order to bring down the degeneracy per probe, three probes were designed. The probes were derived from three CoV groups, as described in Section 2. Probe_I and probe_III were overlapping. The probes had a degeneracy of 8, 8 and 6, respectively, and LNA nucleotides were introduced in order to increase the T m of two of the probes, to achieve approximately same T m for all three probes. When they were tested singly, they gave high relative signal strength, around 0.5, and detected most, but not all (TGEV, FCoV type I, IBV, CCoV type I, PRCoV, SARS, CoV-NL63, HCoV-OC43, BcoV11 and BcoV22), coronaviral strains. However, a NQPCR with a 1 + 1 + 1 mixture with 200 nM of each FAM-labeled probes_I, II and III gave somewhat lower signal strength (0.3), but detected all coronaviral strains. However, when the three probes were labeled with separate fluorophores, signal strengths varied from 0.25 up to 1. This probe mixture was predicted to bind to most, possibly all, known CoV strains (Fig. 1a and b) and was used in the further experiments. A varying pattern of predicted binding was obtained when reference and divergent target CoV sequences were tested in VOMP. No strain came out completely negative in the analysis (Fig. 1b). The predicted hybridization values were largely corroborated when strains were tested experimentally (Fig. 1c).
Twenty-two human and animal strains representative of the four CoV groups, including SARS-CoV, were analysed and detected. The spectrum of detection of the assay was further verified by amplifying cDNA synthesized from RNA extracted from 10 CoV strains; CCoV, PRCoV, HCoV-229E, IBV 297, FCoV, TGEV, Bcov-22, SARS, IBV, BCoV-11, and HCoV-NL63. All of them were detected by the three-probe NQPCR system (Table 1).
3.3. Analysis of human respiratory samples
RNA from 77 human nasopharyngeal aspirates was analysed by both the NQPCR and the SYBRgreen QPCRs. Both techniques indicated that eight samples contained CoV. The SYBRgreen QPCR products of the eight CoV positive samples were sequenced, and aligned with reference sequences. Four of the seven sequenced amplicons turned out to belong to CoV group 2, being OC43/MHV-like (Fig. 2). One belonged to CoV group 1 and was HCoV-NL63-like (Suppl. Info, Fig. S1a). However, it, and two more was incompletely sequenced. They were not included in the trees.
No signal was obtained in the following RNA controls; 3 enterovirus, 2 calicivirus and 2 influenza A positive samples established in previous work. Another sign of specificity was that among the 77 respiratory samples there were 5 RSV positive (by IFA) samples. All 5 were negative in NQPCR.
3.4. Analysis of avian fecal samples
RNA from 35 pooled duck fecal samples in total representing 100 wild birds, 24 pools from spring and 11 from autumn was analysed by both the NQPCR and the SYBRgreen QPCRs. Of the 35 pools, 5/11 autumn pools (Mallard_Ottenby_Autum 2, 3, 4, 5 and 11) and 2/11 spring pools (Mallard_Ottenby_Spring 3 and 13) were CoV positive by NQPCR. Two of the CoV positive pools (one of the spring and one of the autumn pools) were also positive by SYBRgreen QPCR, but five of the seven CoV positive pools were missed by the SYBRgreen QPCR. The PCR products of the seven CoV positive samples were sequenced (from both forward and reverse strands). The sequencing of one pool came out incomplete. All six fully sequenced amplicons were aligned with reference sequences. They resembled the peafowl CoV, accession number AY641576 and avian infectious bronchitis virus, accession number NC_001451, with sequence identities ranging from 83 to 85% and 84 to 86% (Suppl. Info, Fig. S1b). The sequences clustered together, indicating the existence of new CoVs in ducks. The seventh, incompletely sequenced, sample yielded a similar sequence. A caveat is the conserved nature of the targeted stretch. It is not suitable for fine discrimination of coronaviral strains. The major groups do however cluster approximately as expected. Neighbour joining bootstrap consensus (Fig. 2) and minimum evolution, bootstrap consensus and maximum parsimony, bootstrap consensus (Suppl. Info, Fig. S2a and b) trees were created from the alignment. The branching pattern was almost identical in all three trees. However, further sequencing of longer stretches is necessary to establish the duck corona sequences as “new” avian coronaviruses, cf. (Woo et al., 2009). Sequencing of a longer stretch (559 nt without primer sequences, which include the stretch used by Woo et al. (2009) of the polymerase gene of one of the isolates was performed (Mallard_Ottenby_Spring 13). A comparison of the percent identity of Mallard_Ottenby_Spring 13 nucleotide sequence with published sequences of IBV strains and other CoVs is presented in Suppl. Info, Fig. S3. The similarity matrix revealed a percentage identity of Mallard_Ottenby_Spring 13 to IBV strains ranging from 72 to 74%. The IBV strains possess a nucleotide identity of 88 to 92% within the group 3 (IBV proper) of CoVs, and nucleotide identity 60% to other CoVs. This implies that the Mallard_Ottenby_Spring 13 is an IBV-like sequence which is relatively dissimilar from established IBV strains. A more comprehensive analysis will be given in a forthcoming manuscript.
3.5. Calculation of specificity and sensitivity of NQPCR
The molecular limit of detection of NQPCR was 1–10 DNA copies of synthetic DNA nucleotides from a consensus CoV sequence. Serial 10-fold dilutions, from 106 to 100, of the synthetic target was prepared in RNAse free water including 20 ng/μl of yeast tRNA (Ambion, Huntingdon, United Kingdom) as carrier, to determine the lower limit of detection of the assay. The amplification efficiency of a 10-fold serial dilution of BCoV RNA sample with an unknown concentration down to negativity was also determined. The standard curves displayed a linear relationship for NQPCR for the assay, correlation coefficient (R 2) of 0.99 and efficiency of 93%, and an R 2 of 0.99 and efficiency of 83% (Fig. 3a and b). Thus, the CoV DNA and CoV RNA targets gave similar efficiencies.
Fig. 3.

(a) Amplification plot and standard curve, describing sensitivity tests. Primers 11-FW/13-RV were used in combination with FAM-labeled probes_I, II and III. The curves represent, from left to right, 107–100 copies of synthetic corona consensus 130 oligonucleotide, with an efficiency of 0.93 and R2 0.99. No signals were obtained in the negative control. (b) Amplification plot and standard curve, describing an amplification efficiency test. Primers 11-FW/13-RV were used in combination with FAM-labeled probes_I, II and III, and run on 10-fold serial dilutions of BCov RNA, with an efficiency of 0.83 and R2 = 0.99. Thus, amplifying from CoV RNA gave a similar, but somewhat lower, efficiency.
The sensitivity of the NQPCR was studied further by end-point titration of cDNA from BCoV with 6 observations of each dilution step, into the stochastic zone of 1–10 copies per NQPCR reaction. The cDNA sample was titrated in 10-fold dilution steps down to complete negativity. This sensitivity test result indicated a sensitivity of 1–10 copies per PCR assay, with a clear stochastic zone of positivity (Table 2 ).
Table 2.
NQPCR quantification result of 10-fold serially diluted BCoV RNA into the stochastic zone (1–10 copies per PCR reaction).
| Estimated copy number per PCR reaction |
||||
|---|---|---|---|---|
| 100 | 10 | 1a | 0 | |
| Dilution 1 | 107 | 0 | 0 | 0 |
| Dilution 2 | 95 | 10 | 0 | 0 |
| Dilution 3 | 119 | 0 | 0 | 0 |
| Dilution 4 | 67 | 16 | 0 | 0 |
| Dilution 5 | 63 | 0 | 0 | 0 |
| Dilution 6 | 109 | 0 | 13 | 0 |
| S.D.b | 23.27 | 6.98 | 5.31 | 0 |
Six different tubes were analysed per dilution. The estimated RNA concentration in copies per PCR reaction, in relation to the Threshold cycle (Ct) is shown.
At the estimated concentration of one copy per PCR reaction one of six samples were positive.
Standard deviation.
The sensitivity of NQPCR was the same regardless of whether separately labeled or commonly FAM-labeled probes were used (data not shown).
3.6. Reproducibility of the assay
The reproducibility of the NQPCR was evaluated both inter-assay and intra-assay. The intra-assay variability was assessed using high and low amounts of cDNA from 10 different CoV strains. The 10 samples were run in three replicates in same NQPCR run; the intra-assay variability of number of equivalents derived from interpolation in the standard curve had a standard deviation of 0.54 and a standard error of the mean (S.E.M.) of 10%. Inter-assay variability was determined by testing the same set of cDNA samples in three different runs, giving a standard deviation of 0.45 and a S.E.M. of 8%. Judging from these results, at least 1 C t (two standard deviations) is needed to differentiate between the values of two samples.
3.7. RNA extraction and RNA integrity controls
As control for errors occurring after the receipt of samples (post-receipt errors), 11 CoV negative samples from ducks (belonging to the same series as the CoV positive samples) were spiked with a known concentration of IBV. They gave a C t value of 8.0 S.D. 0.3. When the same amount of IBV sample was added to three samples of phosphate buffered saline, a signal with the same C t value 8.0 S.D. 0.1 was obtained. Thus, there was no detectable inhibition of the PCR in the CoV negative samples.
To monitor pre-receipt sample quality, all 35 duck fecal pool samples from Ottenby samples were analysed utilizing a histone 3.3 PCR, which is based on evolutionarily conserved target sequences in histone 3.3, and can be run on samples from most vertebrates, including birds (Andersson et al., 2005, Forsman et al., 2005, Muradrasoli et al., 2006). The histone 3.3 PCR was run with (+rt) and without (−rt) the presence of reverse transcriptase. The −rt results signify the presence of eukaryotic (presumably from duck) DNA, whereas the difference between +rt and −rt results signifies the presence of eukaryotic RNA. The −rt reaction frequently came out negative, while the +rt reaction was positive in most samples. This means that histone 3.3 RNA was more abundant than histone 3.3 DNA, which is logical. RNA often occurs in great excess over DNA in eukaryotic cells. The means and standard deviation (mean ± S.D.) of the C t-value of histone 3.3 PCRs were calculated. The 28 CoV negative, histone 3.3 RNA positive samples yielded an average C t-value of 37.5 ± 1.6, whereas the seven CoV positive and histone 3.3 RNA positive samples yielded an average C t-value of 37.2 ± 0.5. Student's t-test did not detect significantly deviating histone 3.3 +rt and −rt C t values between CoV positive and CoV negative samples (p-value = 0.758). Moreover, one of seven CoV positive samples, and four of 28 CoV negative samples were histone 3.3 +rt negative. The difference between 1/7 vs. 4/28 is not significant (Fisher's exact test; 2-tailed; p-value = 1). Thus, there were approximately equal amounts of eukaryotic RNA in the CoV positive and CoV negative duck fecal samples, making it unlikely that the CoV negativity of some samples was due to RNA degradation.
3.8. Comparison of touchdown and non-touchdown NQPCR
Formation of non-specific products might interfere with amplification of the specific target in low-copy-number samples. An advantage of the presented NQPCR is that the initial high annealing temperature reduces amplification of non-specific PCR, and increases the specificity and sensitivity of the assay (Hecker and Roux, 1996, Mohamed et al., 2006). In developing the NQPCR assay, a standard non-touchdown PCR assay was first used. However, to further increase the sensitivity for aberrant targets a touchdown procedure was designed and assessed side by side with the non-touchdown PCR, by amplifying cDNA synthesized from RNA extracted from 10 CoV strains; CCoV, PRCoV, HCoV-229E, IBV 297, FCoV, TGEV, BCoV-22, HCoV-229E, SARS, IBV, BCoV-11, and HCoV-NL63 (Bustin, 2004, Mohamed et al., 2006). The amplification efficiency was increased with the touch down procedure compared to the non-touchdown procedure. Non-touchdown had an efficiency of 70% (R 2 0.99) while touchdown gave an efficiency of 90% (R 2 0.99). Gel electrophoresis analysis of the PCR product indicated a higher yield compared to the non-touchdown procedure.
4. Discussion
CoV came into the searchlight when the SARS pandemic occurred in 2003. The intense researched spurred then has revealed an unexpected variety of CoV in various host species. However, many questions are still unanswered in CoV biology, including important aspects of viral evolution, subpopulations, hosts, variation in pathogenicity and tissue tropism and regarding reservoirs, which are important in viral “survival” and transmission in nature. Bats are now recognised as reservoirs of a broad range of mammalian CoVs (Poon et al., 2005, Wang and Eaton, 2007). Recently, several findings of CoV in wild birds have been reported (Barr et al., 1988, Jonassen et al., 2005, Liu et al., 2005). The results of this report extend these observations. They indicate that birds, another flying vertebrate, can act as reservoirs for avian CoVs. Considering the many unanswered questions regarding this and other aspects of coronaviral epidemiology, there is a need for novel methods, which are broadly detecting the various CoV variants in different hosts and which are also sensitive, simple and specific. Setting up a generic PCR requires an extensive analysis of variability, a time-consuming evaluation of candidate degenerate primers and probes, and a comprehensive collection of samples. In this study, a one-step real-time RT-PCR, was developed targeting a conserved region of the CoV ORF1b used previously in a SYBRgreen QPCR (Escutenaire et al., 2007). The Consort© program (Blomberg et al., to be published) was used. The new technique was based on nuclease probe degradation (TaqMan®) chemistry, degenerate primers and a low annealing temperature. The assay enabled detection of 22 CoV reference strains, covering a large portion of Coronaviridae (Table 1). The coverage of the new bat CoVs is demonstrated by using synthetic target sequences of three most deviating HKU-CoV: HKU3-CoV, HKU5-CoV and HKU9-CoV) (Fig. 1c). They were all detected by the probe combination.
Non-coronaviral controls were uniformly negative. A major problem with TaqMan® QPCR probes is the sensitivity to mismatches. A nondegenerate probe based on the consensus of the intervening sequence between the primers was first synthesized. It did not detect all coronaviral strains. A degenerate probe encompassing the whole variation, with a degeneracy of 768, was then tested. Not unexpectedly, it gave a very low signal. In our experience (contrary to primers which can work even with a degeneracy of over 100) probes with degeneracy above 10 rarely give sufficient signal strength. In order to bring the number of variants down, the CoV target variation was analysed. It was found possible to spread it onto three overlapping TaqMan® QPCR probes of much lower degeneracy, having 8, 8 and 6 variants each. LNA residues were integrated in two of the probes (Mohrle et al., 2005, Nielsen et al., 2004), to enhance the T m. The introduction of LNA allowed shortening of them, to focus on a short, relatively conserved, stretch (probe_I: 27 nt, 8 LNA, probe_III: 17 nt, 6 LNA). Probe_II could be 28 nt long despite a low degeneracy. As demonstrated here, the probe mixture had a remarkable tolerance to mismatches. The free energy for hybridization was predicted using Visual OMP™. It demonstrated that hybridization between the three probes and a broad range of CoV targets was likely. This was further verified by experimental data that agreed with the result obtained using the Visual OMP™ (Fig. 1b and c). Interestingly, competition between the probes was not an appreciable problem. Although several of the probes sometimes gave a signal, the pattern of positivity indicated which group of CoV that gave rise to the signal. The mismatch tolerance was tested by synthetic target sequences with different degree of mismatch to the probes. Signals from synthetic DNA targets with 1, 2 and 5 mismatches were easily detectable (data not shown). The probes were tested both with the same fluorophore (FAM), and with separate fluorophores. The sensitivity was the same for these alternatives. Although based on a short stretch in the conserved RNA polymerase gene, the latter alternative provided a preliminary identification of the CoV.
PCR efficiency can be affected by numerous factors, inhibition in the sample, secondary structure interference, how well primers and probes fit to their targets, and how optimal the PCR conditions are. It is acknowledged that the efficiency calculation from the slope of a calibration curve often overestimates the PCR efficiency. We therefore determined the amplification efficiency of the NQPCR both for a 10-fold serial dilution of synthetic target and a 10-fold serial dilution of BCoV RNA samples with an unknown concentration down to negativity. Both standard curves displayed a linear relationship for NQPCR, with a correlation coefficient (R 2) of 0.99 and efficiency of 93%, and an R 2 of 0.99 and efficiency of 83%, for the synthetic and natural, imperfectly fitting (BCoV), targets, respectively. This calls for caution regarding quantitative interpretation of C t values. Differences in efficiency of the PCR could also be due to the fact that RNA-to-cDNA conversion is dependent on template abundance. The possibility that mishandling of samples (RNA losses; pre-receipt errors) and PCR inhibition (post-receipt errors) contributed significantly to the results reported here was made unlikely by two control experiments. Signals from RNA of a highly conserved host housekeeping gene (histone 3.3) were the same in CoV positive and CoV negative duck samples. This is a novel application of a control PCR which was earlier developed in the group (Andersson et al., 2005, Forsman et al., 2005, Muradrasoli et al., 2006). The difference between +rt and −rt signal levels from duck fecal samples were relatively even, indicating a rather constant admixture of eukaryotic RNA in them.
The detection of HCoV-OC43-like and HCoV-NL63 sequences in 8 out of 77 nasopharyngeal specimens of human origin, and duck CoV sequences among seven of the pooled wild bird fecal samples, provides evidence of the efficiency of the technique with clinical and field material. The pan-CoV SYBRgreen QPCR missed five duck samples that the NQPCR detected, since the cutoff (threshold) was set higher because of primer dimer signal. Primer dimers do not give a signal in a nuclease and probe-based (TaqMan®) technique like NQPCR. The NQPCR results from the human nasopharyngeal samples were concordant with those of the pan-CoV SYBRgreen QPCR technique used as a control. The 8 NQPCR CoV positive samples were negative in the respiratory virus IFA, and in a nested influenza A + B PCR (data courtesy of dr Björn Herrmann), both of which do not detect CoVs. The method thus should be suitable for diagnostic purposes.
Stephensen et al. (1999) described initially a set of consensus primers targeting a 251-bp fragment of the ORF1b. These primer sequences were subsequently modified for better reactivity, notably to the then newly identified HCoV-NL63 (Moes et al., 2005). Another generic coronaviral PCR was recently published (Vijgen et al., 2008). Furthermore, (Sampath et al., 2005) reported a recently a broad-range PCR targeting the same conserved region of the ORF1b followed by electrospray ionisation mass spectrometry and base composition analysis for viral identification. Viral diagnostic techniques are undergoing a transition. Immunoassays are still valid for some purposes, but real-time PCR and microarray techniques are becoming more important. The technique presented above is simple, sensitive and is able to detect a wide variety of CoV. The extension of the assay by using three different fluorophores, with one for each of them, is a valuable modification. As expected from the alignment and predicted ΔG values, probe 3 was primarily active in positive avian (IBV-like) samples while probe 2 was positive in a SARS sample (data not shown). Although not studied, probe 3 should primarily detect CoV groups 1 and 2.
5. Conclusions
In conclusion, a QPCR with a novel strategy, which uses a mixture of LNA and non-LNA containing probes with an optimised low number of degenerations, is reported here. To the best of our knowledge, this is the first report describing such a strategy to reduce probe degeneracy while maintaining a broad sequence targeting. It should have a wide applicability. High sensitivity was indicated by titrations of a CoV consensus synthetic target DNA content and viral cDNA. The new NQPCR assay, positive with 22 highly diverse strains of CoV, is a useful tool for demonstrating the role of human and wild bird CoVs in infections of the respiratory and digestive tract. This NQPCR thus proved its suitability also for the discovery of new, or so far unknown variants of CoVs.
Acknowledgements
The financial support from the FORMAS grants 2003-1059 and, 2004-2698, from the Ivar and Elsa Sandberg foundation Research 2003 for new trends for safe food: “Improved detection of enteric viruses in foods and beverages by genetic methods”, and from the Swedish Medical Research Council are gratefully acknowledged. We are grateful to ass. prof. Maria Brytting for critical reading of the manuscript, and to the personnel at Ottenby Bird Observatory for collecting the wild bird samples. This is message no. 56 from the Ottenby Bird Observatory. We are indebted to L. van der Hoek (Academic Medical Center, University of Amsterdam, The Netherlands), C. Buonavoglia (Faculty of Veterinary Medicine, University of Bari, Italy), P. Rottier (Department of Infectious Diseases and Immunology, University of Utrecht, The Netherlands), D. Cavanagh (Institute for Animal Health, Compton, United Kingdom) and A. Kheyar (Armand Frappier Institute, Laval, Canada) for contribution to the collection of CoV strains. The SARS RNA was kindly provided by M. Niedrig, Robert Koch-Institute, Berlin.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jviromet.2009.04.022.
Appendix A. Supplementary data
References
- Adzhar A.B., Shaw K., Britton P., Cavanagh D. Avian infectious bronchitis virus: differences between 793/B and other strains. Vet. Rec. 1995;136:548. doi: 10.1136/vr.136.21.548-a. [DOI] [PubMed] [Google Scholar]
- Andersson A.C., Yun Z., Sperber G.O., Larsson E., Blomberg J. ERV3 and related sequences in humans: structure and RNA expression. J. Virol. 2005;79:9270–9284. doi: 10.1128/JVI.79.14.9270-9284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr D.A., Reece R.L., O’Rourke D., Button C., Faragher J.T. Isolation of infectious bronchitis virus from a flock of racing pigeons. Aust. Vet. J. 1988;65:228. doi: 10.1111/j.1751-0813.1988.tb14468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl E.H., Gupta R.K., McCloskey L.W., Saif L. Immunology of transmissible gastroenteritis. J. Am. Vet. Med. Assoc. 1972;160:543–549. [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A. International University Line; California: 2004. A–Z of Quantitative PCR. [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Brian D.A., Brinton M.A., Enjuanes L., Holmes K.V., Horzinek M.C., Lai M.M., Laude H., Plagemann P.G., Siddell S.G. The Coronaviridae now comprises two genera, coronavirus and torovirus: report of the Coronaviridae Study Group. Adv. Exp. Med. Biol. 1993;342:255–257. doi: 10.1007/978-1-4615-2996-5_39. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Sequence analysis of strains of avian infectious bronchitis coronavirus isolated during the 1960s in the UK. Arch. Virol. 1993;130:471–476. doi: 10.1007/BF01309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S.K., Caul E.O., Egglestone S.I. The human enteric coronaviruses. Postgrad. Med. J. 1979;55:135–142. doi: 10.1136/pgmj.55.640.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbyshire J.H., Rowell J.G., Cook J.K., Peters R.W. Taxonomic studies on strains of avian infectious bronchitis virus using neutralisation tests in tracheal organ cultures. Arch. Virol. 1979;61:227–238. doi: 10.1007/BF01318057. [DOI] [PubMed] [Google Scholar]
- Davelaar F.G., Kouwenhoven B., Burger A.G. Occurrence and significance of infectious bronchitis virus variant strains in egg and broiler production in the Netherlands. Vet. Q. 1984;6:114–120. doi: 10.1080/01652176.1984.9693924. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Escutenaire S., Mohamed N., Isaksson M., Thoren P., Klingeborn B., Belak S., Berg M., Blomberg J. SYBR green real-time reverse transcription-polymerase chain reaction assay for the generic detection of coronaviruses. Arch. Virol. 2007;152:41–58. doi: 10.1007/s00705-006-0840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evermann J.F., Baumgartener L., Ott R.L., Davis E.V., McKeirnan A.J. Characterization of a feline infectious peritonitis virus isolate. Vet. Pathol. 1981;18:256–265. doi: 10.1177/030098588101800214. [DOI] [PubMed] [Google Scholar]
- Fields B.N., Knipe D.M., Howley P.M. Lippincott Williams & Wilkins; Philadelphia, London: 2001. Fields Virology. [Google Scholar]
- Fields D.B. Arkansas 99, a new infectious bronchitis serotype. Avian Dis. 1973;17:659–661. [PubMed] [Google Scholar]
- Flewett T.H., Beards G.M., Brown D.W., Sanders R.C. The diagnostic gap in diarrhoeal aetiology. Ciba Found. Symp. 1987;128:238–249. doi: 10.1002/9780470513460.ch14. [DOI] [PubMed] [Google Scholar]
- Foley J.E., Leutenegger C. A review of coronavirus infection in the central nervous system of cats and mice. J. Vet. Intern. Med. 2001;15:438–444. doi: 10.1111/j.1939-1676.2001.tb01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman A., Yun Z., Hu L., Uzhameckis D., Jern P., Blomberg J. Development of broadly targeted human endogenous gammaretroviral pol-based real time PCRs quantitation of RNA expression in human tissues. J. Virol. Methods. 2005;129:16–30. doi: 10.1016/j.jviromet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Gerna G., Passarani N., Battaglia M., Rondanelli E.G. Human enteric coronaviruses: antigenic relatedness to human coronavirus OC43 and possible etiologic role in viral gastroenteritis. J. Infect. Dis. 1985;151:796–803. doi: 10.1093/infdis/151.5.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Passarani N., Cereda P.M., Battaglia M. Antigenic relatedness of human enteric coronavirus strains to human coronavirus OC43: a preliminary report. J. Infect. Dis. 1984;150:618–619. doi: 10.1093/infdis/150.4.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Han M.G., Cheon D.S., Zhang X., Saif L.J. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. J. Virol. 2006;80:12350–12356. doi: 10.1128/JVI.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker K.H., Roux K.H. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- Heikkinen T., Jarvinen A. The common cold. Lancet. 2003;361:51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B., Larsson C., Zweygberg B.W. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription-PCR: comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA) J. Clin. Microbiol. 2001;39:134–138. doi: 10.1128/JCM.39.1.134-138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W. The relationship of severe acute respiratory syndrome coronavirus with avian and other coronaviruses. Avian Dis. 2006;50:315–320. doi: 10.1637/7612-042006R.1. [DOI] [PubMed] [Google Scholar]
- Jacomy H., Fragoso G., Almazan G., Mushynski W.E., Talbot P.J. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology. 2006;349:335–346. doi: 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen C.M., Kofstad T., Larsen I.L., Lovland A., Handeland K., Follestad A., Lillehaug A. Molecular identification and characterization of novel coronaviruses infecting graylag geese (Anser anser), feral pigeons (Columbia livia) and mallards (Anas platyrhynchos) J. Gen. Virol. 2005;86:1597–1607. doi: 10.1099/vir.0.80927-0. [DOI] [PubMed] [Google Scholar]
- Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24:S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. (Discussion S226) [DOI] [PubMed] [Google Scholar]
- Kourtesis A.B., Gelinas A.M., Dea S. Genomic and antigenic variations of the HE glycoprotein of bovine coronaviruses associated with neonatal calf diarrhea and winter dysentery. Arch. Virol. 2001;146:1219–1230. doi: 10.1007/s007050170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lavi E., Gilden D.H., Wroblewska Z., Rorke L.B., Weiss S.R. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology. 1984;34:597–603. doi: 10.1212/wnl.34.5.597. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Luby J.P., Clinton R., Kurtz S. Adaptation of human enteric coronavirus to growth in cell lines. J. Clin. Virol. 1999;12:43–51. doi: 10.1016/S0928-0197(98)00067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milane G., Kourtesis A.B., Dea S. Characterization of monoclonal antibodies to the hemagglutinin-esterase glycoprotein of a bovine coronavirus associated with winter dysentery and cross-reactivity to field isolates. J. Clin. Microbiol. 1997;35:33–40. doi: 10.1128/jcm.35.1.33-40.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P., Pyrc K., Berkhout B., van der Hoek L., Van Ranst M. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005;5:6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N., Belak S., Hedlund K.O., Blomberg J. Experience from the development of a diagnostic single tube real-time PCR for human caliciviruses, Norovirus genogroups I and II. J. Virol. Methods. 2006;132:69–76. doi: 10.1016/j.jviromet.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Mohrle B.P., Kumpf M., Gauglitz G. Determination of affinity constants of locked nucleic acid (LNA) and DNA duplex formation using label free sensor technology. Analyst. 2005;130:1634–1638. doi: 10.1039/b507728a. [DOI] [PubMed] [Google Scholar]
- Muradrasoli S., Forsman A., Hu L., Blikstad V., Blomberg J. Development of real-time PCRs for detection and quantitation of human MMTV-like (HML) sequences HML expression in human tissues. J. Virol. Methods. 2006;136:83–92. doi: 10.1016/j.jviromet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Myint S., Johnston S., Sanderson G., Simpson H. Evaluation of nested polymerase chain methods for the detection of human coronaviruses 229E and OC43. Mol. Cell Probes. 1994;8:357–364. doi: 10.1006/mcpr.1994.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K.E., Rasmussen J., Kumar R., Wengel J., Jacobsen J.P., Petersen M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjug. Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Spackman E., Day J.M., Rives D. Periodic monitoring of commercial Turkeys for enteric viruses indicates continuous presence of astrovirus and rotavirus on the farms. Avian Dis. 2007;51:674–680. doi: 10.1637/0005-2086(2007)51[674:PMOCTF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Decaro N., Tinelli A., Camero M., Cirone F., Elia G., Cavalli A., Corrente M., Greco G., Buonavoglia D., Gentile M., Tempesta M., Buonavoglia C. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Pistello M., Elia G., Decaro N., Buonavoglia D., Camero M., Tempesta M., Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta S., Luby J.P., Rosenfeld C.R., Siegel J.D. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Technol. 2004;23:643–660. doi: 10.20506/rst.23.2.1513. [DOI] [PubMed] [Google Scholar]
- Sampath R., Hofstadler S.A., Blyn L.B., Eshoo M.W., Hall T.A., Massire C., Levene H.M., Hannis J.C., Harrell P.M., Neuman B., Buchmeier M.J., Jiang Y., Ranken R., Drader J.J., Samant V., Grifey R.H., McNeil J.A., Crooke S.T., Ecker D.J. Rapid identification of emerging pathogens: coronavirus. Emerg. Infect. Dis. 2005;11:373–379. doi: 10.3201/eid1103.040629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr. Physical principles and visual-OMP software for optimal PCR design. Methods Mol. Biol. 2007;402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- Schnagl R.D., Foti R., Brookes S., Bucens M. Serum antibodies to human enteric coronavirus-like particles in Australia, South Africa, Indonesia, Niue, and Papua New Guinea. Acta Virol. 1990;34:239–245. [PubMed] [Google Scholar]
- Schnagl R.D., Greco T., Morey F. Antibody prevalence to human enteric coronavirus-like particles and indications of antigenic differences between particles from different areas. Brief report. Arch. Virol. 1986;87:331–337. doi: 10.1007/BF01315312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw K., Britton P., Cavanagh D. Sequence of the spike protein of the Belgian B164S isolate of nephropathogenic infectious bronchitis virus. Avian Pathol. 1996;25:607–611. doi: 10.1080/03079459608419165. [DOI] [PubMed] [Google Scholar]
- Siddell S.G., Anderson R., Cavanagh D., Fujiwara K., Klenk H.D., Macnaughton M.R., Pensaert M., Stohlman S.A., Sturman L., van der Zeijst B.A. Coronaviridae. Intervirology. 1983;20:181–189. doi: 10.1159/000149390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon M. Human-enteric-coronaviruslike particles (CVLP) with different epidemiological characteristics. J. Med. Virol. 1985;16:67–76. doi: 10.1002/jmv.1890160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman D., Cameron I.R. Isolation of infectious bronchitis virus from pheasants. Vet. Rec. 1983;113:354–355. doi: 10.1136/vr.113.15.354. [DOI] [PubMed] [Google Scholar]
- Spackman E., Kapczynski D., Sellers H. Multiplex real-time reverse transcription-polymerase chain reaction for the detection of three viruses associated with poult enteritis complex: turkey astrovirus, turkey coronavirus, and turkey reovirus. Avian Dis. 2005;49:86–91. doi: 10.1637/7265-082304R. [DOI] [PubMed] [Google Scholar]
- Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephensen C.B., Casebolt D.B., Gangopadhyay N.N. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E., Suarez D.L., Spackman E., Tumpey T.M., Beck J.R., Erdman D., Rollin P.E., Ksiazek T.G. Domestic poultry and SARS coronavirus, southern China. Emerg. Infect. Dis. 2004;10:914–916. doi: 10.3201/eid1005.030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vabret A., Dina J., Gouarin S., Petitjean J., Corbet S., Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin. Infect. Dis. 2006;42:634–639. doi: 10.1086/500136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A., Mouthon F., Mourez T., Gouarin S., Petitjean J., Freymuth F. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J. Virol. Methods. 2001;97:59–66. doi: 10.1016/S0166-0934(01)00343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden L.J., van Loon A.M., van Alphen F., Hendriksen K.A., Hoepelman A.I., van Kraaij M.G., Oosterheert J.J., Schipper P., Schuurman R., Nijhuis M. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J. Infect. Dis. 2004;189:652–657. doi: 10.1086/381207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Moes E., Keyaerts E., Li S., Van Ranst M. A pancoronavirus RT-PCR assay for detection of all known coronaviruses. Methods Mol. Biol. 2008;454:3–12. doi: 10.1007/978-1-59745-181-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. Curr. Top. Microbiol. Immunol. 2007;315:325–344. doi: 10.1007/978-3-540-70962-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tu X., Humphrey C., McClure H., Jiang X., Qin C., Glass R.I., Jiang B. Detection of viral agents in fecal specimens of monkeys with diarrhea. J. Med. Primatol. 2007;36:101–107. doi: 10.1111/j.1600-0684.2006.00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S.P., To S.S., Leung P.H., Cheung T.S., Cheng P.K., Lim W.W. Use of dual TaqMan probes to increase the sensitivity of 1-step quantitative reverse transcription-PCR: application to the detection of SARS coronavirus. Clin. Chem. 2005;51:1885–1888. doi: 10.1373/clinchem.2005.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Liu Y., Cai Y., Yu D., Pu Y., Harmon L., Zhang X. Toward the development of an infectious cDNA clone of a human enteric coronavirus. Adv. Exp. Med. Biol. 2006;581:527–530. doi: 10.1007/978-0-387-33012-9_95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


