Highlights
-
•
We discuss progress in aptamer-based detection of viruses.
-
•
We consider the use of aptasensors for point-of-care diagnostics of viruses.
-
•
Aptamers have distinct advantages over antibodies for virus recognition.
-
•
There is strong demand for multiplexed diagnostic measurement of pathogens.
Keywords: Antibody, Aptamer, Assay, Detection, Diagnostics, Pathogen, Point of care, Sensor, Serotype, Virus
Abstract
Novel viral diagnostic tools need to be affordable, fast, accurate and easy to use with sensitivity and specificity equivalent or superior to current standards. At present, viral diagnostics are based on direct detection of viral components or indirect detection by measuring antibodies generated in response to viral infection. While sensitivity of detection and quantification are still important challenges, we expect major advances from new assay formats and synthetic binding molecules, such as aptamers. Compared to traditional antibody-based detection, aptamers could provide faster adaptation to continuously evolving virus strains and higher discriminating capacity between specific virus serotypes. Aptamers are very stable and easily modifiable, so are ideal molecules for detection and chemical sensing applications. Here, we review the use of aptasensors for detection of viral pathogens and consider the feasibility of aptasensors to become standard devices for point-of-care diagnostics of viruses.
1. Introduction
In general, viral diagnostic tests can be divided into two groups; they are either developed to detect the virus directly or indirectly; by determining the host response in particular virus-specific antibodies that are induced upon infection. While the indirect approach is probably the most common in current diagnostics, it cannot be used for hemodialysed and immunocompromised patients. Furthermore, the time lag between virus infection and the ability to detect specific antibodies varies from patient to patient and from virus to virus. For certain viruses, seroconversion occurs only months after infection, leading to false-negative results that can have dramatic consequences for blood donation or screening of populations at risk. In other cases, virus-specific antibodies are circulating in the blood long after the clearance of the virus infection, resulting in false-positive tests. These limitations can be avoided by direct detection of the virus (i.e., virus cultivation, antigen detection and nucleic acid-based detection) (Table 1 ).
Table 1.
Comparison of the available direct methods for the detection of viruses
| Method | Assay time | Assay limit | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|
| Virus isolation | |||||
| Conventional cell culturing | 3–10 days | 1 EID50/mL | Sensitive, accurate, broad detection range, viral isolate available | Time consuming, expertise required, not sensitive enough for all viruses | [1], [2] |
| Shell vial culturing + immunostaining | 1–3 days | NA | Faster than cell culture, detects viruses that replicate poorly in cell culture | Expertise and special equipment required, less sensitive for RSV and adenovirus, detection limited to viruses tested by pre-CPE staining | [1], [2] |
| Antigen detection | |||||
| Direct immunofluorescence Assay (DFA) | 3 h | NA | Sensitive, fast | In general, less sensitive than culturing, expertise and special equipment required | [3] |
| Immunochromatography lateral flow | <10 min | NA | Fast, specific, cheaper than PCR | Poor and variable sensitivity | [4] |
| Membrane-based enzyme immunoassays | 3 h | 1.0 ng, 103.5–105 VP | Rapid | High rate of false positives, less sensitive than DFA | [5] |
| Flow cytometry | <1 h | 2.8 × 106 VP/mL | Rapid | Less sensitive than culturing, sample needs purification | [6] |
| Nucleic acid-based detection | |||||
| Reverse transcriptase PCR (RT-PCR) | 5 h | 0.0256 HAU | Specific and sensitive | Expensive, expertise required, hardly applicable for POCT testing | [7] |
| Real-time quantitative PCR (qPCR) | 3 h | 10 copies/reaction | Rapid, specific and sensitive | Expensive, expertise required, not applicable for POCT | [8] |
| Nucleic acid sequence-based amplification (NASBA) | 6 h | 10 copies/µL | Specific and sensitive | High rate of false positives | [9], [10] |
| Reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) | 40–50 min | 0.1 pg total RNA | Simple, sensitive, rapid, visual identification, POC testing | High rate of false positives | [11], [12] |
NA, Not available; EID50, 50% egg infective dose – 1 EID50 is the amount of virus that will infect 50% of inoculated eggs; VP, Virus particles; HAU, Hemagglutination units – 1 HAU is the amount of virus needed to agglutinate an equal volume of standardized red blood cells.
Virus cultivation is highly specific and can be sensitive, but is labor-intensive and time consuming (it can take up to 10 days) [1]. Development of the shell-vial-culturing protocol led to a reduction in turn-around time as a result of the low-speed centrifugation procedure that facilitates viral entry into cells, but it still takes at least a day and the sensitivity is limited [2]. In contrast, viral antigen-detection methods already enable point-of-care tests (POCTs) with an analysis time of less than 1 h. However, the use of antibodies is costly and some tests are even less sensitive than virus cultivation or have high false-positive rates [5].
Nucleic-acid amplification-based detection is currently the standard diagnostic methodology in many hospitals. The main advantages are high sensitivity and specificity [8]. In addition to the ability to quantify the number of viruses using real-time quantitative PCR (qPCR), isolation of nucleic acids also allows additional analyses, such as sequencing, to gather information on sensitivity to anti-viral treatment and on the origin of the virus, which is important for outbreak management. Quantification of pathogens, in particular of viruses, is often used to stage disease activity, prognosticate disease progression, monitor efficacy of therapy and prevent transmission. For example, in the case of HIV, the risk of progression to AIDS is directly related to the magnitude of the viral load in plasma. In addition, the number of viruses of an HIV-infected mother is the strongest risk factor for transmission to the child during delivery. Another important example is reactivation of cytomegalovirus (CMV) infections after transplantation, which negatively impacts the transplantation outcomes leading to graft versus host disease. In these two examples, quantitative PCR methods are frequently used to monitor viral load. Although the PCR is relatively fast, generally 2–4 h, it is commonly performed in batch in a centralized laboratory and cannot be done near the patient that leads to increased turn-around time. The clinical relevance of qPCR sensitivity for viral detection is questionable, because typical viral infections result in large numbers of viral particles. The quest therefore continues for the ideal diagnostic method that is affordable, fast, accurate and easy to use outside of centralized laboratories.
In this respect, recent progress in the development of aptasensors, using aptamers for selective molecular recognition and application to viral-antigen detection is very promising. In this review, we describe the trends of the emerging field of aptasensors for detection of human viral pathogens in clinical samples in the context of the current practice in virus detection (Fig. 1 ).
Fig. 1.
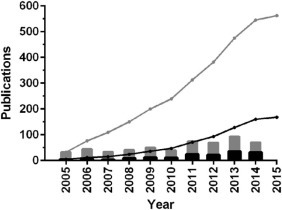
Trends in the analytical applications of aptamers for virus detection (vir* AND aptamer AND detect*) [Black] and in the broader field of aptamers selected for viruses (vir* AND aptamer) [Grey] according to Web of Science (on 3 April 2015). Columns denote yearly publications, lines represent their cumulative number starting from 1 January 2005].
2. Requirements for viral diagnostics
Currently, most of the viral diagnostic assays are still conducted in centralized laboratories and processed in batches. This is an important bottleneck, since it is pivotal to detect the viral agent as early as possible to guide adequate treatment and to take preventive measures. Determining the cause of infection solely based on clinical parameters is quite impossible, while the treatment of viral and bacterial infections differs significantly. Assay requirements for viral diagnostics, such as sensitivity and the possibility for multiplexing and quantification all depend on the purpose of the application.
In the case of a viral outbreak, sensitivity outweighs any other assay specification and false negatives should be avoided at any cost. Also, accuracy and rapidity of diagnosis are crucial for the control of an outbreak. A recent example is the Ebola virus epidemic in West Africa; accurate diagnosis in this context is especially important since early symptoms of Ebola virus disease mimic those of many other diseases commonly seen in this region, including symptoms associated with malaria, typhoid fever, and Lassa fever [13]. A multiplex capacity is, for example, required in the context of respiratory infections that can commonly be caused by a broad range of different viruses. For human immunodeficiency virus (HIV) and cytomegalovirus (CMV) infections, the viral load is strongly related to disease and clinical outcome, thus quantification is important to monitor treatment effectiveness.
The ideal diagnostic method, should be fast, accurate and easy to use, irrespective of the virus(es) or assay requirements. POCTs could comply with these unmet needs. Furthermore, it is important that the assay is compatible with a large variety of sample matrices, such as for human diagnostics, nasopharyngeal wash, oropharyngeal swabs, cerebrospinal fluid, stool, urine, and blood. The design of a POCT starts with the selection of the target. To select the most suitable viral target, it is vital to understand the anatomy and the replication cycle of the virus. The virus anatomy can be divided in three parts:
-
(1)
the genetic material, either RNA or DNA, inside the virus;
-
(2)
a coat consisting of proteins on which surface epitopes essential for attachment and infection are localized; and, for some viruses,
-
(3)
a lipid bilayer or envelope to protect the protein coat.
In viral diagnostics, the components targeted to detect the virus are whole virus (virion), nucleic acids and/or viral (Fig. 2 ). The envelope is not targeted, as it has hardly any particular features to enable selective recognition. Virus-particle quantification is difficult, due to the lack of proper standards; this problem was recently alleviated by emerging particle-characterization methods that can provide virion concentrations in a calibration-free manner by using the resistive pulse technique [14], [15] or optical methods [16]. These methods have been used to quantify poliovirus, which is one of the smallest viruses (~26 nm diameter).
Fig. 2.
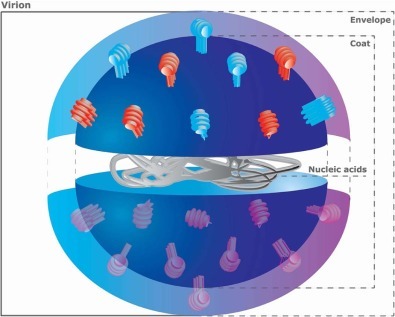
Virus anatomy.
During viral reproduction, virions are produced by the host cell, which expresses every component in excess and not all are integrated in a complete infectious virus. The viral components are therefore far more abundant than complete virions, so they are the preferred targets. The viral proteins seem to be better targets, since nucleic acids are rapidly degraded in a clinical matrix.
3. Virus-specific aptamer selection
Aptamers are short, single-stranded oligonucleotides with a length of 20–100 nucleotides that adopt unique sequence-dependent conformations conferring high affinity and specificity in binding a wide range of non-nucleic acid targets. The conformation of aptamers relies on base-pair stacking and contains hairpins, bulges, interior and multi-branch loops, but can also form complex 3D structures (e.g., the G-quadruplex conformation) [17]. The binding affinity and specificity of aptamers is similar and, in many cases, superior to monoclonal antibodies, but aptamers can be more rapidly, reproducibly and cost-effectively generated by a fully in vitro selection procedure [i.e., Systematic Evolution of Ligands by Exponential Enrichment (SELEX)] [18]. Since the SELEX method does not depend on living organisms, aptamers can be selected against non-immunogenic and toxic agents, and they can detect their targets even in non-physiological conditions [19]. Their fast generation is particularly attractive to anticipate continuously evolving virus strains. Moreover, by using counter-selection steps during SELEX, cross-reacting aptamers can be eliminated so as to result in aptamers with high discriminating capacity between different virus genotypes. Other advantages of aptamers can be attributed to their physical-chemical properties (i.e., they are chemically stable over a wide range of pH, storage conditions and temperature, and are not as sensitive to organic solvents as antibodies) [20]. Aptamers can be directionally, most often terminally, labeled with functional [21], [22], [23], [24], [25] or analytical [26], [27] probes without loss of function, while oriented labeling options for antibodies are limited and can lead to decreased affinity [19]. Further chemical modifications were reported to generate aptamer probes for diagnostic purposes that are fully resistant to nuclease enzymes [28]. All these are important steps towards the development of a new generation of viral diagnostic POCTs.
Over the years, starting in 1990, the SELEX procedure has been improved with many variations. Suggested improvements or alterations include increased variability of targets, target-binding conditions, different amplification methods or better partitioning of the non-binders. The success of the aptamer-generation procedure is primarily determined by the applied selection conditions and the target molecules chosen for selection. The first criterion must be met because, though aptamers can recognize their targets even under non-physiological conditions, ligand-aptamer interactions are very sensitive to the prevailing conditions. To evade this pitfall, the composition of selection buffer should be as close as possible to the matrix of real life samples in which the diagnostic exploitation of the aptamer is intended. Overlooking this condition could lead to a meaningless selection procedure (i.e., the isolated aptamer works ideally under optimal, laboratory conditions but is inadequate for practical application). Seemingly, this factor was ignored during the selection of many published aptamers, which could limit their diagnostic potential.
To raise virus-specific aptamers, the target of selection can be either a virus-derived protein or the inactivated virus particle. Traditionally, SELEX uses purified single proteins as ligand for selection and application of this approach resulted in several virus-selective aptamers. The protein-ligand-based SELEX methods were extensively discussed in recent reviews [29], [30]. When using inactivated virus particles as the target, isolation (purification) and inactivation of viruses are generally more challenging than production of pure proteins and require dedicated laboratories with strict safety control. These hurdles might explain why only a handful of whole virus-selected aptamers has been published so far. Nevertheless, one of the earliest studies demonstrated the feasibility of this approach, i.e., the virus-specific aptamer selection resulted in RNA oligonucleotides having the capacity to discriminate between closely-related human-influenza viruses [31]. The authors incubated the RNA pool with an A/Panama strain and separated the virus-selective oligonucleotides by filtration through a protein-binding nitrocellulose-acetate membrane. To increase the specificity of the isolated oligonucleotides, counter selection steps were also introduced using A/Aichi virus preparation. The aptamer obtained displayed over 15-fold-higher affinity to hemagglutinin compared with the monoclonal antibody and distinguished the various influenza viruses. The same procedure (i.e., isolation of aptamer candidates by passing the virus-oligonucleotide mixture through nitrocellulose membrane and removing the non–specific binders by counter selection with related virus) was effectively applied to raising DNA and RNA aptamers against murine norovirus and human influenza viruses, respectively [32], [33].
Although separation using a membrane is the traditional technique of SELEX and its usefulness was corroborated by several studies, immobilization of targets onto beads has become the method of choice for isolation of selective oligonucleotides. Two recent publications reported generation of aptamers against magnetic bead-immobilized human norovirus strains and influenza A/H1N1 virus [34], [35]. Both groups applied viruses captured by monoclonal antibody cross-linked paramagnetic beads as targets of SELEX and increased the selection pressure by introducing counter-selection steps with human-stool suspension and close relatives of the target virus, respectively. The performed enzyme-linked aptamer sorbent assay (ELASA) demonstrated that the norovirus-specific aptamers are selective for various strains of this virus. Importantly, the isolated aptamers could detect their target virus even in the clinical matrix feces [34]. Using this state-of-the-art procedure, influenza-virus aptamer selection was fully implemented in an integrated microfluidic chip [35].
Developments in microfluidics brought about the appearance of SELEX chips in recent years. Due to their high speed, low reagent need, ease of tuning, and excellent partitioning efficiency, these devices are expected to become standards in aptamer selection.
Another bead-immobilization-based method, MonoLEX, was published for selecting aptamers against a whole virus [36]. The technique involves an affinity-chromatography step of oligonucleotide library with immobilized vaccinia virus followed by subsequent physical partitioning of the virus-coated resin and PCR amplification of the bound aptamers. In contrast to conventional SELEX, MonoLEX is accomplished in one step instead of iterative selection cycles.
Immobilization of the targets of aptamer selection has two adverse consequences:
-
•
stringent counter-selection steps are needed to screen out those oligonucleotides that bind to the support; and,
-
•
the target immobilization may lead to shielding of desired epitopes.
Homogeneous SELEX approaches evade these shortcomings, since they do not require the coupling of the target on a solid support. The homogenous aptamer-generating protocols are dominated by capillary electrophoresis-based SELEX (CE-SELEX) and its modifications [30]. CE-SELEX was applied for the isolation of avian influenza H9N2 virus-specific aptamers using purified hemagglutinin protein as target molecule, but conversion of this method for whole-virus-based selection has not yet been demonstrated [37]. The published, whole-virus-based aptamer-selection protocols highlight the necessity of extensive counter-selection steps with the pertinent viruses and the matrices of prospective analytes. Counter selection is technically not feasible by CE, so successful CE-SELEX demands target molecules with extremely high purity. This requirement is hard to fulfil with virus preparation, which limits the applicability of CE-SELEX for whole-virus selection.
Recently, a unique reverse manner immobilization method was described for production of aptamers against whole bovine viral diarrhea virus (BVDV). It is known that single-stranded DNA (ssDNA) can get adsorbed on graphene oxide (GO) sheets and the desorption can be triggered by conformational change of ssDNA upon target protein binding [38]. Leveraging this phenomenon, so-called GO-SELEX was invented and applied successfully for generation of highly-selective BVDV-specific aptamers (Fig. 3 ) [39]. In the first step of the selection, the incubation of oligonucleotide library with the classical swine-fever virus was followed by addition of GO. The oligonucleotides that did not bind to the counter targets were adsorbed to the surface of GO and separated by centrifugation, while oligonucleotides that bound to the counter target were suspended in the binding buffer. Then, the ssDNA was recovered from the GO with the addition of the target BVDV and amplified by PCR. Following five rounds of this very stringent selection, highly-selective aptamers were obtained and used in a surface-plasmon resonance (SPR)-based sandwich-type assay.
Fig. 3.
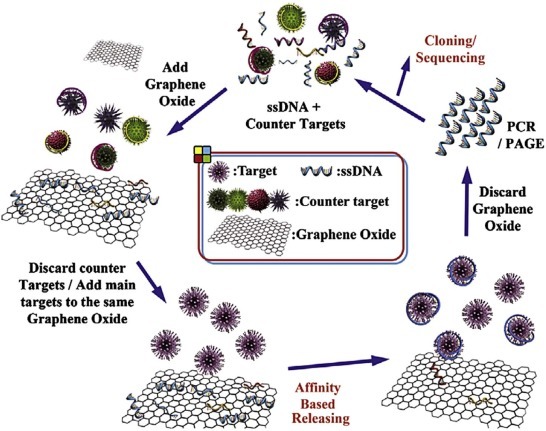
Aptamer selection by GO-SELEX.
{Reprinted with permission from Elsevier from [39], ©2014}.
4. Application of aptamers for virus detection
Recently, a whole range of virus-specific aptamers were generated against vaccinia virus [36], [40], dengue virus [41], severe acute respiratory syndrome (SARS) [42], [43], [44], [45], hepatitis C [46], [47], [48], [49], [50], [51], human immunodeficiency virus (HIV) [52], [53], [54], [55], apple stem pitting virus [56], [57], [58], bovine viral diarrhea virus [39], norovirus [34], [59], rabies virus [60], hepatitis B [61], Ebola [62] and influenza [31], [33], [35], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74].
The use of aptamers in aptasensors often requires immobilization of the aptamer strands as well as their labeling to enable detection. Due to their adaptive recognition mechanism and small size, aptamers require precaution in their immobilization. The surface density of aptamer monolayers is often critical and depends on the target size, as the binding affinity of the surface-confined aptamers may be influenced by steric hindrance or folding interference, which may ultimately reduce the sensitivity. Generally, one of the two ends of the aptamers (3′ or 5′) is modified with functionalities (e.g., -HS, NH2, or biotin) that allows convenient covalent attachment to different material transducers. The position (3′ or 5′) of the functional handle seems to affect the sensitivity, but in a hardly predictable way, as the best position will depend on the aptamer sequence [75]. Most often a linker, such as strand inserts consisting of several thymidine (T) units, known to exhibit low, non-specific adsorption, is incorporated between the recognition unit and the solid support.
For diagnostic applications, targets will be measured in bodily fluids containing a vast number of nucleases. To avoid degradation, a broad range of minor modifications to the oligonucleotides were shown to increase nuclease resistance [76], [77]. Beside all the modifications for stabilization and immobilization, an extensive range of molecular labels can be attached to RNA and DNA aptamers, sometimes with linker oligonucleotides or spacer molecules in between to reduce interference with target recognition. Such molecules are normally used for detection purposes and include fluorophores, quenchers and quantum dots [78].
Reliable, sensitive transduction of an aptasensor is pivotal for its functioning. There are many ways of transduction, but the vast majority transforms bio-recognition processes via a physicochemical transducer into measurable electrochemical, mass, or optical signal. Table 2 gives an overview of the recently developed aptamers and aptasensors in the context of viral pathogens.
Table 2.
Characteristics of aptasensors for human viral diagnostic purposes
| Target | Aptamer | Kd | Detection method | Transducer | Assay time | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| SARS coronavirus | |||||||
| Nucleocapsid protein (C-terminal) | RNA | 1.65 nM | Chemiluminescence immunosorbent assay (CLISA) | Optical | ±8 hours | 20 pg/mL | [45] |
| Nucleocapsid protein (C-terminal) | RNA | 1.65 nM | Nanoarray aptamer chip | Mass | ±6 hours | 2 pg/mL | [45] |
| Nucleocapsid protein | DNA | 4.93 ± 0.30 nM | Western blot | Optical | NA | NA | [44] |
| Influenza virus | |||||||
| Hemagglutinin + virion H5N1 | DNA | 4.65 nM | Surface plasmon resonance (SPR) | Optical | 1.5 hours | 0.128 HAU | [74] |
| Hemagglutinin H5N1 | DNA | 4.65 nM | Hydrogel-based quartz crystal microbalance (QCM) | Mass | 30 minutes | 0.0128 HAU | [64] |
| Hemagglutinin H5N1 | DNA | NA | Bio-nanogate | Electrochemical | 1 hour | 2.10−3 HAU | [79] |
| Polyvalent anti-influenza nucleoprotein | DNA | NA | Surface enhanced Raman spectroscopy (SERS) | Optical | >8 hours | 0.1 µg/mL HA | [68] |
| Hemagglutinin type B | RNA | 28 ± 3 nM | Sedimentation | Optical | >10 minutes | 3.108 VP | [69] |
| Hemagglutinin type A | RNA | 1.6 nM | Sedimentation | Optical | >10 minutes | 3.108 VP | [69] |
| Hemagglutinin type B | RNA | 0.7 and 1.2 ± 0.2 µg mL−1 | NA | Optical | NA | NA | [33] |
| Hemagglutinin H1N1 | DNA | 55.14 ± 22.4 nM | Polymerase chain reaction | Optical | NA | 6.4 × 10−3 HA | [35] |
| Rec. Hemagglutinin I H5N1 | DNA | 15.3 nM | Metal-enhanced fluorescence (MEF) | Optical | 30 minutes | 2–3.5 ng/mL | [67] |
| Hemagglutinin H1N1 | DNA | NA | Functionalized conductive polymer | Electrochemical | <15 minutes | 103 PFU/mL | [70] |
| Human norovirus | |||||||
| Capsid protein VP1 | DNA | NA | Surface plasmon resonance (SPR) | Optical | NA | NA | [59] |
| Hepatitis C virus | |||||||
| Envelop protein E2 | DNA | 1.05 ± 1 nM | Enzyme linked aptamer sorbent assay (ELASA) | Optical | NA | NA | [51] |
| Envelop protein E2 | DNA | 0.8–4.0 nM | Enzyme linked aptamer sorbent assay (ELASA) | Optical | NA | 1.25–2.50 × 103 FFU/mL | [47] |
| Envelop protein E2 | DNA | 0.8–4.0 nM | Enzyme linked aptamer sorbent assay (ELASA) | Optical | NA | 3.13–6.25 × 102 FFU/mL | [47] |
| Helicase | RNA | NA | Microcantilever | Mass | NA | 100 pg/mL | [49] |
| Core antigen | DNA | NA | Nucleic acid lateral flow strip | Optical | 10 minutes | 10 pg/mL | [80] |
| Core antigen, NS5 | RNA | 100 nM | Sol–gel chips | Optical | NA | NA | [48] |
| RNA polymerase | RNA | NA | Microbead-based affinity chromatography chip (µ-BACC) | Mass | NA | 9.6 fmol | [50] |
| Core antigen, NS5B | RNA | 6.3 nM | BioLayer Interferometry (BLI) | Optical | NA | 700 pg/mL | [46] |
| Hepatitis B | |||||||
| Surface antigen | RNA | NA | Fluorescence Resonance Energy Transfer (FRET) | Optical | NA | 1.25 mIU/mL | [61] |
| Vaccinia virus | |||||||
| Virion | DNA | NA | Surface plasmon resonance (SPR) | Optical | NA | NA | [36] |
| Virion | DNA | NA | Electrochemical impedance spectrometry (EIS) | Electrochemical | >1 hour | 330 PFU | [81] |
| Dengue virus | |||||||
| Genomic nucleic acids | DNA | NA | Fluorescence | Optical | NA | NA | [41] |
| Human immunodeficiency virus | |||||||
| HIV-1 Tat protein | RNA | NA | Surface plasmon resonance (SPR) | Optical | NA | NA | [52] |
| HIV-1 Tat protein | RNA | NA | Quartz crystal microbalance (QCM) | Mass | NA | 0.25 ppm | [52] |
| HIV-1 Reverse transcriptase | DNA | NA | Affinity capillary electrophoresis/laser-induced fluorescence (CE/LIF) | Optical | <5 minutes | 50 nM | [54] |
| HIV-1 NCp7 | RNA | NA | Solid-state nanopores | Electrochemical | NA | NA | [82] |
| HIV-1 Tat protein | RNA | NA | Diamond field-effect transistor (FET) | Electrochemical | NA | 1 nM | [53] |
NA, Not available; VP, Virus particles; HAU, Hemagglutination units; HA, Hemagglutinin; PFU, Plaque-forming units; FFU, Focus-forming units; mIU, milli-international units; ppm, parts per million.
4.1. Aptasensors with optical transduction
At present, the field of aptasensors for viral detection is very much dominated by optical transduction (e.g., based on measuring fluorescence, as well as colorimetric or refraction index related changes).
Fluorescent detection works with a signal-ON or a signal-OFF mechanism, which relies respectively on increasing or decreasing the fluorescent signal upon target binding. Signal-ON methods are much more sensitive, but fluorophores are also very sensitive to environmental changes and most methods were tested in aqueous buffers instead of bodily fluids. The nucleic-acid aptamers offer inherent fluorescent transduction mechanisms that are hardly possible with antibody-based recognition, such as the use of molecular beacon aptamers [83]. It was shown that RNA-aptamer beacons can be made specific to detect the presence of HIV-1 Tat proteins or its peptides (i.e., enabling them to discriminate RNA-binding proteins) by using split oligomers derived from RNA Tat aptamer. In the presence of the target, the two oligomers form a duplex that leads to the removal of the fluorophore from the vicinity of the quencher, and consequently to an increase in the fluorescence signal [83].
One-step quantification of viruses was successful by a similar fluorescence resonance-energy transfer (FRET)-based competitive binding assay, as demonstrated by the determination of hepatitis B virus [61]. Cy3 as a FRET donor was covalently attached to the anti-hepatitis B virus RNA aptamer and Cy5 labeled hepatitis B virus was the FRET acceptor. The presence of unlabeled target removed the acceptor from the vicinity of the donor leading to a shift in fluorescence in a dose-dependent fashion, which enabled the determination of the target concentration. This method appeared 40-fold more sensitive than the Abbott Architect assay, which is the current method for hepatitis B quantification [61].
The need for labeled aptamers, which is a significant cost-increasing factor of the assay, can be avoided if external signal reporters are used. A relevant example for such a system was based on the use of thiazole orange (TO) that fluoresces upon binding to the G-quadruplexes stabilized by target-aptamer interactions. This enabled the fluorescence detection of H5N1 influenza virus in human serum by using a guanine-rich aptamer for the recombinant hemagglutinin protein of the virus. An additional sensitivity enhancement was obtained by immobilizing the aptamers on Ag@SiO2 core-shell nanoparticles, which acted as a metal-enhanced fluorescence (MEF)-sensing platform. Thus, a limit of detection (LOD) of 3.5 ng/mL of the protein target could be realized with an assay time ~30 min in human serum [67]. A dramatically lower LOD was reported by Ahn et al. by microarray-based fluorescent detection, though tested in a less demanding matrix [45]. They developed an RNA aptamer against the C-terminal region of SARS coronavirus (SARS-CoV) nucleocapsid protein (with a KD of 1.65 nM) and implemented it in aptamer–antibody hybrid immunoassays [i.e., chemiluminescence immunosorbent assay (CLISA) and a nanoarray aptamer chip]. CLISA had an LOD of 20 pg/mL purified nucleocapsid protein, which was similar to ELISA [84], while the nanoarray aptamer chip had an even lower LOD of 2 pg/mL. However, the detection of the SARS-CoV nucleocapsid was still antibody mediated, and, similar to other immunosorbent assays published [47], [51], a relatively long assay time due to the required labeling made these assays less suitable for POCT.
SPR has also been often examined for its POCT potential [36], [52], [59]. Recently, Bai et al. [74] developed a portable biosensor based on SPR for the detection of avian influenza virus (AIV), H5N1, in poultry swabs. They used hemagglutinin as the target to select a DNA aptamer and subsequently used whole AIV H5N1 particles as a target to increase the specificity. The DNA aptamer with the lowest Kd (4.65 nM) was selected and subsequently immobilized on the gold surface of the sensor. The detection range for poultry-swab samples containing the virus was 1.28–12.8 hemagglutination units (HAU). The SPR biosensor was very specific for AIV H5N2 and displayed only very low signals (<4%) when exposed to six other non-targeted AIVs. A great advantage of this portable aptasensor, when compared to more conventional methods, is the relatively short detection time of 1.5 h. At the same time, the system is label-free, lacks the use of antibodies and can be reused 5–7 times.
Aptamer selection can be performed to obtain a number of aptamers that bind different proteins or epitopes on a virus, enabling the use of sandwich assays to increase the sensitivity and the selectivity of virus detection. Park et al. have shown that a pair of aptamers can be selected to whole bovine viral diarrhea virus type 1 so that the sensitivity of SPR detection can be enhanced by decorating the surface-bound virus in a subsequent step with a secondary, gold-nanoparticle (AuNP)-labeled aptamer [39].
SERS was also used as a label-free optical method for the identification of influenza viruses [68] through detecting the binding of nucleoproteins of different influenza A and B strains to a polyvalent anti-influenza DNA aptamer. Incubation with three different influenza strains (A/Uruguay, A/Brisbane and B/Brisbane) altered the spectrum in a very distinguishable ways due to the conformational changes of the aptamer.
A common issue with many optical sensing methodologies, such as FRET [61], sol-gel chips [48], MEF [67], affinity CE with laser-induced fluorescence (CE-LIF) [54], and bio-layer interferometry (BLI) [46], is that they depend on sensitive, but bulky and expensive, read-out equipment. By contrast, colorimetric aptasensors or lateral-flow assays that would be more suitable for POCT can be evaluated even with the naked eye. Apparently, this aspect has so far received less attention in terms of using aptamers as selective recognition reagents. Colorimetric detection based on aggregation of aptamer-modified AuNPs by virions was explored for influenza virus. The aggregation was combined with a simple centrifugation step to enhance visual perception by precipitation of the aggregates. However, the LOD obtained (108 VP/mL) is at the upper end of the relevant virus load [69].
Published work on the application of lateral-flow assays with aptamer-based reagents is also scarce. The few reports available solely involved the measurement of viral coat proteins [80], [85]. For example, Wang et al. published a competitive lateral-flow assay in which the amount of gold-conjugated aptamer bound to complementary DNA in the test line was reduced by competitive binding to the target, hepatitis C core antigen. Such a test had an LOD of 10 pg/mL when using a scanner for detection and 100 pg/mL with the naked eye [80].
4.2. Mass-sensitive aptasensors
Mass-dependent transduction offers label-free detection if the transducers are modified by selective receptors. Mass-based transduction was realized through the use of quartz-crystal microbalances (QCMs) with aptamer-modified surfaces, as well as micromechanical sensors based on resonant microcantilevers [49].
The application of QCMs for aptamer-based detection of viral components was initiated by Minuni et al. [52], [55] through the detection of HIV-1 Tat protein. A comparison with a Tat antibody-based sensor resulted in higher sensitivity of the aptamer-based sensors, and good selectivity for both types of sensors, but neither was adequate to detect Tat protein in real samples.
Wang and Li [64] took advantage of the unique properties of aptamers over antibodies and created a hydrogel-based QCM aptasensor for the detection of AIV H5N1. When exposed to the viral antigen HA of AIV H5N1, the aptamer changed its conformation by disrupting the hybridization to bind the virus. The QCM sensor detected a decrease of the fundamental frequency of the quartz crystal, as a result of the mass change on the surface of the resonator [64]. In the proposed construction, the hydrogel-based QCM aptasensor proved to have an LOD about an order of magnitude lower than a conventional QCM set-up – 0.0128 HAU compared to 0.128 HAU, respectively. These results made this sensor a promising POCT, suitable for rapid testing (30 min), easy to use, sensitive and specific for the diagnosis of AIV H5N1, and able to compete with RT-PCR in terms of sensitivity.
Micromechanical sensing by using resonating cantilevers was applied for the detection of hepatitis C virus (HCV) with a sensitivity of 100 pg/mL [16], [49]. This method exploited immobilized aptamers on the top surface of the cantilever and their steric crowding upon binding to their bulkier targets. The latter created a surface stress that was detected through the change in the resonant frequency of the microcantilevers. The small size of the microcantilevers made it easy to facilitate high-throughput multiplex screening, which was advantageous for the POCT of multiple viruses. However, due to their susceptibility to environmental conditions, the measurements on the microcantilever are completely executed in a temperature- and relative humidity-controlled chamber, which hinders their use for POCT.
4.3. Electrochemical aptasensors
Label-free electrochemical transducers act upon the formation of the aptamer-target-recognition complex on conductive or semi-conductive surfaces, which changes the resistance and the capacitance of the solution-electrode interface. They are easy to miniaturize and offers LODs in the femtomolar to micromolar range. The sensitivity of these devices increases when the electrode separation decreases and nanochips with multiple aptamer sensors would enable the simultaneous detection of several viruses [86]. A drawback of decreasing the recognition interface is the reduced number of aptamers that can lead to a reduced dynamic range of the assay. However, in viral diagnostics, information on viral load is not always essential to assign proper treatment.
An example of an electrochemical sensor for label-free virus detection was the aptamer-based viability impedimetric sensor for viruses published by Labib et al. [81]. Using electrochemical-impedance spectroscopy (EIS) for detection, the sensor had an LOD of 330 PFU of vaccinia virus. Its unique feature was the capacity to distinguish viable from non-viable virus particles, but the small dynamic range, up to 3000 PFU, assay time of more than 1 h, and, most importantly, requirement for temperature-regulated incubation, made this method less suitable for POCT [81].
Solid-state nanopores can also make use of label-free electrochemical transduction [82]. This label-free real-time detection method with a single interaction sensitivity relies on formation of complexes between target and RNA aptamer, which are measured by the resistive-pulse technique. The principle of this technique was shown by the detection of HIV-1 virus nucleocapsid protein 7. However, its main application is foreseen as the characterization of the relevant biomolecular interactions rather than as a diagnostic tool.
Ruslinda et al. published a diamond field-effect transistor (FET)-based aptasensor for the detection of HIV-1 Tat protein, demonstrating a stable, reusable platform for POCT [53]. Diamond-FET measures the binding between aptamers and target by detecting changes in the charge distribution above the diamond surface. In general, FET-based sensors are simple to fabricate, but miniaturization often leads to fundamental problems in sensing capacity. In diamond-FET-based techniques, the scalability problems are overcome and show potential for a handheld device [87]. The results of the diamond-FET-based aptasensor display stable results measuring the Tat protein in clinical matrices with sensitivity up to 1 nM. These are impressive, first-time results, and very suitable in situations with abundant target molecules.
A competition-based electrochemical-detection method is the bio-nanogate-based biosensor to measure influenza H5N1, published by Wang et al. [79]. Lactate dehydrogenase (LDH) enzymes are coated on a glassy-carbon electrode, which is covered by a gold surface containing bio-nanogates. These bio-nanogates will bind the specific H5N1 aptamers when present, thereby blocking the access of the substrate to the immobilized LDH enzyme. However, in viral presence, the aptamers will interact with the virion, leaving the bio-nanogates open and inducing substrate conversion. The substrate conversion was electrochemically measured and resulted in a LOD of ~2 × 10−3 HAU. Although the method is highly sensitive, the use of LDH for signal conversion can be greatly influenced by the type of clinical matrix, as LDH itself is present in most human tissue.
Aptamer-functionalized conductive polymer (PEDOT-OH:TsO) microelectrodes, published by Kiilerich-Pedersen et al. [70], have high sensitivity; up to 10 plaque-forming units (pfu) of influenza A (H1N1) per mL were detected. The detection mechanism is based on measuring changes in the impedance when virions are captured by immobilized aptamers. The polymer fluidic device consists of four layers with electrodes and electrical connection patches patterned in PEDOT-OH:TsO, and is inserted in a circular device of ~50 mm in diameter [88]. Testing in the clinical matrix, saliva, led to a reduction in sensitivity (103 pfu/mL), but it is still well within the relevant spectrum of the viral load [89]. Since the sample measured was only 200 µL, the absolute interactions measured can be as low as 1–2 pfu. In summary, low-cost manufacture, handheld size, label-free, high sensitivity and assay-time below 15 min make the functionalized conductive polymer aptasensor promising for use as a POCT.
5. Outlook
Since the discovery of aptamers, expectations for clinical application, in particular, for diagnostic aptasensor development, were high. However, the use of aptamers in practice is still rare. Probably the most important scientific reason relates to difficulties in the selection of highly-selective aptamers for widely different targets and measurement conditions. Recently, the first promising aptasensors made their debut in viral diagnostics, which will hopefully increase interest in diagnostic aptasensors in the near future.
User parameters, such as a turn-round time of less than 15 min, portability, robustness and ease of use, are fundamental requirements of aptasensor-based POCT. However, the sensitivity, specificity, and selectivity, as well as quantitative and multiplex capacities, all depend on the clinical need. There is a strong demand for multiplexed diagnostic measurement of pathogens, as disease is often caused by co-infections.
In this respect several multiplex PCR methods have been developed for diagnosis of sepsis [90], and respiratory [91] and gastro-intestinal infections [92]. So far, only a few aptamer-based applications have shown potential for multiplex measurement of mixtures of bacteria [93], [94]. However, no aptamer-based detection method has been developed to measure multiple viruses. Therefore, it is important to define clearly these aspects per case and to follow an iterative approach to continue redefining assay requirements and selecting the most appropriate targets, aptamers and aptasensors, all affecting the development of the POCT (Fig. 4 ).
Fig. 4.
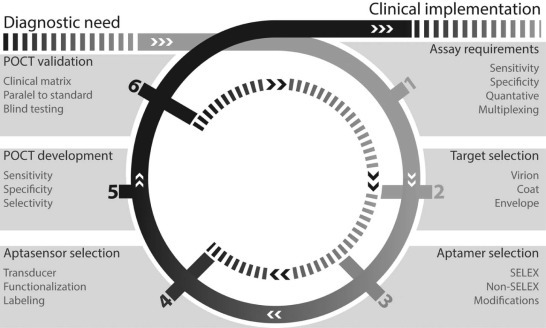
The design-build-test iterative process of aptasensor-based point-of-care testing (POCT).
Despite the seemingly unlimited possibilities for improvement of sensing technologies, the ultimate goal is to create an accurate, fast, affordable and easy-to-use detection system suitable for POCT that is able to become the standard for viral diagnostics. In order to accomplish this goal, it is important to perform comparative studies using different aptasensors targeting similar viruses in different matrices.
Acknowledgements
This work was supported by Agentschap NL, module IV grant PNU10A23, the Lendület program of the Hungarian Academy of Sciences (LP2013-63/2013), and the Technology Foundation STW (FES0901 and FES HTSM).
References
- 1.Ginocchio C.C. Detection of respiratory viruses using non-molecular based methods. J. Clin. Virol. 2007;40:S11–S14. doi: 10.1016/S1386-6532(07)70004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007;20:49–78. doi: 10.1128/CMR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shetty A.K., Treynor E., Hill D.W., Gutierrez K.M., Warford A., Baron E.J. Comparison of conventional viral cultures with direct fluorescent antibody stains for diagnosis of community-acquired respiratory virus infections in hospitalized children. Pediatr. Infect. Dis. J. 2003;22:789–794. doi: 10.1097/01.inf.0000083823.43526.97. [DOI] [PubMed] [Google Scholar]
- 4.Ngom B., Guo Y.C., Wang X.L., Bi D.R. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: a review. Anal. Bioanal. Chem. 2010;397:1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- 5.Luo Q.P., Huang H.L., Zou W., Dan H.B., Guo X.B., Zhang A.D. An indirect sandwich ELISA for the detection of avian influenza H5 subtype viruses using anti-hemagglutinin protein monoclonal antibody. Vet. Microbiol. 2009;137:24–30. doi: 10.1016/j.vetmic.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Rossi C., Kearney B., Olschner S., Williams P., Robinson C., Heinrich M. Evaluation of ViroCyt® Virus Counter for rapid filovirus quantitation. Viruses. 2015;7:857–872. doi: 10.3390/v7030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhumpa R., Handberg K.J., Jorgensen P.H., Yi S., Wolff A., Bang D.D. Rapid detection of avian influenza virus in chicken fecal samples by immunomagnetic capture reverse transcriptase-polymerase chain reaction assay. Diagn. Microbiol. Infect. Dis. 2011;69:258–265. doi: 10.1016/j.diagmicrobio.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Sidoti F., Rizzo F., Costa C., Astegiano S., Curtoni A., Mandola M.L. Development of real time RT-PCR assays for detection of type A influenza virus and for subtyping of avian H5 and H7 hemagglutinin subtypes. Mol. Biotechnol. 2010;44:41–50. doi: 10.1007/s12033-009-9211-7. [DOI] [PubMed] [Google Scholar]
- 9.Lau L.T., Banks J., Aherne R., Brown I.H., Dillon N., Collins R.A. Nucleic acid sequence-based amplification methods to detect avian influenza virus. Biochem. Biophys. Res. Commun. 2004;313:336–342. doi: 10.1016/j.bbrc.2003.11.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore C., Telles J.N., Corden S., Gao R.B., Vernet G., Van Aarle P. Development and validation of a commercial real-time NASBA assay for the rapid confirmation of influenza A H5N1 virus in clinical samples. J. Virol. Methods. 2010;170:173–176. doi: 10.1016/j.jviromet.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Peng Y., Xie Z.X., Liu J.B., Pang Y.S., Deng X.W., Xie Z.Q. Visual detection of H3 subtype avian influenza viruses by reverse transcription loop-mediated isothermal amplification assay. Virol. J. 2011;8:337. doi: 10.1186/1743-422X-8-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shigemoto N., Fukuda S., Takao S., Shimazu Y., Tanizawa Y., Kuwayama M. [Rapid detection of novel influenza A virus and seasonal influenza A (H1N1, H3N2) viruses by reverse transcription-loop-mediated isothermal amplification (RT-LAMP)] Kansenshogaku Zasshi. 2010;84:431–436. doi: 10.11150/kansenshogakuzasshi.84.431. [DOI] [PubMed] [Google Scholar]
- 13.WHO . World Health Organization; 2014. Barriers to Rapid Containment of the Ebola Outbreak, Global Alert and Response (GAR)http://www.who.int/csr/disease/ebola/overview-august-2014/en/ [Google Scholar]
- 14.Roberts G.S., Yu S., Zeng Q., Chan L.C.L., Anderson W., Colby A.H. Tunable pores for measuring concentrations of synthetic and biological nanoparticle dispersions. Biosens. Bioelectron. 2012;31:17–25. doi: 10.1016/j.bios.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 15.Terejánszky P., Makra I., Fürjes P., Gyurcsányi R.E. Calibration-less sizing and quantitation of polymeric nanoparticles and viruses with quartz nanopipets. Anal. Chem. 2014;86:4688–4697. doi: 10.1021/ac500184z. [DOI] [PubMed] [Google Scholar]
- 16.Makra I., Terejánszky P., Gyurcsányi R.E. A method based on light scattering to estimate the concentration of virus particles without the need for virus particle standards. MethodsX. 2015;2:91–99. doi: 10.1016/j.mex.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoinka J., Zotenko E., Friedman A., Sauna Z.E., Przytycka T.M. Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers. Bioinformatics. 2012;28:I215–I223. doi: 10.1093/bioinformatics/bts210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment – RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.O'Sullivan C.K. Aptasensors – the future of biosensing? Anal. Bioanal. Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 20.Janas T., Janas T. The selection of aptamers specific for membrane molecular targets. Cell. Mol. Biol. Lett. 2011;16:25–39. doi: 10.2478/s11658-010-0023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith J.E., Medley C.D., Tang Z.W., Shangguan D., Lofton C., Tan W.H. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 22.Kim K.S., Lee H.S., Yang J.A., Jo M.H., Hahn S.K. The fabrication, characterization and application of aptamer-functionalized Si-nanowire FET biosensors. Nanotechnology. 2009;20:235501. doi: 10.1088/0957-4484/20/23/235501. [DOI] [PubMed] [Google Scholar]
- 23.Kang W.J., Chae J.R., Cho Y.L., Lee J.D., Kim S. Multiplex imaging of single tumor cells using quantum-dot-conjugated aptamers. Small. 2009;5:2519–2522. doi: 10.1002/smll.200900848. [DOI] [PubMed] [Google Scholar]
- 24.Da Pieve C., Williams P., Haddleton D.M., Palmer R.M.J., Missailidis S. Modification of thiol functionalized aptamers by conjugation of synthetic polymers. Bioconjug. Chem. 2010;21:169–174. doi: 10.1021/bc900397s. [DOI] [PubMed] [Google Scholar]
- 25.Bugaut A., Toulme J.J., Rayner B. SELEX and dynamic combinatorial chemistry interplay for the selection of conjugated RNA aptamers. Org. Biomol. Chem. 2006;4:4082–4088. doi: 10.1039/b610890c. [DOI] [PubMed] [Google Scholar]
- 26.Ulrich H., Wrenger C. Disease-specific biomarker discovery by aptamers. Cytometry A. 2009;75A:727–733. doi: 10.1002/cyto.a.20766. [DOI] [PubMed] [Google Scholar]
- 27.Temur E., Zengin A., Boyaci I.H., Dudak F.C., Torul H., Tamer U. Attomole sensitivity of staphylococcal enterotoxin B detection using an aptamer-modified surface-enhanced raman scattering probe. Anal. Chem. 2012;84:10600–10606. doi: 10.1021/ac301924f. [DOI] [PubMed] [Google Scholar]
- 28.Szeitner Z., Lautner G., Nagy S.K., Gyurcsányi R.E., Mészáros T. A rational approach for generating cardiac troponin I selective Spiegelmers. Chem. Commun. 2014;50:6801–6804. doi: 10.1039/c4cc00447g. [DOI] [PubMed] [Google Scholar]
- 29.Blind M., Blank M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szeitner Z., András J., Gyurcsányi R.E., Mészáros T. Is less more? Lessons from aptamer selection strategies. J. Pharm. Biomed. Anal. 2014;101:58–65. doi: 10.1016/j.jpba.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 31.Gopinath S.C.B., Misono T.S., Kawasaki K., Mizuno T., Imai M., Odagiri T. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006;87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 32.Giamberardino A., Labib M., Hassan E.M., Tetro J.A., Springthorpe S., Sattar S.A. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE. 2013;8:e79087. doi: 10.1371/journal.pone.0079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakshmipriya T., Fujimaki M., Gopinath S.C.B., Awazu K. Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir. 2013;29:15107–15115. doi: 10.1021/la4027283. [DOI] [PubMed] [Google Scholar]
- 34.Escudero-Abarca B.I., Suh S.H., Moore M.D., Dwivedi H.P., Jaykus L.A. Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PLoS ONE. 2014;9:e106805. doi: 10.1371/journal.pone.0106805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai H.C., Wang C.H., Liou T.M., Lee G.B. Influenza A virus-specific aptamers screened by using an integrated microfluidic system. Lab Chip. 2014;14:2002–2013. doi: 10.1039/c4lc00187g. [DOI] [PubMed] [Google Scholar]
- 36.Nitsche A., Kurth A., Dunkhorst A., Panke O., Sielaff H., Junge W. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7:48. doi: 10.1186/1472-6750-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Yu Z., Jiang F., Fu P., Shen J., Wu W. Aptamers against Avian Influenza H9N2 Virus Prevent Viral Infection in Cells. PLoS ONE. 2015;10:e0123060. doi: 10.1371/journal.pone.0123060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C.H., Li J., Lin M.H., Wang Y.W., Yang H.H., Chen X. Amplified aptamer-based assay through catalytic recycling of the analyte. Angew. Chem. Int. Ed. Engl. 2010;49:8454–8457. doi: 10.1002/anie.201002822. [DOI] [PubMed] [Google Scholar]
- 39.Park J.W., Lee S.J., Choi E.J., Kim J., Song J.Y., Gu M.B. An ultra-sensitive detection of a whole virus using dual aptamers developed by immobilization-free screening. Biosens. Bioelectron. 2014;51:324–329. doi: 10.1016/j.bios.2013.07.052. [DOI] [PubMed] [Google Scholar]
- 40.Parekh P., Tang Z., Turner P.C., Moyer R.W., Tan W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 2010;82:8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fletcher S.J., Phillips L.W., Milligan A.S., Rodda S.J. Toward specific detection of Dengue virus serotypes using a novel modular biosensor. Biosens. Bioelectron. 2010;26:1696–1700. doi: 10.1016/j.bios.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 42.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho S.J., Woo H.M., Kim K.S., Oh J.W., Jeong Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn D.G., Jeon I.J., Kim J.D., Song M.S., Han S.R., Lee S.W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst. 2009;134:1896–1901. doi: 10.1039/b906788d. [DOI] [PubMed] [Google Scholar]
- 46.Roh C., Kim S.E., Jo S.K. Label free inhibitor screening of hepatitis C virus (HCV) NS5B viral protein using RNA oligonucleotide. Sensors (Basel) 2011;11:6685–6696. doi: 10.3390/s110706685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park J.H., Jee M.H., Kwon O.S., Keum S.J., Jang S.K. Infectivity of hepatitis C virus correlates with the amount of envelope protein E2: development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology. 2013;439:13–22. doi: 10.1016/j.virol.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Lee S., Kim Y.S., Jo M., Jin M., Lee D.K., Kim S. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 2007;358:47–52. doi: 10.1016/j.bbrc.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 49.Hwang K.S., Lee S.M., Eom K., Lee J.H., Lee Y.S., Park J.H. Nanomechanical microcantilever operated in vibration modes with use of RNA aptamer as receptor molecules for label-free detection of HCV helicase. Biosens. Bioelectron. 2007;23:459–465. doi: 10.1016/j.bios.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Cho S., Lee S.H., Chung W.J., Kim Y.K., Lee Y.S., Kim B.G. Microbead-based affinity chromatography chip using RNA aptamer modified with photocleavable linker. Electrophoresis. 2004;25:3730–3739. doi: 10.1002/elps.200406103. [DOI] [PubMed] [Google Scholar]
- 51.Chen F., Hu Y.L., Li D.Q., Chen H.D., Zhang X.L. CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for hepatitis C virus envelope glycoprotein E2. PLoS ONE. 2009;4:e8142. doi: 10.1371/journal.pone.0008142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tombelli S., Minunni M., Luzi E., Mascini M. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry. 2005;67:135–141. doi: 10.1016/j.bioelechem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 53.Rahim Ruslinda A., Tanabe K., Ibori S., Wang X., Kawarada H. Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 Tat protein. Biosens. Bioelectron. 2013;40:277–282. doi: 10.1016/j.bios.2012.07.048. [DOI] [PubMed] [Google Scholar]
- 54.Pavski V., Le X.C. Detection of human immunodeficiency virus type 1 reverse transcriptase using aptamers as probes in affinity capillary electrophoresis. Anal. Chem. 2001;73:6070–6076. doi: 10.1021/ac0107305. [DOI] [PubMed] [Google Scholar]
- 55.Minunni M., Tombelli S., Gullotto A., Luzi E., Mascini M. Development of biosensors with aptamers as bio-recognition element: the case of HIV-1 Tat protein. Biosens. Bioelectron. 2004;20:1149–1156. doi: 10.1016/j.bios.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 56.Balogh Z., Lautner G., Bardóczy V., Komorowska B., Gyurcsányi R.E., Mészáros T. Selection and versatile application of virus-specific aptamers. FASEB J. 2010;24:4187–4195. doi: 10.1096/fj.09-144246. [DOI] [PubMed] [Google Scholar]
- 57.Bai W., Spivak D.A. A double-imprinted diffraction-grating sensor based on a virus-responsive super-aptamer hydrogel derived from an impure extract. Angew. Chem. Int. Ed. Engl. 2014;53:2095–2098. doi: 10.1002/anie.201309462. [DOI] [PubMed] [Google Scholar]
- 58.Lautner G., Balogh Z., Bardóczy V., Mészáros T., Gyurcsányi R.E. Aptamer-based biochips for label-free detection of plant virus coat proteins by SPR imaging. Analyst. 2010;135:918–926. doi: 10.1039/b922829b. [DOI] [PubMed] [Google Scholar]
- 59.Beier R., Pahlke C., Quenzel P., Henseleit A., Boschke E., Cuniberti G. Selection of a DNA aptamer against norovirus capsid protein VP1. FEMS Microbiol. Lett. 2014;351:162–169. doi: 10.1111/1574-6968.12366. [DOI] [PubMed] [Google Scholar]
- 60.Liang H.R., Hu G.Q., Xue X.H., Li L., Zheng X.X., Gao Y.W. Selection of an aptamer against rabies virus: a new class of molecules with antiviral activity. Virus Res. 2014;184:7–13. doi: 10.1016/j.virusres.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 61.Suh S.K., Song S., Oh H.B., Hwang S.H., Hah S.S. Aptamer-based competitive binding assay for one-step quantitation of hepatitis B surface antigen. Analyst. 2014;139:4310–4314. doi: 10.1039/c4an00619d. [DOI] [PubMed] [Google Scholar]
- 62.Binning J.M., Wang T.J., Luthra P., Shabman R.S., Borek D.M., Liu G. Development of RNA aptamers targeting Ebola virus VP35. Biochemistry. 2013;52:8406–8419. doi: 10.1021/bi400704d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wongphatcharachai M., Wang P., Enomoto S., Webby R.J., Gramer M.R., Amonsin A. Neutralizing DNA aptamers against swine influenza H3N2 viruses. J. Clin. Microbiol. 2013;51:46–54. doi: 10.1128/JCM.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R.H., Li Y.B. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013;42:148–155. doi: 10.1016/j.bios.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 65.Shiratori I., Akitomi J., Boltz D.A., Horii K., Furuichi M., Waga I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014;443:37–41. doi: 10.1016/j.bbrc.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 66.Park S.Y., Kim S., Yoon H., Kim K.B., Kalme S.S., Oh S. Selection of an antiviral RNA aptamer against hemagglutinin of the subtype H5 avian influenza virus. Nucleic Acid Ther. 2011;21:395–402. doi: 10.1089/nat.2011.0321. [DOI] [PubMed] [Google Scholar]
- 67.Pang Y., Rong Z., Wang J., Xiao R., Wang S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF) Biosens. Bioelectron. 2015;66:527–532. doi: 10.1016/j.bios.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 68.Negri P., Chen G.J., Kage A., Nitsche A., Naumann D., Xu B.Q. Direct optical detection of viral nucleoprotein binding to an anti-influenza aptamer. Anal. Chem. 2012;84:5501–5508. doi: 10.1021/ac202427e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le T.T., Adamiak B., Benton D.J., Johnson C.J., Sharma S., Fenton R. Aptamer-based biosensors for the rapid visual detection of flu viruses. Chem. Commun. 2014;50:15533–15536. doi: 10.1039/c4cc07888h. [DOI] [PubMed] [Google Scholar]
- 70.Kiilerich-Pedersen K., Dapra J., Cherre S., Rozlosnik N. High sensitivity point-of-care device for direct virus diagnostics. Biosens. Bioelectron. 2013;49:374–379. doi: 10.1016/j.bios.2013.05.046. [DOI] [PubMed] [Google Scholar]
- 71.Jeon S.H., Kayhan B., Ben-Yedidia T., Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 2004;279:48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 72.Gopinath S.C.B., Sakamaki Y., Kawasaki K., Kumar P.K.R. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 2006;139:837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 73.Cheng C.S., Dong J., Yao L.H., Chen A.J., Jia R.Q., Huan L.F. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008;366:670–674. doi: 10.1016/j.bbrc.2007.11.183. [DOI] [PubMed] [Google Scholar]
- 74.Bai H., Wang R.H., Hargis B., Lu H.G., Li Y.B. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors (Basel) 2012;12:12506–12518. doi: 10.3390/s120912506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cho E.J., Collett J.R., Szafranska A.E., Ellington A.D. Optimization of aptamer microarray technology for multiple protein targets. Anal. Chim. Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 76.Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S.M., McGee D. Characterization of Fully 2′-Modified Oligoribonucleotide Heteroduplex and Homoduplex Hybridization and Nuclease Sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 78.Nutiu R., Li Y.F. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 79.Wang R.H., Xu L.Z., Li Y.B. Bio-nanogate controlled enzymatic reaction for virus sensing. Biosens. Bioelectron. 2015;67:400–407. doi: 10.1016/j.bios.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 80.Wang C., Zhang L., Shen X. Development of a nucleic acid lateral flow strip for detection of hepatitis C virus (HCV) core antigen. Nucleosides Nucleotides Nucleic Acids. 2013;32:59–68. doi: 10.1080/15257770.2013.763976. [DOI] [PubMed] [Google Scholar]
- 81.Labib M., Zamay A.S., Muharemagic D., Chechik A.V., Bell J.C., Berezoyski M.V. Aptamer-based viability impedimetric sensor for viruses. Anal. Chem. 2012;84:1813–1816. doi: 10.1021/ac203412m. [DOI] [PubMed] [Google Scholar]
- 82.Niedzwiecki D.J., Iyer R., Borer P.N., Movileanu L. Sampling a biomarker of the human immunodeficiency virus across a synthetic nanopore. ACS Nano. 2013;7:3341–3350. doi: 10.1021/nn400125c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamamoto R., Kumar P.K.R. Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells. 2000;5:389–396. doi: 10.1046/j.1365-2443.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 84.He Q.G., Du Q.Y., Lau S.L., Manopo I., Lu L.Q., Chan S.W. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods. 2005;127:46–53. doi: 10.1016/j.jviromet.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bruno J.G., Carrillo M.P., Richarte A.M., Phillips T., Andrews C., Lee J.S. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes. 2012;5:633. doi: 10.1186/1756-0500-5-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de-los-Santos-Alvarez N., Jesus Lobo-Castanon M., Miranda-Ordieres A.J., Tunon-Blanco P. Aptamers as recognition elements for label-free analytical devices. Trends Analyt. Chem. 2008;27:437–446. [Google Scholar]
- 87.Kuga S., Yang J.H., Takahashi H., Hirama K., Iwasaki T., Kawarada H. Detection of mismatched DNA on partially negatively charged diamond surfaces by optical and potentiometric methods. J. Am. Chem. Soc. 2008;130:13251–13263. doi: 10.1021/ja710167z. [DOI] [PubMed] [Google Scholar]
- 88.Kiilerich-Pedersen K., Poulsen C.R., Jain T., Rozlosnik N. Polymer based biosensor for rapid electrochemical detection of virus infection of human cells. Biosens. Bioelectron. 2011;28:386–392. doi: 10.1016/j.bios.2011.07.053. [DOI] [PubMed] [Google Scholar]
- 89.To K.K., Chan K.H., Li I.W., Tsang T.Y., Tse H., Chan J.F. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J. Med. Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van den Brand M., Peters R.P., Catsburg A., Rubenjan A., Broeke F.J., van den Dungen F.A. Development of a multiplex real-time PCR assay for the rapid diagnosis of neonatal late onset sepsis. J. Microbiol. Methods. 2014;106:8–15. doi: 10.1016/j.mimet.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 91.Mengelle C., Mansuy J.M., Pierre A., Claudet I., Grouteau E., Micheau P. The use of a multiplex real-time PCR assay for diagnosing acute respiratory viral infections in children attending an emergency unit. J. Clin. Virol. 2014;61:411–417. doi: 10.1016/j.jcv.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khare R., Espy M.J., Cebelinski E., Boxrud D., Sloan L.M., Cunningham S.A. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J. Clin. Microbiol. 2014;52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu S., Duan N., Shi Z., Fang C., Wang Z. Simultaneous aptasensor for multiplex pathogenic bacteria detection based on multicolor upconversion nanoparticles labels. Anal. Chem. 2014;86:3100–3107. doi: 10.1021/ac404205c. [DOI] [PubMed] [Google Scholar]
- 94.Duan N.O., Wu S.J., Ma X.Y., Xia Y., Wang Z.P. A universal fluorescent aptasensor based on AccuBlue dye for the detection of pathogenic bacteria. Anal. Biochem. 2014;454:1–6. doi: 10.1016/j.ab.2014.03.005. [DOI] [PubMed] [Google Scholar]


