Abstract
Nipah virus (NiV) is a new zoonotic paramyxovirus that emerged in 1998 and is now classified in the genus Henipavirus along with the closely related Hendra virus (HeV). NiV is highly pathogenic in several vertebrate species including humans, and the lack of available vaccines or specific treatment restricts it to biosafety level 4 (BSL4) containment. A serum neutralization test was developed for measuring NiV neutralizing antibodies under BSL2 conditions using a recombinant vesicular stomatitis virus (VSV) expressing green fluorescent protein (GFP) and bearing the F and G proteins of NiV (VSV–NiV–GFP). The neutralization titers were obtained by counting GFP-expressing cells or by measuring fluorescence. The performance of this new assay was compared against the conventional test using live NiV with panels of sera from several mammalian species, including sera from NiV outbreaks, experimental infections, as well as HeV-specific sera. The results obtained with the VSV–NiV–GFP based test correlated with those obtained using live NiV. Using a 50% reduction in VSV–NiV–GFP infected cells as the cut-off for neutralization, this new assay demonstrated its potential as an effective tool for detecting NiV neutralizing antibodies under BSL2 containment with greater speed, sensitivity and safety as compared to the conventional NiV serum neutralization test.
Keywords: Nipah, Henipavirus, Pseudotype, Neutralization, Diagnosis, Biosafety
1. Introduction
Nipah virus (NiV) is a highly pathogenic paramyxovirus that emerged as the causal agent of respiratory disease in pigs and an acute febrile encephalitis in humans working in the pig industry in Malaysia and Singapore in 1998 (Chua et al., 2000). NiV is classified in the new genus Henipavirus, based on unique genetic characteristics distinct from other paramyxoviruses (Eaton et al., 2006, Eaton et al., 2007), together with Hendra virus (HeV) which has caused repeated incidences of severe respiratory and neurological diseases among horses and humans in Australia since 1994 (Bishop and Broder, 2008, Murray et al., 1995). Natural NiV infections were also confirmed in dogs, cats and horses. Further NiV outbreaks have occurred in Bangladesh and India almost every year since 2001 (Bishop and Broder, 2008, Chadha et al., 2006, Harcourt et al., 2005, Hsu et al., 2004). Henipavirus reservoirs appear to be several species of fruit bats, primarily of the genus Pteropus. During the NiV outbreak on the Malay Peninsula, humans and other species are presumed to be infected through an intermediate species such as pigs (Eaton et al., 2006). On the other hand, in the NiV outbreak in Bangladesh in 2004, involvement of animals other than fruit bats was not confirmed, indicating direct transmission from bats to humans (Hsu et al., 2004, Luby et al., 2006). Human-to-human transmission has also been suggested based on the epidemiological studies conducted in Bangladesh (Gurley et al., 2007a, Gurley et al., 2007b). The presence of antibodies against NiV or NiV-like viruses in fruit bats have been reported in Thailand (Wacharapluesadee et al., 2005), Cambodia (Olson et al., 2002, Reynes et al., 2005), Indonesia (Sendow et al., 2006), Madagascar (Lehle et al., 2007), Ghana (Hayman et al., 2008) and China (Li et al., 2008).
For safe and rapid serological diagnosis of NiV infection, several enzyme-linked immunosorbent assays (ELISAs) have been developed (Daniels et al., 2001). Although the specificities of these ELISAs are fairly high, they still yield false positive results. This means that confirmatory diagnosis is required when an animal is suspected of being infected with NiV by ELISA. The serum neutralization test, which detects neutralizing antibodies against the viral fusion (F) and/or attachment (G) glycoproteins is regarded currently as the gold standard for serological diagnosis, and is accepted as the reference standard of the World Organization for Animal Health (OIE) (OIE, 2008). Although cross-reactivity exists between NiV and HeV by serum neutralization test, they can be differentiated when sera are tested against both viruses simultaneously (Daniels et al., 2001).
Due to the highly pathogenic nature of NiV and HeV and the absence of any therapeutic or prophylactic measures, both are classified as biosafety level 4 (BSL4) pathogens and serum neutralization test using live virus must be conducted under BSL4 conditions (OIE, 2008). These safety concerns have prompted the development of a system for detecting neutralizing antibodies against NiV without using live infectious virus. One of the most promising systems is based on vesicular stomatitis virus (VSV) ΔG*, a recombinant VSV whose G gene was replaced by the green fluorescent protein (GFP) gene (Lawson et al., 1995, Takada et al., 1997).
In this study, a serum neutralization test using VSV-based pseudotyped virus possessing NiV-F and G was developed. The results of the neutralization test using VSV–NiV–GFP was found to be well correlated with those obtained using live NiV. Using a 50% reduction in VSV–NiV–GFP infected cells as the cut-off for neutralization, this assay demonstrated its potential as a more rapid and sensitive method than the standard serum neutralization test using live-NiV. Furthermore, the VSV–NiV–GFP based neutralization assay could be carried under BSL2 conditions, eliminating the use of infectious NiV and requirement for BSL4 containment and also improving safety.
2. Materials and methods
2.1. Cells and viruses
HEK293T and Vero cells were maintained in Eagle's minimum essential medium (EMEM) including 10% fetal calf serum (FCS). NiV was isolated from human brain tissue during the 1998/1999 outbreak in Malaysia (Chua et al., 2000), propagated in Vero cells and stored at −70 °C until use. All the experiments using live-NiV were conducted under strict bio-containment procedures in a BSL4 laboratory of the Australian Animal Health Laboratory.
2.2. Plasmids
To construct expression plasmids encoding NiV-F and G genes, coding regions for the F and G proteins were amplified from viral cDNA. The F gene was amplified from randomly primed cDNA (forward primer: 5′-CGC GGA TCC TCG ACA ATG GTA GTT ATA CTT G-3′; reverse primer: 5′-GGT TGA AGC TTC AAT CTG AAT ACA CTA TGT CC-3′) and first cloned into the pRSET-A vector (Invitrogen, Carlsbad, CA, USA), followed by subcloning into the pCAGGS vector (a kind gift from Dr J. Miyazaki, Osaka University, Japan) (Niwa et al., 1991). The cDNA of G gene was cloned into the same vector, pCAGGS, as described elsewhere (Patch et al., 2007).
2.3. Sera
Sera from various species used in this study are shown in Table 1 . Rabbit αNiV-F and αNiV-G were obtained by immunization with plasmid DNA, NiV-F/pCAGGS and NiV-G/pCAGGS (see above). Briefly, rabbits received 100 μg of plasmid DNA in each injection a total of eight times with 2-week-intervals. Before use in the serum neutralization test in this study, all sera were screened for αNiV or αHeV antibodies by ELISA using inactivated viral antigens as described elsewhere (Daniels et al., 2001).
Table 1.
The comparison of neutralization titers between VSV–NiV–GFP and live-NiV based assays.
| Species | No. | Origin | Ref. | ELISA | Neutralization titer |
|
|---|---|---|---|---|---|---|
| VSV–NiV–GFP | Live-NiV | |||||
| Human | 1 | NiV outbreak | 1 | + | >1,280 | 15 |
| 2 | HeV outbreak | 2 | + | 1,280 | 20 | |
| 3 | + | 1,280 | 20 | |||
| 4 | Negative control | – | − | <10 | <10 | |
| 5 | − | <10 | <10 | |||
| Fruit bat | 1 | NiV experimental infection | 3 | + | >640 | 240 |
| 2 | + | >640 | 80 | |||
| 3 | + | >640 | 160 | |||
| 4 | Negative control | − | <10 | <10 | ||
| Horse | 1 | HeV outbreak | 2 | + | >1,280 | 240 |
| 2 | + | >1,280 | 240 | |||
| 3 | + | >1,280 | 480 | |||
| 4 | + | 160 | 15 | |||
| 5 | + | >1,280 | 240 | |||
| 6 | + | >1,280 | 240 | |||
| 7 | + | >1,280 | 160 | |||
| 8 | Negative control | – | − | <10 | <10 | |
| Cat | 1 | NiV experimental infection | 4 | + | 160 | <10 |
| 2 | + | >640 | 640 | |||
| 3 | + | >640 | 640 | |||
| 4 | Negative control | 4 | − | <10 | <10 | |
| Rabbit | 1 | NiV-F DNA immunization | 5 | + | 40,960 | 320 |
| 2 | + | 164,840 | 480 | |||
| 3 | NiV-G DNA immunization | 5 | + | 40,960 | 960 | |
| 4 | + | 40,960 | 1280 | |||
| 5 | Negative control | 5 | − | <10 | NT | |
| 6 | − | <10 | NT | |||
| 7 | − | <10 | NT | |||
| 8 | − | <10 | NT | |||
| Pig | 1 | NiV experimental infection | 6 | + | >1,280 | 60 |
| 2 | + | >1,280 | 240 | |||
| 3 | + | >1,280 | 240 | |||
| 4 | Negative control | 6 | − | <10 | <10 | |
| 5 | − | <10 | <10 | |||
| 6 | NiV outbreak in Malaysia | 1 | + | 80 | <20 | |
| 7 | + | 640 | 60 | |||
| 8 | + | >1,280 | 240 | |||
| 9 | + | >1,280 | 80 | |||
| 10 | + | 640 | 80 | |||
| 11 | + | 640 | 40 | |||
| 12 | + | 640 | 80 | |||
| 13 | + | 640 | 30 | |||
| 14 | + | 640 | 60 | |||
| 15 | + | 320 | 20 | |||
| 16 | + | 160 | 30 | |||
| 17 | − | <10 | <20 | |||
| 18 | Japanese field-A αJEV (−) | 7 | − | 5 | NT | |
| 19 | − | <5 | NT | |||
| 20 | − | 5 | NT | |||
| 21 | − | <5 | NT | |||
| 22 | − | 5 | NT | |||
| 23 | − | 5 | NT | |||
| 24 | − | 5 | NT | |||
| 25 | − | 5 | NT | |||
| 26 | Japanese field-B αJEV (+) | 7 | − | 40 | <10 | |
| 27 | − | 40 | <10 | |||
| 28 | − | 10 | <10 | |||
| 29 | − | 20 | <10 | |||
| 30 | JEV experimental infection | 8 | − | <10 | <10 | |
| 31 | − | <10 | <10 | |||
| 32 | − | <10 | <10 | |||
| 33 | − | <10 | <10 | |||
| 34 | − | <10 | <10 | |||
(1) Chua et al. (2000), (2) Murray et al. (1995), (3) Middleton et al. (2007), (4) McEachern et al. (2008), (5) see Section 2, (6) Middleton et al. (2002), (7) provided by Dr. T. Takasaki (Department of Virology I, National Institute of Infectious Diseases, Japan, (8) Williams et al. (2001).
2.4. Generation of pseudotyped virus, VSV–NiV–GFP
Generation of VSV pseudotyped with NiV-F/G was performed according to Fukushi et al. (2008) with several modifications. The NiV-F/pCAGGS and NiV-G/pCAGGS DNA were co-transfected into 80–90% confluent HEK293T cells using Fugene HD (Roche, Basel, Switzerland) according to the manufacturer's instructions. The cells were incubated at 37 °C in a CO2 incubator for 48 h, with the replacement of medium 4–24 hrs post-transfection, followed by infection with VSVΔG* (a kind gift from Dr. M.A. Whitt, GTx Inc., TN, USA) at a multiplicity of infection (MOI) of 1. After 24 h, the culture supernatants containing VSV pseudotyped with NiV-F/G (VSV–NiV–GFP) were collected. The supernatants were centrifuged to remove cell debris, filtered through a 0.45 μm pore size filter, and then stored at −80 °C.
2.5. Titration of VSV–NiV–GFP
Serial 10-fold dilutions of VSV–NiV–GFP in EMEM-2% FCS were inoculated onto Vero cell monolayers seeded in 96-well culture plates. After 20–24 h incubation, the number of GFP-positive cells in each well was determined using fluorescent microscopy. The titer of VSV–NiV–GFP was defined as the reciprocal of the highest dilution which gave rise to positive fluorescence and expressed as infectious units. The virus stock was determined to contain 1.0 × 106 infectious units/ml.
2.6. Serum neutralization test with VSV–NiV–GFP
Serial twofold dilutions (160 μl) of the sera were mixed with an equal volume of VSV–NiV–GFP supernatant containing 5000 infectious units of pseudotyped viruses. After incubation at 37 °C for 1 h, 100 μl of each mixture were inoculated onto Vero cell monolayers in the 96-well plate in triplicate. The cultures were incubated at 37 °C for 20–24 h, and the neutralization titers were determined. The number of cells emitting fluorescence was counted using a fluorescent microscope IX71 (OLYMPUS, Tokyo, Japan) or BZ-8000 “Bio-zero” (KEYENCE, Osaka, Japan). Alternatively, the fluorescent intensity of individual wells was measured using the Fluoroskan Ascent Luminometer (Thermo Fisher Scientific, Waltham, MA, USA) under the conditions of 20 ms integration with excitation and emission of 485 nm and 538 nm, respectively.
2.7. Serum neutralization test with live NiV
Serial twofold dilutions of the test sera were made using EMEM without FCS. 50 μl aliquots of serially diluted antibody were mixed with an equal volume of EMEM containing 100 TCID50 of live NiV and incubated for 30 min at 37 °C. 100 μl of Vero cell suspension containing 5 × 105 cells/ml were added and the mixture was incubated at 37 °C in a CO2 incubator for 3 days. The neutralization titers were expressed as the reciprocal of the dilution of antibody that completely blocked development of CPE.
3. Results
3.1. Serum neutralization test
To test whether VSV–NiV–GFP could be used in place of live virus in a serum neutralization test for NiV, rabbit αNiV-F and αNiV-G sera and other NiV- or HeV-reactive sera (Table 1) were examined for their ability to neutralize VSV–NiV–GFP. In this study, the level of pseudotyped virus infection was determined primarily by counting the GFP-expressing cells. Pre-incubation with the αNiV-F and αNiV-G rabbit sera neutralized the infectivity of VSV–NiV–GFP in a dose-dependent manner (Fig. 1a), while no neutralization was observed with sera from naive rabbits (Fig. 1b). Alternatively, the fluorescent intensity of individual wells was measured using a plate fluorometer to indicate the VSV–NiV–GFP infectivity. Although the latter method gave non-specific fluorescence in several wells due to the presence of debris, the results obtained by both methods correlated well when wells showing extraordinarily high fluorescent intensity were omitted (data not shown).
Fig. 1.
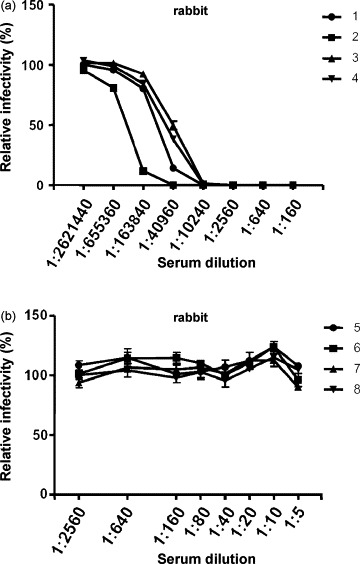
The specificity of VSV–NiV–GFP infection. VSV–NiV–GFP was pre-incubated with serially diluted rabbit αNiV-F and αNiV-G sera (a) and non-immunized sera (b). The infectivity of VSV–NiV–GFP was measured by counting GFP-expressing cells. The number of GFP-expressing cells in the absence of sample serum was set as 100%. The results are shown as mean ± SD for at least three independent assays.
Subsequently, serum neutralization tests were performed using sera from several species of animals (Table 1). VSV–NiV–GFP was neutralized by sera from various animals including humans which were positive by NiV-specific ELISA. Except for one serum (cat No. 1 in Fig. 2d), no neutralizing activity was observed in those sera that were negative by ELISA. However, naive sera from a cat (No. 4 in Fig. 2d), a negative control pig in the experimental infection with NiV (No. 4 in Fig. 2e) and a Malaysian field pig (No. 17 in Fig. 2f) that were ELISA negative showed some VSV–NiV–GFP neutralization at lower dilutions (<1:40), and this was likely due to a low level of non-specific inhibition of pseudotyped virus infection.
Fig. 2.
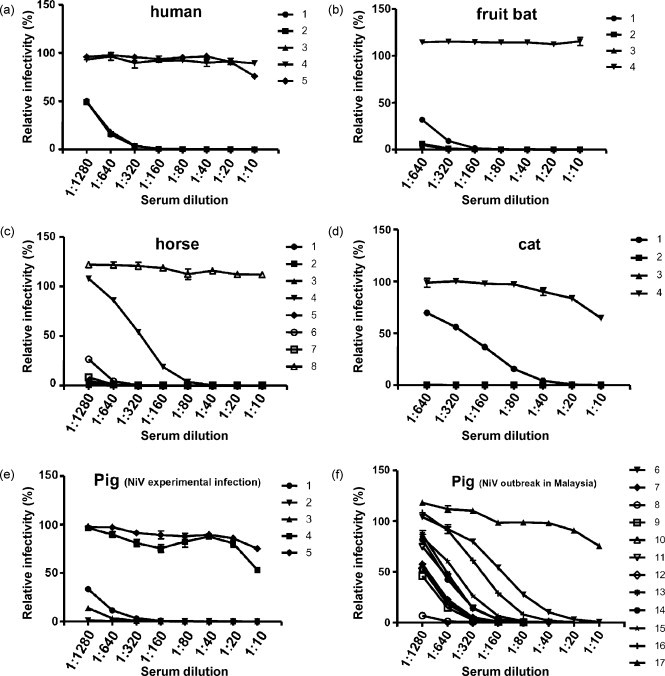
The reduction curves of VSV–NiV–GFP infectivity obtained in the serum neutralization test against the sera of various species; human (a), fruit bat (b), horse (c), cat (d), NiV-experimentally-infected pig (e) and Malaysian field pig (f). The infectivity of VSV–NiV–GFP was measured by counting the number of GFP-expressing cells as described in Fig. 1.
3.2. Estimating the non-specific inhibition of pseudotyped virus infection by pig sera
Since some ELISA negative sera from animals, pigs in particular, appeared to show slight inhibition of VSV–NiV–GFP infectivity when lower dilutions of sera were examined, 12 other pig sera from Japan, where no NiV infection has been detected, were examined for the presence of non-specific inhibition of pseudotyped virus infection. To confirm whether this inhibition was related to pre-exposure to other pathogens, the 12 pigs included 8 pigs (group A) that had been confirmed to have no antibodies against Japanese encephalitis virus (JEV), and four pigs (group B) that were JEV antibody-positive.
As shown in Fig. 3a, all the sera exhibited varying levels of non-specific inhibition at dilutions less than 1:40 or 1:80. Since the sera from group B which were αJEV-antibody positive showed slightly higher inhibition of VSV–NiV–GFP, another serum neutralization test was conducted using sera obtained from five pigs in a previous experiment that had been infected experimentally with JEV (Williams et al., 2001). Interestingly, these five sera showed no inhibition against VSV–NiV–GFP infection even at the minimum serum dilution (1:10) (Fig. 3b). This indicated that the non-specific inhibition seen in pig group B (Fig. 3a) was not attributable to the existence of αJEV-antibody.
Fig. 3.
The reduction curve of VSV–NiV–GFP infectivity obtained in the serum neutralization test against the sera of Japanese field pigs (a) and JEV-experimentally-infected pigs (b). The infectivity of VSV–NiV–GFP was measured by counting the number of GFP-expressing cells as described in Fig. 1.
3.3. Setting the cut-off value and threshold serum concentration
A cut-off value to define positive versus negative test outcomes and threshold serum concentration to be analyzed were determined next. For this purpose, the sera obtained from Japanese field pigs (Fig. 3a), which were thought to have no previous exposure to NiV, were used. In previous reports of serum neutralization test using pseudotyped viruses expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus (SARS-CoV) (Fukushi et al., 2006, Fukushi et al., 2005), 50% reduction of infectivity compared to the no-serum control was used as the cut-off value to define a positive neutralization outcome. In this study, a similar approach was used with the neutralization titer defined as the reciprocal of the highest serum dilution resulting in at least 50% reduction in VSV–NiV–GFP infectivity. Analysis of the data presented in Fig. 3a from sero-negative pigs demonstrates that non-specific inhibition may occur in serum samples up to 1:80. Diluting the sample sera greater than 1:80 as a starting dilution should avoid potential non-specific inhibition and allow testing of samples where only small amounts of sera are available. If testing only a 1:80 dilution of sera as a preliminary screen for sero-positive animals, all sera tested as shown in Fig. 2 would have been correctly characterized as either positive or negative. This supports the use of 1:80 as the threshold serum concentration for determining specific neutralization with high confidence.
3.4. Comparison of neutralization titers between serum neutralization tests using VSV–NiV–GFP and live-NiV
Using the neutralization test parameters defined above, the neutralization titers of all the sera were determined by new serum neutralization test with VSV–NiV–GFP and compared with those obtained by conventional test with live-NiV (Table 1). In all the sera, the neutralization titers using the VSV–NiV–GFP based assay were significantly higher than those obtained using the conventional neutralization test employing live-NiV. There appeared to be positive correlation between the neutralization titer obtained by both methods within respective species and antigens (in the case of rabbit monospecific antisera). However, the exact correlation coefficient could not be obtained since the neutralization titers of several sera exceeded the highest dilution (1:640 or 1:1280) used in this study.
In serum neutralization test with live-NiV, two sera from Malaysian pigs (Nos. 6 and 17) showed cell toxicity when they were used at the lowest dilution (<1:20).
4. Discussion
In this study, a serum neutralization test using pseudotyped viruses possessing NiV-F/G proteins (VSV–NiV–GFP) for detection of NiV infection was developed. Due to their inability to produce infectious progeny virus and quantitative nature, pseudotyped viruses would provide a safe diagnostic tool for conducting serum neutralization tests and facilitate research on virus entry and cell tropism. One of the pseudotyping system used most commonly is based on vesicular stomatitis virus (VSV) and the VSVΔG* system, in which the VSV G gene is replaced by the green fluorescent protein (GFP) gene. So far, the VSVΔG* system has been reported to produce several pseudotyped viruses incorporating the envelope glycoproteins of RNA viruses, such as measles virus (Tatsuo et al., 2000), hantavirus (Ogino et al., 2003), Ebola virus (Takada et al., 1997), hepatitis C virus (Matsuura et al., 2001) and severe acute respiratory syndrome coronavirus (SARS-CoV) (Fukushi et al., 2005). For diagnostics, a neutralization test, using SARS-CoV-S proteins-bearing pseudotyped viruses was reported to be more sensitive than a conventional virus neutralization test using live SARS-CoV (Fukushi et al., 2006). Recently, pseudotyped viruses possessing NiV-F/G were developed to examine the virus entry (Negrete et al., 2005), however, they have not yet been examined for their potential in diagnostic applications. Using the serum neutralization test in this study, the infectivity of VSV–NiV–GFP was measured by counting the number of GFP-expressing cells or by measuring the fluorescent intensity using a plate fluorometer. This new assay is significant on several levels and will afford rapid and high-throughput diagnosis, facilitate epidemiological monitoring during outbreak investigations or surveillance, and can be conducted under BSL2 conditions with a greater level of safety and substantial cost savings.
The neutralization titers of sera from various species were compared between this method and the conventional test using live-NiV. Although there seemed to be positive correlation between the results obtained by the two methods, the former appeared far more sensitive than the latter (Table 1). This remarkably high sensitivity is likely due to the inability of VSV–NiV–GFP to undergo multi-cycle replication due to lack of its G gene. Indeed, neutralization of VSV–NiV–GFP can, therefore, be achieved with neutralizing antibodies that just inhibit the initial binding of pseudotyped viruses to the cells. Whereas, in the case of a serum neutralization test employing live-NiV, any virus which successfully enters the cell will replicate and multiply, and typically greater levels of neutralizing antibodies are required to inhibit further viral spread, often resulting in an apparently lower neutralization titer. A further advantage of the VSV–NiV–GFP system was its ability to use far less serum, hence reduced the potential effect of cell toxicity (Table 1). In addition, the data also demonstrated that NiV-neutralizing antibodies below the detection level of the conventional neutralization test could be detected using this assay system, for example cat No.1 (Table 1).
The non-specific inhibition of pseudotyped virus infection, seen particularly in pig sera in this study, was also observed in a previous study (Fukushi et al., 2006). In general, field sera seem to be more problematic in exhibiting non-specific reactivity in serum diagnosis (such as ELISA and virus neutralization tests) than sera from animals infected experimentally. The results shown in Fig. 3 indicated that the non-specific inhibition was not the result of αJEV antibody. One possibility is that the non-specific reactivity may result from differences in nutritional or immunological conditions of the animals, the degree of hemolysis or in the preparation of samples. However, by setting the appropriate threshold serum concentration to minimize the influence of non-specific activities, reliable diagnosis was achievable (Fig. 3).
Antisera to NiV and HeV have been shown to cross-neutralize, making it impossible to define viral etiology based on serum neutralization test using a single virus, NiV or HeV. To differentiate better between these two viral infections, development of pseudotyped viruses possessing HeV-F/G will be required. However, since current epidemiological information seems to indicate a non-overlapping geographical distribution of NiV and HeV, the VSV-based assay system could serve as a useful tool to carry out sero-epidemiological investigations on either virus until a HeV-specific system is established.
In conclusion, a serum neutralization test using pseudotyped virus for NiV possessing several advantages over conventional assays using live virus has been developed. This assay does not require the use of live virus and can be conducted safely under BSL2 conditions, providing a new diagnostic tool that will help facilitate studies on the epidemiology of NiV and one that can be deployed readily in countries where BSL4 containment laboratories are not available.
Acknowledgements
We thank Dr. M.A. Whitt for providing the VSVΔG*. We also thank Dr. T. Takasaki (Department of Virology I, National Institute of Infectious Diseases, Japan) for providing the sera of Japanese field pigs. A part of this study was performed through Special Coordination Fund for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government.
References
- Bishop K.A., Broder C.C. Hendra and Nipah viruses: lethal zoonotic paramyxoviruses. In: Sheld W.M., Hammer S.M., Hughes J.M., editors. vol. 8. ASM Press; Washington, DC, USA: 2008. pp. 155–187. (Emerging Infections). [Google Scholar]
- Chadha M.S., Comer J.A., Lowe L., Rota P.A., Rollin P.E., Bellini W.J., Ksiazek T.G., Mishra A. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 2006;12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., Goldsmith C.S., Gubler D.J., Roehrig J.T., Eaton B., Gould A.R., Olson J., Field H., Daniels P., Ling A.E., Peters C.J., Anderson L.J., Mahy B.W. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Daniels P., Ksiazek T., Eaton B.T. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect. 2001;3:289–295. doi: 10.1016/s1286-4579(01)01382-x. [DOI] [PubMed] [Google Scholar]
- Eaton B.T., Broder C.C., Middleton D., Wang L.F. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton B.T., Mackenzie J.S., Wang L.-F. Henipaviruses. In: Knipe D.E.G.D.M., Lamb R.A., Straus S.E., Howley P.M., Martin M.A., Roizman B., editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; Philadelphia, USA: 2007. pp. 1587–1600. [Google Scholar]
- Fukushi S., Mizutani T., Saijo M., Kurane I., Taguchi F., Tashiro M., Morikawa S. Evaluation of a novel vesicular stomatitis virus pseudotype-based assay for detection of neutralizing antibody responses to SARS-CoV. J. Med. Virol. 2006;78:1509–1512. doi: 10.1002/jmv.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S., Mizutani T., Saijo M., Matsuyama S., Miyajima N., Taguchi F., Itamura S., Kurane I., Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86:2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Watanabe R., Taguchi F. Pseudotyped vesicular stomatitis virus for analysis of virus entry mediated by SARS coronavirus spike proteins. Methods Mol. Biol. 2008;454:331–338. doi: 10.1007/978-1-59745-181-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley E.S., Montgomery J.M., Hossain M.J., Bell M., Azad A.K., Islam M.R., Molla M.A., Carroll D.S., Ksiazek T.G., Rota P.A., Lowe L., Comer J.A., Rollin P., Czub M., Grolla A., Feldmann H., Luby S.P., Woodward J.L., Breiman R.F. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 2007;13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley E.S., Montgomery J.M., Hossain M.J., Islam M.R., Molla M.A., Shamsuzzaman S.M., Akram K., Zaman K., Asgari N., Comer J.A., Azad A.K., Rollin P.E., Ksiazek T.G., Breiman R.F. Risk of nosocomial transmission of Nipah virus in a Bangladesh hospital. Infect. Control Hosp. Epidemiol. 2007;28:740–742. doi: 10.1086/516665. [DOI] [PubMed] [Google Scholar]
- Harcourt B.H., Lowe L., Tamin A., Liu X., Bankamp B., Bowden N., Rollin P.E., Comer J.A., Ksiazek T.G., Hossain M.J., Gurley E.S., Breiman R.F., Bellini W.J., Rota P.A. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005;11:1594–1597. doi: 10.3201/eid1110.050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman D.T., Suu-Ire R., Breed A.C., McEachern J.A., Wang L., Wood J.L., Cunningham A.A. Evidence of henipavirus infection in West African fruit bats. PLoS ONE. 2008;3:e2739. doi: 10.1371/journal.pone.0002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu V.P., Hossain M.J., Parashar U.D., Ali M.M., Ksiazek T.G., Kuzmin I., Niezgoda M., Rupprecht C., Bresee J., Breiman R.F. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N.D., Stillman E.A., Whitt M.A., Rose J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle C., Razafitrimo G., Razainirina J., Andriaholinirina N., Goodman S.M., Faure C., Georges-Courbot M.C., Rousset D., Reynes J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007;13:159–161. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang J., Hickey A.C., Zhang Y., Li Y., Wu Y., Zhang H., Yuan J., Han Z., McEachern J., Broder C.C., Wang L.F., Shi Z. Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg. Infect. Dis. 2008;14:1974–1976. doi: 10.3201/eid1412.080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby S.P., Rahman M., Hossain M.J., Blum L.S., Husain M.M., Gurley E., Khan R., Ahmed B.N., Rahman S., Nahar N., Kenah E., Comer J.A., Ksiazek T.G. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Tani H., Suzuki K., Kimura-Someya T., Suzuki R., Aizaki H., Ishii K., Moriishi K., Robison C.S., Whitt M.A., Miyamura T. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology. 2001;286:263–275. doi: 10.1006/viro.2001.0971. [DOI] [PubMed] [Google Scholar]
- McEachern J.A., Bingham J., Crameri G., Green D.J., Hancock T.J., Middleton D., Feng Y.R., Broder C.C., Wang L.F., Bossart K.N. A recombinant subunit vaccine formulation protects against lethal Nipah virus challenge in cats. Vaccine. 2008;26:3842–3852. doi: 10.1016/j.vaccine.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D.J., Morrissy C.J., van der Heide B.M., Russell G.M., Braun M.A., Westbury H.A., Halpin K., Daniels P.W. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus) J. Comp. Pathol. 2007;136:266–272. doi: 10.1016/j.jcpa.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Middleton D.J., Westbury H.A., Morrissy C.J., van der Heide B.M., Russell G.M., Braun M.A., Hyatt A.D. Experimental Nipah virus infection in pigs and cats. J. Comp. Pathol. 2002;126:124–136. doi: 10.1053/jcpa.2001.0532. [DOI] [PubMed] [Google Scholar]
- Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- Negrete O.A., Levroney E.L., Aguilar H.C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Ogino M., Ebihara H., Lee B.H., Araki K., Lundkvist A., Kawaoka Y., Yoshimatsu K., Arikawa J. Use of vesicular stomatitis virus pseudotypes bearing hantaan or seoul virus envelope proteins in a rapid and safe neutralization test. Clin. Diagn. Lab. Immunol. 2003;10:154–160. doi: 10.1128/CDLI.10.1.154-160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE, 2008. Hendra and Nipah virus diseases, Manual of Diagnostic Tests and Vaccines for Terrestrial Animal, pp. 1227–1238.
- Olson J.G., Rupprecht C., Rollin P.E., An U.S., Niezgoda M., Clemins T., Walston J., Ksiazek T.G. Antibodies to Nipah-like virus in bats (Pteropus lylei), Cambodia. Emerg. Infect. Dis. 2002;8:987–988. doi: 10.3201/eid0809.010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch J.R., Crameri G., Wang L.F., Eaton B.T., Broder C.C. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 2007;4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J.M., Counor D., Ong S., Faure C., Seng V., Molia S., Walston J., Georges-Courbot M.C., Deubel V., Sarthou J.L. Nipah virus in Lyle's flying foxes, Cambodia. Emerg. Infect. Dis. 2005;11:1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendow I., Field H.E., Curran J., Darminto, Morrissy C., Meehan G., Buick T., Daniels P. Henipavirus in Pteropus vampyrus bats, Indonesia. Emerg. Infect. Dis. 2006;12:711–712. doi: 10.3201/eid1204.051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Robison C., Goto H., Sanchez A., Murti K.G., Whitt M.A., Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuo H., Okuma K., Tanaka K., Ono N., Minagawa H., Takade A., Matsuura Y., Yanagi Y. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J. Virol. 2000;74:4139–4145. doi: 10.1128/jvi.74.9.4139-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Lumlertdacha B., Boongird K., Wanghongsa S., Chanhome L., Rollin P., Stockton P., Rupprecht C.E., Ksiazek T.G., Hemachudha T. Bat Nipah virus, Thailand. Emerg. Infect. Dis. 2005;11:1949–1951. doi: 10.3201/eid1112.050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.T., Daniels P.W., Lunt R.A., Wang L.F., Newberry K.M., Mackenzie J.S. Experimental infections of pigs with Japanese encephalitis virus and closely related Australian flaviviruses. Am. J. Trop. Med. Hyg. 2001;65:379–387. doi: 10.4269/ajtmh.2001.65.379. [DOI] [PubMed] [Google Scholar]


