Abstract
Severe acute respiratory syndrome (SARS) is a serious infectious threat to public health. To create a novel trial vaccine and evaluate its potency, we attempted to generate a SARS inactivated vaccine using SARS coronavirus (SARS-CoV) strain F69 treated with formaldehyde and mixed with Al(OH)3. Three doses of the vaccine were used to challenge three groups of BALB/c mice. We found that the mice exhibited specific IgM on day 4 and IgG on day 8. The peak titers of IgG were at day 47 in low-dose group (1:19,200) and high-dose group (1:38,400) whereas in middle-dose group (1:19,200), the peak was at day 40. On day 63, the IgG levels reached a plateau. Neutralization assay demonstrated that the antisera could protect Vero-E6 cells from SARS-CoV's infection. Analysis of the antibody specificity revealed that the mouse antisera contained a mixture of antibodies specifically against the structure proteins of SARS-CoV. Furthermore, the mouse antisera conferred higher amount of antibodies against protein N, polypeptide S4 and S2 than those of proteins M and 3CL. These findings suggest that the inactivated SARS-CoV could preserve its antigenicity and the inactivated vaccine can stimulate mice to produce high levels of antibodies with neutralization activity. Results also suggest that polypeptides originating from protein N or S might be a potential target for the generation of a recombinant SARS vaccine.
Keywords: SARS, Inactivated vaccine, IgG antibody, Neutralization
1. Introduction
A severe acute respiratory disease of unknown etiology was first reported in Guangdong Province, the People's Republic of China in late 2002. Sequentially broke out in Hong Kong and Northern America in early 2003, this disease was named as “severe acute respiratory syndrome” (SARS) [1]. A novel coronavirus, SARS coronavirus (SARS-CoV), was first identified as an etiological pathogen of SARS. Several SARS-CoV genomes were mapped and proteome research was carried out. These achievements have paved paths for developing a safe and effective SARS vaccine.
A number of studies using coronavirus vaccines have demonstrated that inactivated vaccines are one of the most effective methods to protect against animal coronavirus, including canine coronavirus [2], infectious bronchitis virus [3] and bovine coronavirus [4]. Although the host–pathogen relationship and immunopathogenesis of SARS-CoV have not been fully understood, we postulated that it may be feasible to develop an inactivated vaccine for SARS-CoV. Moreover, the safety and effectiveness of inactivated vaccine in human have also been demonstrated by clinical applications of vaccines against Japanese encephalitis [5], hemorrhagic fever (type I) [6] and rabies [7].
This study was performed with two goals in mind: (i) to prepare a SARS inactivated vaccine, and (ii) to investigate whether this vaccine could stimulate animal models to produce neutralization antibodies. We used SARS-CoV, F69 isolated previously [8] to prepare a trial SARS inactivated vaccine. Our data on immunogenicity studies suggest that this vaccine could stimulate mice to produce high levels of antibodies which could protect Vero-E6 cells from SARS-CoV's challenge. These results may shed lights for the development of SARS vaccine in future.
2. Materials and methods
2.1. Reagents, cells and animals
SARS coronavirus antibody (Ab) diagnostic kits were purchased from Beijing BGI-GBI Biotech. HRP-conjugated goat antimouse IgG was purchased form Bethyl Laboratories. SARS Ab detection and typing kits were obtained from Shanghai Health Digit. Specific pathogen free Balb/c mice, weighted from 18 g to 22 g, were provided by the First Military Medical University of PLA.
2.2. Virus and vaccine preparation
SARS-CoV F69 was isolated from the oropharyngeal swab of a Cantonese patient who had lived in Amoy Gardens. DNA sequence comparison showed that F69 has highly homologous similarity to other strains reported, including HKU, US urbani and TOR2. The titer of F69 was determined to be 1 × 106.7 TCID50/ml with M-U method. Large-scale cultivated viruses were inactivated with 0.4% formaldehyde for 2 days at 37 °C, and then purified by gel filtration. The treated viruses were mixed with equivalent volume of Al(OH)3 adjuvant and used as a trial SARS vaccine.
2.3. Animal immunizations
Balb/c mice were randomly divided into three groups (n = 10 mice for each group) and challenged with the trial vaccine at three doses, low-dose (equivalent to 2.5 × 105 TCID50 of SARS-CoV), middle-dose (5.0 × 105 TCID50) and high-dose (7.5 × 105 TCID50). All grouped mice were challenged at three times (days 0, 14 and 24). The control group was injected with 0.9% NaCl at the same time. Mice were bled from the tail veins on days 4, 8, 12, 19, 26, 34, 40, 47, 56 and 63.
2.4. Antibody detection
IgM or IgG specific to SARS-CoV in mice sera were determined by indirect enzyme-linked immunosorbent assay (ELISA). In brief, polystyrene plates (Corning Costar) coated with lysed SARS-CoV were incubated with serially two-fold diluted sera, and then colorized with HRP-conjugated Ab and O-phenylendiamine (Sigma). The maximum dilution of each serum, which displayed positive reaction, was defined to be the titer of SARS-CoV specific Abs.
2.5. Specificity analysis of antisera
Chemiluminescent enzyme immunoassay (CLEIA) was applied to analyze the strength of mouse Abs binding to SARS-CoV structure proteins [9]. Seven recombinant proteins or polypeptides of SARS-CoV, S1, S2, S3, S4, N, M and 3CL, were immobilized on a solid matrix. Mouse antisera were incubated with the immobilized proteins, followed by enzyme-conjugated antimice IgG and chemiluminescent substance. The light signal of the reaction was detected by HD-2001A Chip Reader (Shanghai Health Digital), and P/N value, ratio of signal strength of antisera and that of negative control, were calculated using the instrument bound software.
Complete ORFs of protein N, M and 3CL of SARS-CoV TOR2 were cloned into plasmid pET22 and produced as a His-tag fused protein in Escherichia coli. The spike gene was divided into four fragments, S1 (nt 52-954), S2 (nt 772-1719), S3 (nt 1564-2799) and S4 (nt 2647-3765). Recombinant polypeptides S1 and S2 were produced using the same strategy as for protein N and 3CL. Polypeptides S2 and S4 were produced as a GST and pentra-His double-tagged fusion protein. After purification, seven recombinant polypeptides were analyzed in 12% SDS–PAGE and confirmed to be single bands with sizes identical to those deduced from their amino acid sequences (data not shown). The antigenicity of these recombinant polypeptides was further confirmed by Western blot analysis using a serum from a convalescent SARS patient.
2.6. Neutralization test
Neutralizing antibody was determined by the inhibition of cytopathic effects (CPE) defined by morphological changes and cellular death mediated by SARS-CoV on Vero-E6 cell monolayers as described [10]. In brief, Vero-E6 cells (3 × 105 cells/ml) were cultured in 24-well microtiter plates. Pretreated serum–virus mixture, containing equivalent volume of diluted serum and SARS-CoV (200 TCID50/ml), was inoculated into microtiter plates (1 ml/well) and incubated for 6 days at 37 °C in a 5% CO2 atmosphere. CPE of each well was recorded every day. The maximum dilution of each serum that completely prevented CPE in 50% of the test-wells was defined as the titer of this serum when the virus control (no serum) showed complete CPE.
3. Results
3.1. Changing of IgM Ab against SARS-CoV
Fig. 1 shows the production of IgM Ab in three groups of mice injected with different doses of vaccine. Specific IgM Ab levels in mice sera were expressed as OD490 of 1:100 diluted sera. We found that IgM Ab in plasma became detectable on day 4 and increased sharply on day 8 in vaccine-challenged groups. On day 19, the levels of IgM Ab remained stable and reached a plateau. On day 26, IgM Ab levels started to decline. Of note, the levels of IgM in different groups were generally correlated with the doses used for immunization.
Fig. 1.
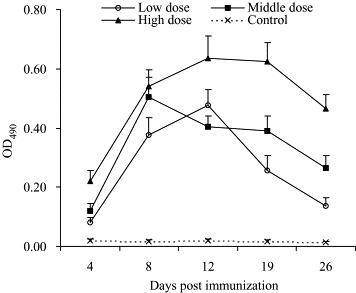
Changing of IgM Ab against SARS-CoV in mice sera. Three doses (equivalent to 2.5 × 105 TCID50, 5 × 105 TCID50 or 7.5 × 105 TCID50 of virus) of SARS inactivated vaccine were challenged in three groups of BALB/c mice on days 0, 14 and 24. Titers of IgM against SARS-CoV were measured by ELISA. The levels of IgM Ab are expressed as OD490 of 1:100 diluted sera.
3.2. Profile of IgG Ab against SARS-CoV
The profile of IgG Ab production in response to SARS vaccine is shown in Fig. 2 . The IgG Ab appeared in mice sera on day 8, reached to the peak on day 47 (middle-dose group was on day 40) and went into platform phase on day 63. The peak titer of IgG was 1:19,200 in low-dose group and middle-dose groups, and 1:38,400 in high-dose group, respectively. On day 63, the titers of three groups were 1:6400, 1:8000 and 1:16,000, respectively.
Fig. 2.
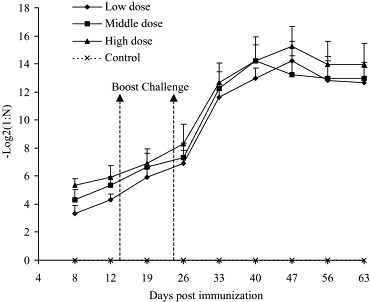
Kinetic changes of SARS-CoV specific IgG in mice sera. SARS-CoV specific IgG Ab in mice sera was measured by ELISA. The maximum dilution displayed positive reaction was defined to be the titer of this serum. Every titer value was transformed into negative log 2 and displayed in the graph. The values are expressed as mean ± S.D.
Comparing the titer among the different groups, we found that the levels of IgG Ab exhibited a clear correlation with the inoculation doses (except day 47). In general, the IgG titer of high-dose group was two–three-fold higher than that of low-dose group.
3.3. Antigen-binding specificity of antisera
Seven recombinant polypeptides originating from four SARS-CoV structure proteins, S, M, N and 3CL, were applied to determine the specificity and strength of Abs in mice antisera. We found that all antisera contained Abs specifically against seven antigens. However, in general, Abs specific to protein N, S4 and S2 tended to be stronger than other proteins M and 3CL (Fig. 3 ).
Fig. 3.
Specificity and the relative strength of antibodies specific to SARS-CoV structure proteins in mice sera on day 63. Protein N, M and 3CL of SARS-CoV TOR2 were produced as full-length protein from recombinant Escherichia coli. Protein S was divided into four fragments: S1, S2, S3 and S4. The seven recombinant polypeptides were immobilized on a solid matrix and chemiluminescent enzyme immunoassay (CLEIA) was applied to analyze the specificity and the relative strength of antibodies in antisera. P/N value, ratio of signal strength of antisera and that of control sera are listed above.
3.4. Neutralization activity of mice antisera
To determine the protective strength of antisera obtained from mice on days 12–63, we performed a neutralization assay using cultured Vero-E6 cells. We found that all antisera showed various ability of inhibiting cytopathic effects mediated by SARS-CoV in Vero-E6 cells (Fig. 4 ). From days 33 to 56, the neutralization Abs remained at the plateau. The peak titer of three vaccine-immunized groups was 1:2560, 1:5120 and 1:10,240, respectively. On day 63, the neutralization titer of low-dose group and middle-dose group was 1:1280; of high-dose group, it was 1:5120.
Fig. 4.
Kinetic changes of neutralization Ab in mice sera from day 12 to day 63. The maximum dilution of each sera sample being able to protect half test-wells from CPE was defined as the neutralization titer, which was transformed into negative log 2 and displayed in the graph.
4. Discussion
In this study, we demonstrated that the SARS inactivated vaccine can stimulate Balb/c mice to produce high levels of specific Abs with neutralization activity. In comparison with convalescent SARS patients, the Ab levels and neutralization activity of immunized mice were about 10 times higher. The curve of IgG Ab in mice indicates that the specific Abs remained at a high level for a long period. These results confirmed the feasibility and efficacy of SARS inactivated vaccine. We also performed the similar experiments using other species such as rats, rabbits, horses and monkeys which also confirmed the effect of this vaccine (Wang et al., in preparation).
Specificity analysis revealed that the IgG Ab in mice antisera was a mixture of Abs specific for structure proteins of SARS-CoV. Recent studies reported that the DNA vaccines targeting protein S or N could induce several events including the production of specific Abs and/or the activation of cytotoxic T lymphocytes, and protective immunity in mice [11], [12], [13]. A highly attenuated modified vaccinia virus Ankara containing the gene encoding full-length SARS-CoV S was also proved to be able to induce protective immunity in mice [14]. Here, we showed for the first time that the Abs specific to proteins N, S4 and S2 are stronger than other structure-protein specific Abs induced by inactivated vaccine. Sui et al. [15] recently reported a potent neutralization McAb, the epitope of which was located within the N-terminal 261–672 amino acids of protein S. In accordance with their observations, we found that the level of S2-specific Ab predominated in mice sera. From these results and other antigenicity analysis of SARS-CoV structure protein, we propose that protein S and/or N may be a potential subunit for an effective vaccine.
SARS-CoV, like HIV, is an RNA virus that has an error-prone replication mechanism. Escaping mutations in response to immune system have been observed and selected point mutations are known to be responsible for major shifts in the pathogenicity, as well as in the tissue specificity of bovine coronavirus [16]. Mutations in the genome of SARS-CoV have recently been described [17]. These mutations may hamper the development of effective vaccine against SARS-CoV; however, during the neutralization test of antisera induced by SARS inactivated vaccine, we found that the antisera could neutralize not only the CPE of SARS-CoV strain F69, but also strain Y3, another SARS-CoV isolated from a female patient who belonged to a “super-spread” event in Guangdong Province [18]. We did not find any significant difference between two strains in terms of the neutralization titer (data not shown). The cross-protection strengthened the urgency of evaluating the protection immunity of inactivated vaccine in primates in future.
Immune response to other coronaviruses suggests that both cell-mediated and humoral immunity contribute to long-term protection. But inactivated vaccine usually induces weak cell-mediated immunity. We also observed that there were no significant changes of CD4, CD8 subset of T lymphocyte in vaccine-challenged mice (data not shown). In the latest study it was found that Abs alone could prevent replication of the SARS coronavirus in the lungs of mice [10]. This observation suggests that the weakness of SARS inactivated vaccine might be overcome by the advantages of the vaccine produced in the current study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China, No. 3037017 and the Natural Science Foundation of Guangdong Province, China, No. 039213.
References
- 1.Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C., Lam B., Ip M.S., Chan J., Yuen K.Y., Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N Eng J Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 2.Pratelli A., Tinelli A., Decaro N., Cirone F., Elia G., Roperto S., Tempesta M., Buonavoglia C. Efficacy of an inactivated canine coronavirus vaccine in pups. New Microbiol. 2003;26:151–155. [PubMed] [Google Scholar]
- 3.Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takamura K., Matsumoto Y., Shimizu Y. Field study of bovine coronavirus vaccine enriched with hemagglutinating antigen for winter dysentery in dairy cows. Can J Vet Res. 2002;66:278–281. [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzuhara S., Nakamura H., Hayashida K., Obata J., Abe M., Sonoda K., Nishiyama K., Sugawara K., Takeda K., Honda T., Matsui H., Shigaki T., Kino Y., Mizokami H., Tanaka M., Mizuno K., Ueda K. Non-clinical and phase I clinical trials of a Vero cell-derived inactivated Japanese encephalitis vaccine. Vaccine. 2003;21:4519–4526. doi: 10.1016/s0264-410x(03)00506-1. [DOI] [PubMed] [Google Scholar]
- 6.Song G., Huang Y.C., Hang C.S., Hao F.Y., Li D.X., Zheng X.L., Liu W.M., Li S.L., Huo Z.W., Huei L.J., Zhang Q.F. Preliminary human trial of inactivated golden hamster kidney cell (GHKC) vaccine against haemorrhagic fever with renal syndrome (HFRS) Vaccine. 1992;10:214–216. doi: 10.1016/0264-410x(92)90154-c. [DOI] [PubMed] [Google Scholar]
- 7.Jones R.L., Froeschle J.E., Atmar R.L., Matthews J.S., Sanders R., Pardalos J., Moeller L., Chin J.E., Famula M., Briggs D.J., Lang J. Immunogenicity, safety and lot consistency in adults of a chromatographically purified Vero-cell rabies vaccine: a randomized, double-blind trial with human diploid cell rabies vaccine. Vaccine. 2001;19:4635–4643. doi: 10.1016/s0264-410x(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X., Li H., Zheng K., Chen Q.X., Wan Z.Y., Huang J.C., Zhong H.J., Zhou H.Q., Huang P., Zhang W.L., Diao L.M., Chen J.D., Zhang Q.F., Cui J.M., Huang X.J., Zhang J.Q. Isolation, identification and variance of a coronavirus from a imputing SARS case. Zhonghua Wei Sheng Wu Xue He Mian Yi Xue Za Zhi. 2003;23:409–413. [Google Scholar]
- 9.Botchkareva A.E., Eremin S.A., Montoya A., Manclus J.J., Mickova B., Rauch P., Fini F., Girotti S. Development of chemiluminescent ELISAs to DDT and its metabolites in food and environmental samples. J Immunol Methods. 2003;283:45–57. doi: 10.1016/j.jim.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing Ab prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu M.S., Pan Y., Chen H.Q., Shen Y., Wang X.C., Sun Y.J., Tao K.H. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol Lett. 2004;92:237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T.W., Lee J.H., Hung C.F., Peng S., Roden R., Wang M.C., Viscidi R., Tsai Y.C., He L., Chen P.J., Boyd D.A., Wu T.C. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78:4638–4645. doi: 10.1128/JVI.78.9.4638-4645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., Moore M.J., Tallarico A.S., Olurinde M., Choe H., Anderson L.J., Bellini W.J., Farzan M., Marasco W.A. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasoksuz M., Sreevatsan S., Cho K.O., Hoet A.E., Saif L.J. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. 2002;84:101–109. doi: 10.1016/S0168-1702(02)00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004;303:1666–9. [DOI] [PubMed]
- 18.Lu J.H., Zheng H.Y., Yan X.G., Wan Z.Y., Zhang R.L., Meng J.X., Guo Z.M., Yu X.B., Cheng S.Y., Ling W.H., Xu J., Wang Y.F., Xiong S., Li J.X., Han W.Y., Wang C.Y., Xiang H., Wu Y.S., Li M., Gu W.W. Establishment of SARS virus vaccine line (Y3) Guangdong Yi Xue. 2003;24(SARS Suppl II):204–205. [Google Scholar]


