Abstract
A rapid one-step reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay targeting the pol-integrase gene was developed to detect human immunodeficiency virus type 1 (HIV-1) group M. This HIV-1 RT-LAMP assay is simple and rapid, and amplification can be completed within 35 min under isothermal conditions at 60 °C. The 100% detection limit of HIV-1 RT-LAMP was determined using a standard strain (WHO HIV-1 [97/656]) in octuplicate and found to be 120 copies/ml. The RT-LAMP assay was evaluated for use for clinical diagnosis using plasma samples collected from 57 HIV-1-infected and 40 uninfected individuals in Cameroon, where highly divergent HIV-1 strains are prevalent. Of the 57 samples from infected individuals, 56 harbored group-M HIV-1 strains, such as subtypes A, B, G, F2, and circulating recombinant forms (CRFs) _01, _02, _09, _11, _13; all were RT-LAMP positive. One sample harboring group-O HIV-1 and the 40 HIV-1-uninfected samples were RT-LAMP negative. These findings indicate that HIV-1 RT-LAMP can detect HIV-1 group-M RNA from plasma samples rapidly and with high sensitivity and specificity. These data also suggest that this RT-LAMP assay can be useful for confirming HIV diagnosis, particularly in resource-limited settings.
Keywords: LAMP, HIV-1 group-M, Confirmatory test
1. Introduction
The number of people living with human immunodeficiency virus (HIV) infection was estimated at 33 million as of December 2007, and over 2.7 million people acquired new HIV infections in 2007 (UNAIDS, in press). HIV testing and counseling have been recognized as entry points for prevention, care, treatment, and support (WHO, 2004). Recently, rapid serological HIV tests have been introduced to facilitate radical scaling up of HIV testing and counseling services in many settings, such as in diagnosing and treating sexually transmitted infections, in services providing and linked to the prevention of mother-to-child transmission, and in general medical settings (WHO, 2004). It has been shown that sequential combinations of two or three antibody (Ab) tests (ELISA and/or rapid tests) are reliable for confirming HIV-positivity (WHO, 2004, Aghokeng et al., 2004, Carvalho et al., 1996, Meda et al., 1999). However, considering that the fourth generation HIV ELISA test, which can detect both HIV P24 antigen and HIV antibody in the same sample simultaneously, has been introduced to detect early-stage HIV infection (Meda et al., 1999) and that a combined antigen–antibody rapid test for diagnosing HIV will be introduced soon (Keren et al., 2008), a method for detecting rapidly HIV-1 RNA and/or proviral DNA to confirm HIV diagnosis in these settings would be a valuable diagnostic aid.
HIV-1 is classified into three groups: M, N, and O. Group M, which accounts for the HIV pandemic, is further classified into nine major clades (A–D, F–H, J, and K) and 42 circulating recombinant forms (CRFs; Heeney et al., 2006, Powell et al., 2007, HIV, 2008). The diverse nature of HIV causes difficulties in nucleotide-based diagnoses of HIV infection. In addition, low HIV DNA burden and low concentrations of HIV RNA in plasma often result in failure to detect HIV RNA or DNA in clinical specimens (Zazzi et al., 1995). These two factors, high diversity and low plasma RNA/proviral DNA concentration, limit the ability to diagnose HIV infection reliably and efficiently.
The reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay developed by Notomi is a simple method for nucleotide-based diagnostics that exhibits high sensitivity and specificity (Notomi et al., 2000). This method relies on auto-cycling strand displacement DNA synthesis by a DNA polymerase with high strand displacement activity and a set of two each of specially designed inner and outer primers. The entire RT-LAMP procedure can be completed in a single step at a constant temperature without a programmed thermal cycler. LAMP provides highly efficient DNA amplification, up to 109–1010 times in 15–60 min, and the concentration of the LAMP product is much higher than that generated by conventional polymerase chain reaction (PCR). Conventional PCR is relatively time consuming (3–4 h) and much more complicated than RT-LAMP, requiring several amplification steps and the use of a high-precision thermal cycler. The RT-LAMP assay has been validated and applied to the rapid detection of a number of RNA viruses, such as rubella virus (Mori et al., 2006), Japanese encephalitis virus (Toriniwa and Komiya, 2006), influenza virus (Ito et al., 2006), mumps virus (Okafuji et al., 2005), West Nile virus (Parida et al., 2004), severe acute respiratory syndrome corona virus (Hong et al., 2004, Poon et al., 2005), measles virus (Fujino et al., 2005), dengue virus (Parida et al., 2005), respiratory syncytial virus (Ushio et al., 2005), and HIV-1 (Curtis et al., 2008).
In the present study, another RT-LAMP assay was developed for the rapid detection of HIV-1 RNA. Its intended application is on-site confirmation of HIV diagnosis.
2. Materials and methods
2.1. Standard serum
WHO standard 97/656 (105 international units (IU) per vial, National Institute for Biological Standards and Control, Herts, UK) was used to determine the detection limit of the RT-LAMP assay (Davis et al., 2003, Holmes et al., 2001). The assay was carried out in octuplicate. The lowest concentration of genome copies with all octuplicate samples confirmed as positive was considered the detection limit.
2.2. Human plasma samples
Plasma samples were collected from 57 HIV-1-infected individuals in eastern Cameroon in 2001 (Ndembi et al., 2004) and 40 HIV-1-uninfected antenatal clinic attendees in western Cameroon in 2003. These samples were used to evaluate the sensitivity and specificity of HIV-1 RT-LAMP. In a previous study (Ndembi et al., 2004), phylogenetic analysis of genomic DNA samples from the 57 infected individuals revealed the presence of highly divergent strains of HIV-1 circulating in eastern Cameroon (Table 1 ). The 40 samples from uninfected individuals collected in 2003 were confirmed HIV-negative by HIV-Ab testing (AxSYM HIV1/2 and/or Determine HIV-1/2; Abbott Japan, Tokyo, Japan) and conventional PCR, as described previously (Ndembi et al., 2004).
Table 1.
HIV-1 genotype data for 57 infected individuals from eastern Cameroon and the results of HIV-1 RT-LAMP.
| Sample ID | Genetic subtypea |
LAMP |
||||
|---|---|---|---|---|---|---|
| gag | pol | env-C2V3 | gp41 | Ttb | EP | |
| 01CM2213 | CRF_01.AE | nac | CRF_01.AEA | na | 19.2d | Pe |
| 01CF2214 | G | U | U | na | 25.8 | P |
| 01CM2215 | CRF_02.AG | na | CRF_02.AG | na | 28.7 | P |
| 01CM2216 | A | na | A | na | 21.2 | P |
| 01CM2217 | CRF_11.cpx | na | CRF_11.cpx | na | 26.5 | P |
| 01CM2218 | CRF_11.cpx | CRF_11.cpx | nd | U | 31.0 | P |
| 01CM2219 | CRF_11.cpx | na | CRF_02.AG | na | No Tt | P |
| 01CM2220 | CRF_02.AG | na | A | na | 29.2 | P |
| 01CM2222 | CRF_02.AG | na | CRF_02.AG | na | 29.2 | P |
| 01CM2223 | CRF_01.AE | na | CRF_02.AG | na | 26.2 | P |
| 01CM2224 | CRF_02.AG | na | CRF_02.AG | na | 28.8 | P |
| 01CM2225 | B | na | A | na | 24.3 | P |
| 01CM2226 | CRF_02.AG | na | CRF_02.AG | na | 26.4 | P |
| 01CM2227 | CRF_02.AG | na | CRF_02.AG | na | 27.2 | P |
| 01CM2228 | CRF_02.AG | na | CRF_02.AG | na | 30.9 | P |
| 01CM2229 | CRF_11.cpx | na | CRF_11.cpx | na | 27.0 | P |
| 01CM2230 | A | na | A | na | 22.7 | P |
| 01CM2231 | CRF_02.AG | na | A | na | 23.4 | P |
| 01CM2232 | B | U | A | U | No Tt | P |
| 01CM2234 | CRF_11.cpx | na | CRF_02.AG | na | 26.0 | P |
| 01CM2235 | B | U | nd | U | 21.9 | P |
| 01CM2236 | CRF_02.AG | na | CRF_02.AG | na | 25.2 | P |
| 01CM2237 | F2 | na | F2 | na | 25.1 | P |
| 01CM2238 | CRF_13.cpx | na | CRF_01.AE | na | 22.2 | P |
| 01CM2239 | CRF_13.cpx | na | CRF_11.cpx | na | 26.2 | P |
| 01CM2240 | CRF_02.AG | na | CRF_13.cpx | na | 29.6 | P |
| 01CM2241 | CRF_01.AE | CRF_11cpx | nd | U | 27.5 | P |
| 01CM2242 | CRF_02.AG | na | CRF_02.AG | na | 24.8 | P |
| 01CM2243 | CRF_11.cpx | CRF_11cpx | nd | CRF_11.cpx | 24.7 | P |
| 01CM2244 | CRF_01.AE | na | CRF_11.cpx | na | 23.1 | P |
| 01CM2246 | B | na | CRF_01.AE | na | 23.6 | P |
| 01CF2247 | CRF_11.cpx | na | CRF_01.AE | na | 24.1 | P |
| 01CM2248 | CRF_01.AE | na | A | na | 21.9 | P |
| 01CM2249 | A | na | A | na | 23.6 | P |
| 01CM2250 | CRF_02.AG | CRF_02.AG | nd | U | 30.5 | P |
| 01CM2252 | CRF_02.AG | U | nd | U | 28.6 | P |
| 01CM2253 | CRF_01.AE | U | nd | A | 21.7 | P |
| 01CM2256 | CRF_01.AE | na | A | na | 21.6 | P |
| 01CM2257 | CRF_01.AE | na | A | na | 21.9 | P |
| 01CM2260 | CRF_13.cpx | U | A | CRF_13.cpx | 23.7 | P |
| 01CM2262 | B | na | CRF_02.AG | na | 27.8 | P |
| 01CF2268 | CRF_02.AG | CRF_02.AG | nd | CRF_02.AG | 32.5 | P |
| 01CM2269 | CRF_11.cpx | CRF_11.cpx | nd | CRF_11.cpx | 26.7 | P |
| 01CM2270 | CRF_02.AG | CRF_02.AG | nd | U | 31.9 | P |
| 01CM2271 | CRF_11.cpx | CRF_02.AG | nd | CRF_11.cpx | 23.9 | P |
| 01CM2272 | CRF_11.cpx | na | CRF_11.cpx | na | 21.2 | P |
| 01CM2273 | CRF_11.cpx | na | CRF_11.cpx | na | 25.5 | P |
| 01CM2274 | CRF_02.AG | na | CRF_02.AG | na | 22.6 | P |
| 01CM2275 | CRF_09.cpx | CRF_02.AG | nd | CRF_09.cpx | 24.5 | P |
| 01CM2276 | CRF_11.cpx | na | CRF_11.cpx | na | 23.9 | P |
| 01CM2277 | CRF_11.cpx | CRF_11.cpx | nd | CRF_11.cpx | 21.4 | P |
| 01CM2278 | B | na | CRF_02.AG | na | 24.2 | P |
| 01CM2280 | CRF_11.cpx | CRF_02.AG | nd | CRF_02.AG | 29.8 | P |
| 01CM2281 | CRF_02.AG | CRF_02.AG | nd | CRF_02.AG | 23.4 | P |
| 01CM2284 | CRF_11.cpx | CRF_11.cpx | nd | CRF_11.cpx | 24.5 | P |
| 01CM2287 | CRF_11.cpx | na | CRF_01.AE | na | 33.2 | P |
| 02CM319 | ndf | Og | nd | O | No Tt | Nh |
Genotyping based on part of gag-p24 (460 bp), env-C2V3 (approximately 550 bp), pol-integrase, and env-gp41 (approximately 405 bp) regions.
Threshold time by LA-200.
Not available.
Agarose gel electrophoresis.
Positive.
Not detected.
Group O.
Negative.
2.3. RNA preparation
HIV RNA was extracted from plasma as follows: 200 μl of plasma was incubated with 400 μl of lysis buffer consisting of 10 mM Tris–HCl (pH 8.0), 68% (w/v) guanidine isothiocyanate, 3% (w/v) dithiothreitol, and 4 μl of co-precipitant (10 mg/ml amylopectin azure) at 25 °C for 10 min. HIV RNA was precipitated by adding 600 μl of isopropanol and centrifuging at 20,000 × g for 15 min. The RNA pellet was washed with 70% ethanol and resuspended in 10 μl of RNAse-free and DNAse-free water.
2.4. Primer design
A set of primers that recognizes eight distinct target sites in the HIV-1 pol-integrase gene, a well-conserved region of HIV-1 genome, was designed based on the HIV-1 genome sequence (GenBank accession number K02013) using a primer-designing software program for LAMP (Primer Explorer ver. 2.0; Net laboratory, Japan, http://venus.netlaboratory.com; Table 2 ). The set consisted of the six following primers: a forward inner primer (FIP), backward inner primer (BIP), two outer primers (F3 and B3), and two loop primers (loop F and loop B). Two additional inner primers comprise the combination of two functionally different primer parts: FIP consists of F1c (complementary to F1) and F2 and BIP consists of B1c (complementary to B1) and B2. The sequences of the two loop primers are complementary to the primers located between regions corresponding to F1 and F2 primer sequences.
Table 2.
Sequences of primers used for HIV-1 RT-LAMP.
| Primer name | Sequence | Genome positiona |
|---|---|---|
| F3 | 5′-GGTAAGAGATCAGGCTGAACATC-3′ | 4721–4743 |
| F2 | 5′-AGACAGCAGTACAAATGGCA-3′ | 4747–4766 |
| Loop F | 5′-TTAAAATTGTGGATGAAT-3′ | 4786–4769 |
| F1c | 5′-CCCCAATCCCCCCTTTTCTT-3′ | 4806–4787 |
| B1c | 5′-AGTGCAGGGGAAAGAATAGTAGAC-3′ | 4812–4835 |
| Loop B | 5′-GCAACAGACATACAAACTAAAG-3′ | 4842–4863 |
| B2 | 5′-CTGCTGTCCCTGTAATAAACCC-3′ | 4921–4900 |
| B3 | 5′-GCTGGTCCTTTCCAAAGTGG-3′ | 4945–4926 |
| FIP | F1c + F2 | |
| BIP | B1c + B2 | |
In HIV-1HXB2.
2.5. RT-LAMP assay
The RT-LAMP reaction was carried out in 25 μl using a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd., Tochigi, Japan) containing FIP (40 pmol), BIP (40 pmol), F3 (5 pmol), B3 (5 pmol), loop F (40 pmol), loop B (40 pmol), Bst DNA polymerase (16 U), AMV reverse transcriptase (2 U), and 5 μl of target RNA. The reaction mixture was incubated at 60 °C for 60 min in a Loopamp real-time turbidimeter (LA-200; Teramecs, Kyoto, Japan; Fig. 1A). A turbidity value of more than 0.1 was considered positive. The amplified products of RT-LAMP were resolved by 2% agarose gel electrophoresis (Agarose S; Wako Pure Chemical Industries, Ltd., Osaka, Japan); the gel was stained with ethidium bromide and visualized using an ultraviolet (UV) transilluminator (Fig. 1B). The turbidity of the amplified products was also ascertained by naked eye. The amplified products were inspected further under UV irradiation with or without adding ethidium bromide, an intercalating dye, when RT-LAMP assay was carried out in the presence of Fluorescent Detection Reagent (Eiken Chemical Co., Ltd., Tokyo, Japan; Fig. 1C).
Fig. 1.
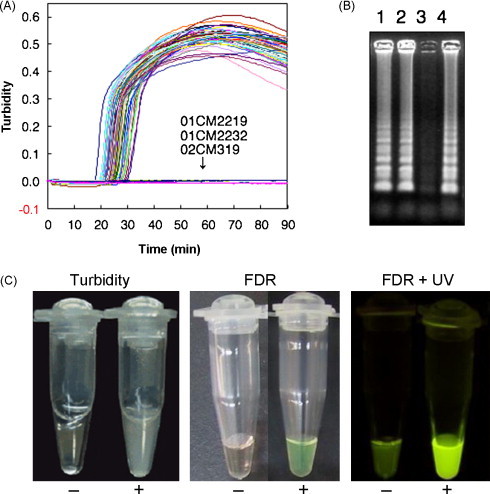
Real-time detection of HIV-1 RT-LAMP products of 57 HIV-1-positive samples from Cameroon by turbidimeter (LA-200). (A) Agarose gel electrophoresis of HIV-1 RT-LAMP products that were undetectable by LA-200. A turbidity value of more than 0.1 was considered positive. Turbidity of three samples (01CM2219, 01CM2232, and 02CM319) was less than 0.1. (B and C) Representative pictures of HIV-1 RT-LAMP products with (B) and without (C) Fluorescent Detection Reagent. (B) Lane 1: 01CM2219; lane 2: 01CM2232; lane 3: 02CM319; and lane 4: 01CM2213 (positive control). (C) HIV-1 RT-LAMP positive (+) and negative (−). FDR: Fluorescent Detection Reagent; UV: ultraviolet irradiation.
3. Results
3.1. Development of the HIV-1 RT-LAMP assay
Using the primer sets targeting the HIV-1 pol-integrase gene (Table 2), a one-step RT-LAMP assay for the rapid detection of HIV-1 RNA was standardized. The success of amplification was assessed using a real-time turbidimeter (LA-200; Fig. 1A). Threshold time (Tt), the time required for the turbidity value to exceed 0.1, is shown in Table 1. Amplification was also detected by the presence of a ladder-like pattern on a 2% agarose gel. The ladder-like pattern results from a mixture of stem-loop DNAs of various stem lengths and cauliflower-like structures with multiple loops (formed by annealing between alternately inverted repeats of the target sequence in the same strand; Fig. 1B). Furthermore, amplification was detected by naked eye inspection of turbidity; visual detection was enhanced further by the addition of Fluorescent Detection Reagent and/or the intercalating dye under UV irradiation (Fig. 1C).
3.2. Sensitivity and specificity of the HIV-1 RT-LAMP assay
The sensitivity of the RT-LAMP assay for detecting HIV-1 RNA was determined using RNA from WHO standard HIV-1 97/656 (105 IU/vial) diluted to 6000, 600, 240, 120, 90, and 60 copies/ml. One IU was reported to be equivalent to 0.62 genome copies (Davis et al., 2003). The assay was carried out in octuplicate using viral RNA extracted from the equivalent of 100 μl of diluted serum. The reproducible 100% detection limit of the RT-LAMP assay was 120 copies/ml.
Of the 57 HIV-1-positive samples, 54 were positive for RT-LAMP in 19.2–33.2 min as assessed by turbidity using the LA-200 detection system (Table 1 and Fig. 1A). HIV-1 RT-LAMP products of the two samples that were not detected by the real-time turbidimeter (01CM2219 and 01CM2232) could be detected by agarose gel electrophoresis (Fig. 1B) and by the naked eye after adding the intercalating dye under UV irradiation in the presence of Fluorescent Detection Reagent (data not shown). The remaining sample (02CM319) containing HIV-1 group-O RNA was RT-LAMP negative (Table 1 and Fig. 1B). Thus, all 56 samples that harbored HIV-1 group-M were positive by HIV-1 RT-LAMP assay.
Plasma specimens obtained from 40 pregnant women without HIV infection were also subjected to RT-LAMP and all were confirmed negative.
4. Discussion
An RT-LAMP assay was developed to detect HIV-1 RNA. This method was simple, rapid, and highly sensitive and specific for group-M HIV-1. Therefore, the HIV-1 RT-LAMP assay can be used as a rapid confirmatory test for HIV-1 group-M infection.
The HIV genome is usually detected by RT-PCR and PCR performed on plasma RNA and proviral DNA, respectively. These methods require at least 2–3 h despite the implementation of real-time PCR. In this study, the HIV-1 RT-LAMP assay was completed within 35 min, considerably faster than by RT-PCR or PCR. In addition, unlike RT-PCR and PCR, a simple apparatus such as a water bath can be used to maintain the constant incubation temperature at 60 °C.
The RT-LAMP reaction yields a white precipitate of magnesium pyrophosphate in the reaction mixture, indicating a positive result. This white precipitate is easily detected by the naked eye (Fig. 1C); thus, the results of the assay can be assessed without a turbidimeter. Although the amount of HIV-1 RT-LAMP products was monitored by a real-time turbidimeter (LA-200) in the current study, the results of visual inspection were consistent with those determined by turbidimeter (data not shown). According to the manufacturer's instructions for the Loopamp DNA amplification kit, visual detection can be enhanced by the addition of Fluorescent Detection Reagent to the reaction mixture. Interestingly, HIV-1 RT-LAMP products of the two samples that were undetectable by LA-200 (01CM2219 and 01CM2232) could be visualized by adding the intercalating dye under UV irradiation, when the assay was carried out in the presence of Fluorescent Detection Reagent. Thus, the HIV-1 RT-LAMP assay has the advantage of enabling the amplification of HIV-1 RNA and/or DNA in resource-limited settings in which sophisticated machines such as the thermal cycler and real-time turbidimeter are unavailable. In the two samples that were not detected by LA200, the production of magnesium pyrophosphate was prevented by unknown inhibitor(s). The cause and frequency of this phenomenon are under investigation.
RT-LAMP assay exhibits high specificity due to its use of multiple primers, including two loop primers, that recognize eight distinct regions of the target sequences. Previous studies in which RT-LAMP was used to detect various viral RNAs have documented the high specificity of RT-LAMP (Mori et al., 2006, Toriniwa and Komiya, 2006, Ito et al., 2006, Okafuji et al., 2005, Parida et al., 2004, Parida et al., 2005, Hong et al., 2004, Poon et al., 2005, Fujino et al., 2005, Ushio et al., 2005). Similarly, HIV-1 RT-LAMP analysis of 40 sero-negative and PCR-negative samples showed 100% specificity, making the RT-LAMP assay ideal for confirming diagnosis.
The 100% detection limit of the HIV-1 RT-LAMP assay was found to be 120 copies/ml (12 copies/100 μl/assay). This sensitivity is inferior to the quantification limit (50 copies/ml) of the UltraSensitive Assay of the COBAS AMPLICOR HIV-1 MONITOR test, v 1.5 (Roche), but superior to the detection limit of the Standard Assay in the kit (400 copies/ml), and typical RT-PCR assays. Furthermore, the sensitivity of the current HIV-1 RT-LAMP could be improved to reach or exceed that of the UltraSensitive Assay by using a larger initial plasma sample (more than 240 μl) for extracting viral RNA.
The HIV-1 RT-LAMP assay was evaluated using 57 HIV-1 strains belonging to nine different group-M subtypes/CRFs and one group O based on gag and pol sequences, respectively (Table 1): subtypes A (n = 3), B (n = 6), F2 (n = 1), G (n = 1), CRF_01AE (n = 8), CRF_02.AG (n = 17), CRF_09.cpx (n = 1), CRF_11.cpx (n = 16), CRF_13.cpx (n = 3), and group O (n = 1; Ndembi et al., 2004). This assay system identified all of the 56 group-M HIV-1 strains despite their diversity, but did not detect the group-O strain, indicating that the primers used in the current HIV-1 RT-LAMP assay were group-M specific. Thus, in order to detect not only all of the HIV-1 groups but also HIV type-2 strains as well, the design of universal primer set will be necessary.
Although the viral RNA extraction method used in this study is relatively easy and cheap as compared to conventional methods, it still requires knowledge and training not usually available in resource-limited settings. Therefore, it will be necessary to revise and simplify the extraction method in order to use this assay as a confirmatory test for HIV diagnosis in the field. Future evaluation of the direct use of plasma or serum after heating as a test material is warranted (Curtis et al., 2008).
In conclusion, a one-step RT-LAMP assay for detecting group-M HIV-1 has been developed. The RT-LAMP assay is simple, rapid, and highly sensitive and specific for group-M HIV-1; therefore, this assay can be used to confirm group-M HIV-1 diagnosis. Once the RNA extraction method is simplified, the group-M HIV-1 RT-LAMP assay will be ideal for use in resource-limited settings.
Acknowledgements
This work was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, MEXT Japan and the Ministry of Health, Labor and Welfare, Japan.
References
- Aghokeng A.F., Ewane L., Awazi B., Nanfack A., Delaporte E., Peeters M., Zekeng L. Evaluation of four simple/rapid assays and two fourth-generation ELISAs for the identification of HIV infection on a serum panel representing the HIV-1 group M genetic diversity in Cameroon. J. Acquir. Immune Defic. Syndr. 2004;37:1632–1640. doi: 10.1097/00126334-200412150-00018. [DOI] [PubMed] [Google Scholar]
- Carvalho M.B., Hammerschlak N., Vaz R.S., Ferreira O.C., Jr. Risk factor analysis and serological diagnosis of HIV-1/HIV-2 infection in a Brazilian blood donor population: validation of the World Health Organization strategy for HIV testing. AIDS. 1996;10:1135–1140. [PubMed] [Google Scholar]
- Curtis K.A., Rudolph D.L., Owen S.M. Rapid detection of HIV-1 by reverse-transcription, loop-medicated isothermal amplification (RT-LAMP) J. Virol. Methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Davis C., Heath A., Best S., Hewlett I., Lelie N., Schuurman R., Holmes H. Calibration of HIV-1 working reagents for nucleic acid amplification techniques against the 1st international standard for HIV-1 RNA. J. Virol. Methods. 2003;107:37–44. doi: 10.1016/s0166-0934(02)00187-8. [DOI] [PubMed] [Google Scholar]
- Fujino M., Yoshida N., Yamaguchi S., Hosaka N., Ota Y., Notomi T., Nakayama T. A simple method for the detection of measles virus genome by loop-mediated isothermal amplification (LAMP) J. Med. Virol. 2005;76:406–413. doi: 10.1002/jmv.20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney J.L., Dalgleish A.G., Weiss R.A. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–466. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- HIV sequence Compendium 2008, Los Alamos HIV Sequence Database, http://www.hiv.lanl.gov/.
- Holmes H., Davis C., Heath A., Hewlett I., Lelie N. An international collaborative study to establish the 1st international standard for HIV-1 RNA for use in nucleic acid-based techniques. J. Virol. Methods. 2001;92:141–150. doi: 10.1016/s0166-0934(00)00283-4. [DOI] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Watanabe M., Nakagawa N., Ihara T., Okuno Y. Rapid detection and typing of influenza A and B by loop-mediated isothermal amplification: comparison with immunochromatography and virus isolation. J. Virol. Methods. 2006;135:272–275. doi: 10.1016/j.jviromet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Keren T., Ruso I., Dwir O., Pessler-Cohen D., Sharon Y., Fish F. XVII International AIDS. 2008. A new fourth-generation rapid test Determine HIV-1/2 Ag/Ab Combo. THPE0052. [Google Scholar]
- Meda N., Gautier-Charpentier L., Soudre R.B., Dahourou H., Ouedraogo-Traore R., Ouangre A., Bambara A., Kpozehouen A., Sanou H., Valea D., Ky F., Cartoux M., Barin F., Van de Perre P. Serological diagnosis of human immunodeficiency virus in Burkina Faso: reliable, practical strategies using less expensive commercial test kits. Bull World Health Organ. 1999;77:731–739. [PMC free article] [PubMed] [Google Scholar]
- Mori N., Motegi Y., Shimamura Y., Ezaki T., Natsumeda T., Yonekawa T., Ota Y., Notomi T., Nakayama T. Development of a new method for diagnosis of rubella virus infection by reverse transcription-loop-mediated isothermal amplification. J. Clin. Microbiol. 2006;44:3268–3273. doi: 10.1128/JCM.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndembi N., Takehhisa J., Zekeng L., Kobayashi E., Ngansop C., Songok E.M., Kageyama S., Takemura T., Ido E., Hayami M., Kaptue L., Ichimura H. Genetic diversity of HIV type 1 in rural eastern Cameroon. J. Acquir. Immune Defic. Syndr. 2004;37:1641–1650. doi: 10.1097/00126334-200412150-00019. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafuji T., Yoshida N., Fujino M., Motegi Y., Ihara T., Ota Y., Notomi T., Nakayama T. Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:1625–1631. doi: 10.1128/JCM.43.4.1625-1631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Horioke K., Ishida H., Dash P.K., Saxene P., Jana A.M., Islam M.A., Inoue S., Hosaka N., Morita K. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Wong B.W., Chan K.H., Ng S.S., Yuen K.Y., Guan Y., Peiris J.S. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J. Clin. Microbiol. 2005;43:3457–3459. doi: 10.1128/JCM.43.7.3457-3459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R.L., Zhao J., Konings F.A., Tang S., Ewane L., Burda S., Urbanski M.M., Saa D.R., Hewlett I., Nyambi P.N. Circulating recombinant form (CRF) CRF37_cpx: an old strain in Cameroon composed of diverse, genetically diverse lineages of subtype A and G. AIDS Res. Hum. Retroviruses. 2007;23:923–933. doi: 10.1089/aid.2007.0040. [DOI] [PubMed] [Google Scholar]
- Toriniwa H., Komiya T. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription loop-mediated isothermal amplification. Microbiol. Immunol. 2006;50:379–387. doi: 10.1111/j.1348-0421.2006.tb03804.x. [DOI] [PubMed] [Google Scholar]
- UNAIDS, 2008. Report on the Global AIDS Epidemic.
- Ushio M., Yui I., Yoshida N., Fujino M., Yonekawa T., Ota Y., Notomi T., Nakayama T. Detection of respiratory syncytial virus genome by subgroups-A B specific reverse transcription loop-mediated isothermal amplification (RT-LAMP) J. Med. Virol. 2005;77:121–127. doi: 10.1002/jmv.20424. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2004. Rapid HIV Tests: Guidelines for Use in HIV Testing and Counseling Services in Resource-Constrained Settings. [Google Scholar]
- Zazzi M., Romano L., Catucci M., De Milito A., Almi P., Gonnelli A., Rubino M., Valensin P.E. Low human immunodeficiency virus type 1 (HIV-1) DNA burden as a major cause for failure to detect HIV-1 DNA in clinical specimens by PCR. J. Clin. Microbiol. 1995;33:205–208. doi: 10.1128/jcm.33.1.205-208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


