Abstract
A reverse transcription multiplex real-time PCR (RT-MRT-PCR) was developed for rapid detection and genotyping of classical swine fever virus (CSFV). The universal primers and specific TaqMan probes for each of the three genotypes, genotypes 1, 2, and 3, were designed within the 3′-UTR of the CSFV. Non-CSFV swine virus and clinical samples from specific pathogen-free (SPF) pigs were both demonstrated to be CSFV-negative by RT-MRT-PCR. The diagnostic sensitivity of RT-MRT-PCR was determined to be 1 viral copy/μl for each genotype of standard plasmid. For the analytical sensitivity experiment, 100 samples of 14 CSFV genotype 1 strains and 86 samples from CSFV outbreak farms were all detected as CSFV-positive by RT-MRT-PCR, and the genotype results were consistent with the results of sequencing from a previous study. The intra-assay and inter-assay variations of RT-MRT-PCR were below 3% in all experiments. The sensitivity of RT-MRT-PCR was the same as the reverse transcription nested PCR (RT-nPCR) and higher than reverse transcription PCR (RT-PCR) and viral isolation from clinical samples. This assay was used further to evaluate the duration of viremia of wild-type CSFV in vaccinated exposed pigs. The results indicated that pigs vaccinated with the E2 subunit vaccine had longer viremia than pigs given the C-strain vaccine, which is compatible with the findings of previous studies. Thus, the new RT-MRT-PCR is a rapid, reproducible, sensitive, and specific genotyping tool for CSFV detection.
Keywords: Classical swine fever virus, Reverse transcription multiplex real-time PCR, Genotyping
1. Introduction
Classical swine fever (CSF) is a highly contagious and multi-systemic hemorrhagic disease that results in economic losses in the swine industry worldwide and is a notifiable disease to the Office International des Epizooties, according to the Terrestrial Animal Health Code (OIE, 2007). The course of disease can be acute, subacute, chronic, or late onset, but CSF can also remain unnoticed in infected pigs (van Oirschot, 2003).
Classical swine fever virus (CSFV), which is an enveloped, positive-sense, single-stranded RNA virus, is classified in the genus Pestivirus within the family Flaviviridae, which also includes bovine viral diarrhea virus (BVDV) and border disease virus (BDV). The genus pestivirus, including CSFV, BVDV, and BDV, can be differentiated by the sequences of their 5′-UTR fragments (Hofmann et al., 1994). At the genetic level, CSFVs can be divided into genotypes 1, 2, and 3, based on the partial sequences of the E2 and NS5B genes. Each genotype can be classified further into three or four sub-genotypes, referred to as 1.1, 1.2, and 1.3; 2.1, 2.2, and 2.3; and 3.1, 3.2, 3.3, and 3.4, respectively (Paton et al., 2000).
Detailed genetic information about CSFV field isolates can be used to form an important database for molecular epidemiology of CSFV, which allows for a greater understanding of the origin of CSFV as well as for improvements in eradication programs against CSF (Vilcek and Nettleton, 2006). Currently, CSF is still spreading gradually and evolving worldwide. In Europe, all CSFV strains were genotype 1 prior to the 1970s, but the various sub-genotype strains 2.1, 2.2, and 2.3 were isolated from different European countries during the 1980s and 1990s (Paton et al., 2000, Stegeman et al., 2000, Biagetti et al., 2001). CSF is still endemic currently in various European countries. In Latin America, only strains of genotype 1 have been reported thus far, and sub-genotypes 1.1, 1.2, and 1.3 are circulating currently within that region (Paton et al., 2000, Pereda et al., 2005, Sabogal et al., 2006). In Asia, CSF epidemics are also fairly ubiquitous and strains of genotypes 1, 2, and 3 have been isolated in different Asian countries (Paton et al., 2000, Blacksell et al., 2005). In Taiwan, all CSFV strains were found to be sub-genotype 3.4 prior to 1996. However, genotype 2 strains of CSFV, identified as sub-genotype 2.1 or 2.2, have been isolated since 1994 and have replaced gradually the sub-genotype 3.4 strain (Pan et al., 2005).
Currently, genotyping of isolated CSFV is based on the amplification of the E2 and NS5B sequences by reverse transcription PCR (RT-PCR), which requires nucleotide sequencing for further confirmation and takes approximately 2–3 days for completion. From the perspective of CSF control, a more rapid and accurate method for viral detection and genotyping is essential for the emergency responses to the disease outbreak. Reverse transcription multiplex real-time PCR (RT-MRT-PCR) has been successfully demonstrated for simultaneous detection of many pathogens, including BVDV (Baxi et al., 2006), dengue virus (Lai et al., 2007), influenza virus (Payungpom et al., 2006), yellow fever virus, Japanese encephalitis virus, West Nile virus, and St. Louis encephalitis virus (Chao et al., 2007). The aim of this study was to develop RT-MRT-PCR for the detection and genotyping of CSFV.
2. Materials and methods
2.1. Cell and viral isolation
The porcine kidney cell line (PK-15) was maintained in Minimal Eagle Medium (MEM) with 5% heat inactivated fetal calf serum, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. The cells were used to isolate CSFV.
Classical swine fever virus, bovine viral diarrhea virus types 1 and 2, Japanese B encephalitis virus, swine influenza virus, porcine reproductive and respiratory syndrome virus, porcine respiratory coronavirus, transmissible gastroenteritis virus, porcine epidemic diarrhea virus, porcine astrovirus, porcine teschovirus type 1, porcine enterovirus types 8 and 9, foot-and-mouth disease virus type O, reovirus, pseudorabies virus, and porcine circovirus types 1 and 2 were used in this study. The classical swine fever virus variants used included 10 strains of C-strain, 4 non-C-strain strains (ALD, GP−, A76, and CAP strain) of genotype 1, the Q90-278 strain of genotype 2, and the 83-19 strain of genotype 3.
2.2. Field samples
Field samples were collected from three different sources: (1) 39 samples, including 32 buffy coats and 7 tissue extracts, were collected from a specific pathogen-free (SPF) farm which were free of hog cholera virus, pseudorabies virus, Actinobacillus, and Toxoplasma gondii, (2) 86 tissue extracts, including 52 samples of genotype 2 and 34 samples of genotype 3, were collected from farms that had a CSFV outbreak during the period 1989–2003, and (3) 44 tissue extracts from different vaccinated farms that had been submitted to the Animal Heath Research Institute for routine detection of CSFV by local animal disease control centers.
2.3. RNA extraction
TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from plasma, buffy coat, tissue extracts, and PK-15 cells inoculated with tissue extracts. To do this, 100 μl of each sample was first mixed with 1 ml of the TRIzol® reagent and incubated for 5 min at room temperature. Then 0.2 ml of chloroform was added and the samples were incubated for 3 min at room temperature. All samples were centrifuged at 12,000 × g for 15 min at 4 °C. The supernatants were transferred to fresh tubes and 0.5 ml of isopropyl alcohol was added to each. Samples were then incubated at room temperature for 10 min. Centrifugation was performed again at 12,000 × g for 10 min at 4 °C. The supernatants were removed and the RNA pellet from each sample was washed with 75% ethanol. Finally, each RNA pellet was air dried and resuspended with 100 μl of DEPC-treated water.
2.4. Primer and probe
The primers and probes used for the study were designed based on the CSFV sequences published by the National Center for Biotechnology Information, and sequence alignment was performed using DNASTAR version 5 (DNASTAR, Madison, WI, USA). Universal CSFV primers, two pairs (CP5, CP6, CP3F, and CP3R), and specific TaqMan probes for genotypes 1, 2, and 3 (G1, G2, and G3) were designed based on the 3′-UTR of CSFV (Table 1 ). The 5′-ends of G1, G2, and G3 were labeled with 5′-hexachloro-fluorescein-CE phosphoramidite (HEX), cyanine 5 (Cy5), and 6-carboxy-flourescein (FAM), respectively. The 3′-ends of G1, G2, and G3 were all labeled with 4,4-bis-(2-butyloctyloxy)-p-quaterphenyl (BBQ). The primers CP5 and CP6 were used in both RT-PCR and RT-MRT-PCR combined with TaqMan probes G1, G2, and G3. The primers, CP3F and CP3R, were used in reverse transcription nested PCR (RT-nPCR).
Table 1.
Primers and probes used.
| Primers/probes | Strains | Genome position | Sequences (5′–3′) |
|---|---|---|---|
| CP5 | Alfort/187 | 11874-11895 | GTAGCAAGACTGGRAAYAGGTA |
| CP6 | Alfort/187 | 12240-12219 | AAAGTGCTGTTAAAAATGAGTG |
| CP3F | Alfort/187 | 12106-12126 | ACCCTRTTGTARATAACACTA |
| CP3R | Alfort/187 | 12234-12209 | GTTAAAAATGAGTGTAGTGTGGTAAC |
| G1 | Alfort/187 | 12094-12118 | HEX-ACCCGCCAGTAGGACCCTATTGTAG-BBQ |
| G2 | 93-TD | 12084-12106 | Cy5-CTTGACCGGGCCCTATCAGTGGA-BBQ |
| G3 | P97 | 12063-12086 | FAM-CACGTGAGTGCGGGTAGCCCAA-BBQ |
Y: C or T. R: A or G. HEX: 5′-hexachloro-fluorescein-CE phosphoramidite. BBQ: 4,4-bis-(2-butyloctyloxy)-p-quaterphenyl. Cy5: cyanine 5. FAM: 6-carboxy-flourescein.
2.5. RT-PCR and RT-nPCR
The extracted RNA from each sample was transcribed reversely and amplified in a one-tube reaction using RT-PCR. The RT-PCR reaction was carried out in a final volume of 50 μl containing 5 μl of extracted RNA, 1× DNA polymerase buffer, 8 units of recombinant RNase inhibitor (Promega, Madison, WI, USA), 2 units of AMV reverse transcriptase (Promega, Madison, WI, USA), 1 unit of DNA polymerase (JMR, UK), 0.2 μM of deoxyNTP mixture, and 0.4 μM of each primer (CP5 and CP6). The reaction was carried out in a 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). The reaction condition for RT-PCR included samples incubation at 42 °C for 40 min followed by 94 °C for 5 min. This incubation was then followed by 35 cycles of denaturation (94 °C for 30 s), annealing (55 °C for 30 s), and extension (72 °C for 30 s). A final extension at 72 °C for 10 min was performed before storing the sample at 4 °C.
For the subsequent nested PCR, 2 μl of RT-PCR product was used as the template. The reaction condition was similar to that of the RT-PCR described previously, and the primer pairs CP3F and CP3R were used.
2.6. Two step RT-MRT-PCR
2.6.1. cDNA synthesis
The extracted RNA from each sample was reverse transcribed into cDNA. The reverse transcription was carried out in a volume of 20 μl containing 5 μl of extracted RNA, 1× AMV reaction buffer (Promega, Madison, WI, USA), 8 units of recombinant RNase inhibitor (Promega, Madison, WI, USA), 2 units of AMV reverse transcriptase (Promega, Madison, WI, USA), 0.5 μM of deoxyNTP mixture, and 0.5 μM of CP6 primer. The reaction was carried out in the 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). The reaction conditions involved incubation at 42 °C for 40 min followed by 94 °C for 5 min. Finally, the reaction was held at 4 °C and the products were used as templates for multiplex real-time PCR.
2.6.2. Multiplex real-time PCR
The multiplex real-time PCR was carried out in the iQ™5 multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) in a reaction volume of 50 μl, containing 5 μl of cDNA, 1× Powermix (Bio-Rad, Hercules, CA, USA), 0.5 mM MgCl2, 0.1 μM G1, G2, and G3 each of the TaqMan probe, and 0.75 μM each of primer CP5 and CP6. The reaction condition involved incubation at 95 °C for 5 min followed by 45 cycles of denaturation (95 °C for 30 s), and annealing/extension (60 °C for 1 min). All samples were simultaneously detected for HEX, Cy5, and FAM fluorescence by the monitoring system. The threshold level was fixed at 70, 80, and 105 RFU in HEX, Cy5, and FAM fluorescence, respectively. Samples were recognized as CSFV-negative when the threshold cycle rose above 42.
2.7. Standard plasmid of RT-MRT-PCR
The RT-PCR products of C-strain (genotype 1), Q91-84 (genotype 2), and 85-12A (genotype 3) were cloned using the pGEM-T Easy vector system (Promega, Madison, WI, USA) and were propagated in E. coli, JM109, according to the manufacturer's instructions. Plasmids were purified by the QIAamp plasmid Maxi Kit (Qiagen, Valencia, CA, USA) and were quantified by measuring OD260 with spectrophotometer DU®640 (Beckman, Palo Alto, CA, USA). The viral copy of the extracted plasmids was calculated using the formula:
Each plasmid was optimized to 3 × 1010 viral copy/μl. Following optimization of concentration, the standard plasmid was equally mixed with plasmids of genotypes 1, 2, and 3. The standard plasmid was then used to develop standard curves and evaluate the diagnostic sensitivity of the RT-MRT-PCR.
2.8. Preliminary test of RT-MRT-PCR
2.8.1. Analytical and diagnostic specificity
The non-CSFV swine viruses were used to determine the analytical specificity of the primers and the TaqMan probes used in the experiment. To evaluate the diagnostic specificity, 39 samples from the SPF farm were used.
2.8.2. Analytical sensitivity
A total of 100 samples, including 14 strains of genotype 1, 52 strains of genotype 2, and 34 strains of genotype 3, were examined by RT-MRT-PCR for the presence of CSFV and their genotypes. For genotype 1, 14 samples comprising 10 C-strain viruses and ALD, GP−, CAP, A76 strains. The samples of genotypes 2 and 3 were collected from farms with CSFV outbreaks. The genotypes of all samples used in this experiment had also been identified by sequencing and described in a previous study (Pan et al., 2005).
2.8.3. Diagnostic sensitivity and standard curve
10-fold serial dilutions of the standard plasmid from 109 to 1 viral copy/μl of each genotype were tested to determine the detection limits of RT-MRT-PCR. The serially diluted plasmid was also used to establish a standard curve for genotypes 1, 2, and 3 by plotting the threshold cycle and the viral copy logarithm.
2.8.4. Reproducibility assay
The standard plasmid, including 101, 104, or 107 viral copy/μl in each genotype, was used to evaluate the inter-assay and intra-assay reproducibility of RT-MRT-PCR. Each concentration was detected in triplicate. The threshold cycle of each concentration was obtained and calculated.
2.9. Correlation between TCID50 and the viral copy of RT-MRT-PCR
C-strain (genotype 1), Q90-278 (genotype 2), and 83-19 (genotype 3) strains were 10-fold serially diluted and their viral copies quantified by RT-MRT-PCR. Linear regression was used to calculate the linear correlations (R 2) between TCID50 and the viral copy logarithm.
2.10. Comparison between viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR
2.10.1. Sensitivity comparison
10-fold serially diluted samples of C-strain, Q90-278, and 83-19 strains were determined to be CSFV-positive by viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR.
2.10.2. Agreement among methods
A total of 169 clinical samples were used to detect CSFV by viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR. These samples included 39 samples collected from SPF pigs, 86 samples from CSFV outbreak farms, and 44 samples from local animal disease control centers. The agreement among tests was based on kappa statistics (Thrusfield, 1995). The agreement was classified by kappa statistic values into five groups: almost perfect (0.81 or higher), substantial (0.61–0.8), moderate (0.41–0.6), fair (0.21–0.4), slight (0.01–0.2), and poor (0).
2.11. Detection of the wild-type CSFV in plasma samples from vaccinated exposed pigs by RT-MRT-PCR
In order to use the RT-MRT-PCR to study the duration of viremia of wild-type CSFV in vaccinated exposed pigs, plasma samples were collected from pigs as described below. Biocontainment facilities at the Animal Health Research Institute, Council of Agriculture, Taiwan, were used to house 11 SPF pigs. These pigs were randomly divided into C-strain, E2 subunit, and control groups with 4, 5, and 2 pigs, respectively. The pigs in the C-strain group were vaccinated once at 8 weeks of age with one dose of attenuated lapinized C-strain vaccine for CSFV produced by the Division of Biologics at Animal Heath Research Institute. The pigs in the E2 subunit group were vaccinated twice with one dose of E2 subunit vaccine (Bayer, Munich, Germany), at 6 and 10 weeks of age. The control pigs were not vaccinated. At 12 weeks of age, all pigs were placed in the same room with a CSFV-infected pig that had been infected with 2 × 107 TCID50 of the ALD strain of CSFV at 2 days prior to contact with the other pigs. Plasma samples were collected from all pigs at 0, 2, 4, 6, 8, 10, 12, 14, and 21 days post-contact (DPC).
3. Results
3.1. Analytical and diagnostic specificity of the RT-MRT-PCR
The standard plasmids were detected simultaneously using the HEX, Cy5, and FAM fluorescence by RT-MRT-PCR, and the fluorescence levels were all higher than the threshold level. The level of the HEX, Cy5, and FAM fluorescence of the non-CSFV swine viruses and the 39 samples from SPF farm were all below the threshold level (Fig. 1 ).
Fig. 1.
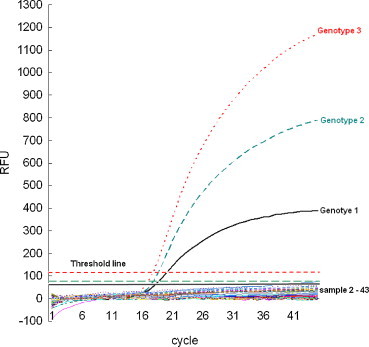
Analytical and diagnostic specificity of reverse transcription multiplex real-time PCR. The HEX, Cy5, and FAM fluorescence were genotype 1 (black real line), 2 (green broken line), and 3 (red broken line), respectively. The x-axis represents cycle number. The y-axis is the amount of fluorescent signal detected. Sample 1: standard plasmid of genotypes 1, 2, and 3; samples 2–17: non-CSFV swine virus; samples 18–43: buffy coats and tissue emulsions from an SPF farm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2. Analytical sensitivity of the RT-MRT-PCR
A total of 100 samples were detected as CSFV-positive by RT-MRT-PCR. The genotyping results of the RT-MRT-PCR were all consistent with the results of the genotypes verified by sequencing of the 100 samples (Table 2 ).
Table 2.
Analytical sensitivity of RT-MRT-PCR.
| Genotype | Numbera | RT-MRT-PCR |
||
|---|---|---|---|---|
| Genotype 1 | Genotype 2 | Genotype 3 | ||
| 1 | 14 | 14 | 0 | 0 |
| 2 | 52 | 0 | 52 | 0 |
| 3 | 34 | 0 | 0 | 34 |
Genotype 1 included 14 samples comprising 10 C-strains virus and ALD, GP−, CAP, A76 strain viruses. The samples of genotypes 2 and 3 were collected from CSFV outbreak farms.
3.3. Diagnostic sensitivity and standard cures of the RT-MRT-PCR
The detection limit of RT-MRT-PCR was determined to be 1 viral copy/μl for each genotype of the standard plasmid. The threshold cycles for standard plasmid, including 1 copy number/μl in each genotype, were 38.67, 39.07, and 39.78 in the genotypes 1, 2, and 3 samples, respectively (Fig. 2 ). The standard curves of RT-MRT-PCR were established from 1 to 109 viral copy/μl. The linear correlations (R 2) between the threshold cycle and the viral copy logarithm were 0.9976, 0.9965, and 0.9993 for genotypes 1, 2, and 3, respectively (Fig. 2).
Fig. 2.
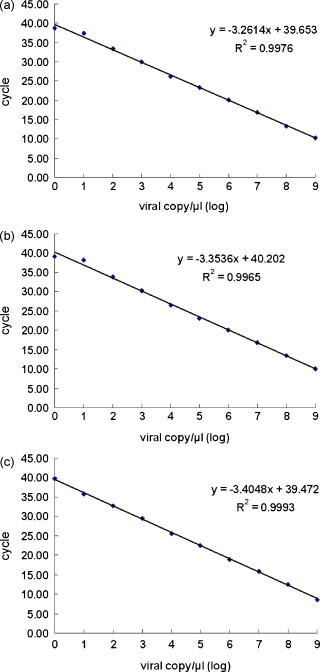
Diagnostic sensitivity and standard curve of reverse transcription multiplex real-time PCR based on a 10-fold serial dilution of standard plasmid, including plasmids of genotypes 1(a), 2(b), and 3(c).
3.4. The reproducibility of the RT-MRT-PCR
The coefficient of variation (CV) of genotype 1 for the threshold cycle values ranged between 0.46% and 1.54% for the intra-assay and 0.44% and 1.6% for inter-assay (Table 3 ). The CVs for both intra-assay and inter-assay of genotype 2 ranged between 0.56% and 2.08% and between 0.4% and 1.76%, respectively (Table 3). The CVs for intra-assay and inter-assay of genotype 3 ranged between 0.19% and 2.17% and between 0.31% and 1.48%, respectively (Table 3).
Table 3.
Reproducibility assay of threshold cycle value quantified by RT-MRT-PCR from the standard plasmid.
| Plasmid | Concentrationa (viral copy/μl) | Intra-assay |
Inter-assay |
||||
|---|---|---|---|---|---|---|---|
| Mean | S.D. | CV (%) | Mean | S.D. | CV (%) | ||
| Genotype 1 | 101 | 39.25 | 0.60 | 1.54 | 39.12 | 0.17 | 0.44 |
| 104 | 27.56 | 0.13 | 0.46 | 27.41 | 0.26 | 0.93 | |
| 107 | 17.82 | 0.18 | 1.00 | 18.13 | 0.29 | 1.60 | |
| Genotype 2 | 101 | 38.25 | 0.31 | 0.82 | 38.33 | 0.45 | 1.17 |
| 104 | 27.02 | 0.15 | 0.56 | 26.98 | 0.11 | 0.40 | |
| 107 | 17.41 | 0.36 | 2.08 | 17.96 | 0.32 | 1.76 | |
| Genotype 3 | 101 | 38.33 | 0.61 | 1.60 | 37.50 | 0.37 | 0.99 |
| 104 | 26.69 | 0.05 | 0.19 | 26.62 | 0.08 | 0.31 | |
| 107 | 16.86 | 0.37 | 2.17 | 17.49 | 0.26 | 1.48 | |
Each concentration of standard plasmid contained plasmids of genotypes 1, 2, and 3 and simultaneously detected three genotypes by RT-MRT-PCR. Each standard plasmid was detected in triplicate by RT-MRT-PCR in the intra-assay and inter-assay.
3.5. Correlation between TCID50 and the viral copy of the RT-MRT-PCR
To investigate the correlation between TCID50 and the viral copy of RT-MRT-PCR, CSFV was diluted serially 10-fold and was assayed by RT-MRT-PCR. The R 2s of C-strain (genotype 1), Q90-278 (genotype 2), and 83-19 (genotype 3) strains between TCID50 and the viral copy were 0.998, 0.9082, 0.9741, respectively (Fig. 3 ).
Fig. 3.
The linear correlations (R2) of C-strain (a), Q90-278 (b), and 83-19 (c) strains was calculated between TCID50 and viral copy number of reverse transcription multiplex real-time PCR.
3.6. Comparison of viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR
3.6.1. Sensitivity comparison
To compare the sensitivity of the different methods, the C-strain, Q90-278, and 83-19 strains of CSFV were diluted serially 10-fold and were examined by viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR. The sensitivities of viral isolation, RT-nPCR, and RT-MRT-PCR were all found to range between 1 and 3.2 TCID50/ml, which was a 10-fold greater sensitivity than RT-PCR (Table 4 ).
Table 4.
Summary of the sensitivity of viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR.
| CSFV straina (genotype) | Dilution | Viral isolation | RT-PCR | RT-nPCR | RT-MRT-PCR |
|---|---|---|---|---|---|
| C-strain (1.1) | 10−1 | 8/8 | + | + | + |
| 10−2 | 8/8 | + | + | + | |
| 10−3 | 8/8 | + | + | + | |
| 10−4 | 8/8 | + | + | + | |
| 10−5 | 8/8 | − | + | + | |
| 10−6 | 4/8 | − | + | + | |
| 10−7 | 1/8 | − | − | − | |
| 10−8 | 0/8 | − | − | − | |
| TCID50: 106/ml | |||||
| Q90-278 (2.1) | 10−1 | 8/8 | + | + | + |
| 10−2 | 8/8 | + | + | + | |
| 10−3 | 8/8 | − | + | + | |
| 10−4 | 0/8 | − | − | − | |
| 10−5 | 0/8 | − | − | − | |
| 10−6 | 0/8 | − | − | − | |
| 10−7 | 0/8 | − | − | − | |
| 10−8 | 0/8 | − | − | − | |
| TCID50: 103.5/ml | |||||
| 83-19 (3.4) | 10−1 | 8/8 | + | + | + |
| 10−2 | 8/8 | + | + | + | |
| 10−3 | 8/8 | + | + | + | |
| 10−4 | 7/8 | + | + | + | |
| 10−5 | 4/8 | − | + | + | |
| 10−6 | 0/8 | − | − | − | |
| 10−7 | 0/8 | − | − | − | |
| 10−8 | 0/8 | − | − | − | |
| TCID50: 105/ml | |||||
CSFV strains were 10-fold serially diluted and CSFV was detected from viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR. The TCID50 of CSFV strains was calculated from result of viral isolation.
3.6.2. Assessment of agreement of methods
Agreement of viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR, 101 of 169 total clinical samples were detected as CSFV-positive, and 68 of these samples were CSFV-negative by viral isolation. All 101 CSFV-positive samples detected by viral isolation were also shown to be CSFV-positive by RT-MRT-PCR, RT-PCR, and RT-nPCR. Among the 68 CSFV-negative samples detected by viral isolation, 11, 7, and 10 were shown to be CSFV-positive by RT-MRT-PCR, RT-PCR, and RT-nPCR, respectively. The kappa values were 0.935 for viral isolation and RT-MRT-PCR, 0.959 for viral isolation and RT-PCR, and 0.941 for viral isolation and RT-nPCR (Table 5 ). The sensitivity values of the three assays were all 100% or equal to viral isolation. Regarding the viral isolation with the three assays, the specificity values were 83.8%, 89.7% and 85.3% in RT-MRT-PCR, RT-PCR, and RT-nPCR, respectively.
Table 5.
The agreement among viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCRa.
| Method | Result | Viral isolation |
||||
|---|---|---|---|---|---|---|
| Positive | Negative | Kappa values | Sensitivity (%) | Specificity (%) | ||
| RT-MRT-PCR | Positive | 101 | 11 | 93.5 | 100 | 83.8 |
| Negative | 0 | 57 | ||||
| RT-PCR | Positive | 101 | 7 | 95.9 | 100 | 89.7 |
| Negative | 0 | 61 | ||||
| RT-nPCR | Positive | 101 | 10 | 94.1 | 100 | 85.3 |
| Negative | 0 | 58 | ||||
A total of 169 clinical samples was detected for the presence of CSFV and the results included and used to evaluate agreement among viral isolation, RT-PCR, RT-nPCR, and RT-MRT-PCR. Agreement among tests was based on kappa statistics (Thrusfield, 1995).
3.7. Detection of viremia of wild-type CSFV in vaccinated exposed pigs by RT-MRT-PCR
The plasma samples of vaccinated exposed pigs were collected and assayed by RT-MRT-PCR (Table 6 ). In the C-strain group, wild-type CSFV was detected from the plasma of vaccinated exposed pigs only at the time point of 8 days post-contact (DPC). Two of the four pigs in this group revealed 3.34 and 3.77 log viral copy number/ml in the plasma samples. In the E2 subunit group, wild-type CSFV was detected between 6 and 12 days post-contact (DPC). All four pigs in this group showed serum viral loads ranging from 3.35 to 5.8 log copy number/ml. In the non-vaccinated control group, CSFV could be detected from 2 to 12 DPC, and the serum viral loads ranged from 3.31 to 10 log viral copy number/ml.
Table 6.
Detection of wild-type CSFV in plasma of vaccinated exposed pigs by RT-MRT-PCRa.
| ID of pigs | Group | 0 DPC | 2 DPC | 4 DPC | 6 DPC | 8 DPC | 10 DPC | 12 DPC | 14 DPC | 21 DPC |
|---|---|---|---|---|---|---|---|---|---|---|
| 1212 | C-strain | −b | − | − | − | 3.34 | − | − | − | − |
| 1214 | C-strain | − | − | − | − | 3.77 | − | − | − | − |
| 1416 | C-strain | − | − | − | − | − | − | − | − | − |
| 1217 | C-strain | − | − | − | − | − | − | − | − | − |
| 1218 | E2 subunit | − | − | − | 5.49 | − | − | − | − | − |
| 1219 | E2 subunit | − | − | − | − | 4.49 | 4.79 | 3.36 | − | − |
| 1220 | E2 subunit | − | − | − | − | 5.8 | 3.35 | − | − | − |
| 1221 | E2 subunit | − | − | − | 3.36 | 3.88 | − | − | − | − |
| 1222 | E2 subunit | − | − | − | − | − | − | − | − | − |
| 1213 | Control | − | 3.4 | 6.3 | 7.3 | 10 | npc | np | np | np |
| 1215 | Control | − | 3.31 | 4.34 | 7.52 | 8.21 | 4.99 | 4.78 | ( | ( |
−, negative; np, not performed.
The pigs in the C-strain group were vaccinated once at 8 weeks of age with one dose of attenuated lapinized C-strain vaccine of CSFV. The pigs in the E2 subunit group were vaccinated twice with one dose of E2 subunit vaccine, at 6 and 10 weeks of age. The control pigs were not vaccinated. At 12 weeks, all pigs were placed in the same room with the wild-type CSFV-infected pig. Plasma samples were collected from all pigs at 0, 2, 4, 6, 8, 10, 12, 14, and 21 days post-contact (DPC).
The unit of concentration of CSFV in plasma determined by RT-MRT-PCR was log viral copy/ml.
The pig 1213 was death at 7 DPC.
4. Discussion
The RT-MRT-PCR developed in the present study is a rapid and highly specific technique for the detection and the genotyping of CSFV. RT-MRT-PCR shows no inter-genotypic cross-reactivity among different CSFV strains or with other swine viral pathogens (Fig. 1 and Table 2). In addition, this method is more rapid for the identification of genotypes than other CSFV-detecting assays, such as RT-PCR, RT-nPCR, or real-time PCR, which require nucleic acid sequencing of the amplified products for further genotype identification (Greiser-Wilke et al., 2006).
In addition, the RT-MRT-PCR can distinguish simultaneously different genotypes of CSFV and can be used further for epidemiological studies of CSFV infection, which cannot be achieved by viral isolation, RT-PCR, and RT-nPCR. RT-MRT-PCR has the ability to identify the genotypes of clinical samples much more quickly than a sequencing assay. This method was used for a retrospective study of the epidemiology of CSFV that occurred in Taiwan from 1989 to 2003, and all results were consistent with the sequencing analysis (Table 2). However, there is difficulty in utilizing RT-MRT-PCR in areas that are endemic for wild-type CSFV genotype 1 virus and are using currently the attenuated CSFV genotype 1 vaccine, such as C-strain, to control CSFV. In this case, the G1 probe is not able to differentiate between the wild-type and attenuated CSFV genotype 1 vaccine virus using the RT-MRT-PCR products, except when further electrophoresis of the RT-MRT-PCR products is applied (Pan et al., 2008). The difference in product size is due to the 3′-end sequences of the attenuated CSFV genotype 1 vaccine strains, which contain a T-rich insertion that the wild-type CSFV lacks (Pan et al., 2008).
The sensitivity of the CSFV detection by RT-MRT-PCR could be equivalent to that of detection using real-time PCR. It has been widely considered that real-time PCR is one of the most sensitive methods for CSFV detection, more sensitive than the methods of viral isolation, antigen ELISA, RT-PCR, and RT-nPCR (Hoffmann et al., 2005, Haegeman et al., 2006, Depner et al., 2007). It was shown that the sensitivity of RT-MRT-PCR is comparable to those of RT-nPCR and viral isolation, and higher than RT-PCR for the samples of CSFV diluted serially (Table 4). However, it was also found that among the 68 CSFV-negative samples of clinical samples tested by viral isolation, 7–11 samples were detected as CSFV-positive by RT-PCR, RT-nPCR, and RT-MRT-PCR (Table 5). The false-positive samples were repeated three times by RT-PCR, RT-nPCR, and RT-MRT-PCR and the sequenced PCR products were all highly homologous to the attenuated lapinized C-strain (data not shown). The results indicated that RT-MRT-PCR, RT-PCR, and RT-nPCR have higher sensitivity than viral isolation for clinical samples. The discrepancy in the sensitivity of those methods between samples of CSFV diluted serially and clinical samples could be due to the different detection targets of each method. The detection targets for viral isolation are infectious CSFV, whereas those for RT-PCR, RT-nPCR, and RT-MRT-PCR are all nucleic acids isolated from CSFV. The detection of infectious CSFV by viral isolation is affected by several factors, including the viral stage, anti-CSFV antibody, storage stage of clinical samples, and stage and type of cell lines used, resulting in lower positive results than expected. However, it must be noted that molecular assays cannot replace completely viral isolation. This is due to the fact that the presence of viral genetic materials detected by molecular assays does not represent viral infectivity. For example, the viral genetic material may be isolated from inactive viruses trapped in, for example, the phagocytic cells or immune complexes, in addition to infectious viruses. On the other hand, the sensitivity of the RT-nPCR was equivalent to the RT-MRT-PCR; however, the RT-nPCR has lower reproducibility than that of the RT-MRT-PCR (data not shown) and requires a longer processing time than RT-MRT-PCR. Thus, the RT-MRT-PCR is a superior method for CSFV detection as compared to virus isolation, RT-PCR, and RT-nPCR.
When RT-MRT-PCR was applied for evaluation of viremia caused by the wild-type CSFV in vaccinated exposed pigs, a different viremic pattern was demonstrated in preliminary results. These data are in agreement with the shorter duration of viremia in the viral challenged-pigs with C-strain group vaccination, as compared to that with the E2 subunit group in this RT-MRT-PCR study and previous studies. Previous studies have demonstrated that vaccination with the C-strain vaccine could induce complete protection against subsequent CSFV challenge (Ferrari, 1992, Uttenthal et al., 2001, van Oirschot, 2003, Suradhat et al., 2007), in which neither viremia nor viral shedding was observed in vaccinated pigs that had been challenged 1 week after vaccination. However, the pigs vaccinated with C-strain in this study showed a further transient viremia in the plasma at 8 DPC (Table 4). It was also found that viremia in E2 subunit vaccinated pigs was similar to C-strain results in that vaccinated exposed pigs in our experiment had longer durations of viremia than those in other studies. A period of wild-type CSFV viremia in pigs treated with the E2 subunit vaccine was detected by RT-MRT-PCR (Table 6). This period was longer than the one reported in the study by Uttenthal et al. (2001) in which either complete protection or only transient viremia at 6 days post-infection was detected. The differences in the viremic periods of wild-type CSFV in this study and previous studies might be due to the different challenge methods used or the different sensitivities of the methods of detection used.
The efficacy of the CSFV vaccines is correlated with the amount of viral particles contained in each dose of the vaccine. It has been shown that pigs vaccinated with a dose of less than 100 TCID50 or 1/100 dose of C-strain viral particles do not develop complete protection and often have viremia and viral shedding after challenge with virulent CSFV (Jong et al., 1988, Jong et al., 1989). In Taiwan, there are two types of commercial C-strain vaccines, which are prepared either from cell cultures or from visceral organs of rabbits. The viral titer of the cell culture-derived C-strain vaccine can be evaluated and titrated during its preparation; however, such a procedure cannot be performed in the preparation of the C-strain vaccine from the visceral organs of rabbit. According to official regulations, the viral load per dose of cell culture-derived C-strain vaccine must contain at least 103.5 of 50% of the fluorescence assay infected-dose (FAID50), but no official regulations have been established for the rabbit visceral organ-derived vaccine. The 103.5 FAID50 is equivalent roughly to 106.6 viral copy, as determined by the RT-MRT-PCR used in this study (data not shown). The lack of any evaluation systems and official regulations on rabbit visceral organ-derived C-strain vaccine may have certain negative impacts on the quality of this kind of vaccine. This may result in further adverse effects on the CSFV prevention program in the field. The RT-MRT-PCR was also used for evaluation of the viral load on all available commercial C-strain vaccines in Taiwan, including two cell culture-derived vaccines and six rabbit visceral organ-derived vaccines. These results indicate that none of the commercial C-strain vaccines have met the minimum requirement of the official regulation. The established RT-MRT-PCR could be utilized to evaluate not only the quality of the rabbit visceral organ-derived C-strain vaccine, but it could also be used as an alternative for monitoring the quality of cell culture-derived C-strain vaccine. However, since real-time PCR does not differentiate between infective and non-infective viral particles, then if the viral copy number in the vaccines tested, determined by RT-MRT-PCR, meets the official regulation, the assay measuring of viral infectivity, such as viral isolation and titration, should be a supplemental analysis to confirm the potency of the tested vaccines.
Real-time PCR may be an alternative to viral isolation for the study of the pathogenesis of CSFV infection, which includes tissue distribution, viral load, and the routes of viral shedding (Kamolsiriprichaiporn et al., 1992, Ophuis et al., 2006, Koenig et al., 2007), as this method was able to quantify the copy numbers of CSFV in the samples examined. The RT-MRT-PCR is capable of simultaneously detecting and quantifying three genotypes of CSFV. Thus, this method should be an effective tool for any of the studies on chronological changes in viral prevalence, such as the gradual replacement of the sub-group 3.4 strains of CSFV with the sub-groups 2.1 and 2.2 strains in Taiwan after 1994.
In summary, the RT-MRT-PCR is a rapid, reproducible, specific, and sensitive assay for the detection, quantitation, and genotype identification of CSFV. Additionally, RT-MRT-PCR can also be used in CSFV studies in various areas, including epidemiology, pathogenesis, and vaccine quality evaluation. RT-MRT-PCR appears to be more functional than all of the existing assays and may be suitable for routine laboratory diagnosis, both for the detection and the genotyping of CSFV. In the future, the two-step RT-MRT-PCR could be improved into a one-step approach in which reactions are performed generally in smaller reaction volumes and a positive control can be included to preclude false negative results.
Acknowledgements
This research was supported in part by grant 95AS-13.2.4-HI-H5 from the Council of Agriculture. We especially thank Drs. Sue-Min Huang (Animal Health Research Institute) for her technical support on the iQ™5 multicolor Real-Time PCR Detection System, Yeou-Liang Lin (Animal Health Research Institute) for his assistance in the animal experiment, and Fan Lee (Animal Health Research Institute) for his valuable suggestions regarding experimental design.
References
- Baxi M., McRae D., Boxi S., Greiser-Wike I., Vilcek S., Amoako K., Deregt D. A one-step multiplex real-time RT-PCR for detection and typing of bovine viral diarrhea viruses. Vet. Microbiol. 2006;116:37–44. doi: 10.1016/j.vetmic.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Biagetti M., Greiser-Wilke I., Rutili D. Molecular epidemiology of classical swine fever in Italy. Vet. Microbiol. 2001;83:205–215. doi: 10.1016/s0378-1135(01)00424-2. [DOI] [PubMed] [Google Scholar]
- Blacksell S.D., Khounsy S., Boyle D.B., Gleeson L.J., Westbury H.A., Mackenzie J.S. Genetic typing of classical swine fever viruses from Lao PDR by analysis of the 5′ non-coding region. Virus Genes. 2005;31:349–355. doi: 10.1007/s11262-005-3253-0. [DOI] [PubMed] [Google Scholar]
- Chao D.Y., Davis B.S., Chang G.J. Development of multiplex real-time reverse transcriptase PCR assays for detecting eight medically important flaviviruses in mosquitoes. J. Clin. Microbiol. 2007;45:584–589. doi: 10.1128/JCM.00842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depner K., Hoffmann B., Beer M. Evaluation of real-time RT-PCR assay for the routine intra vitam diagnosis of classical swine fever. Vet. Microbiol. 2007;121:338–343. doi: 10.1016/j.vetmic.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Ferrari M. A tissue culture vaccine with lapinized Chinese (LC) strain of hog cholera virus (HCV) Comp. Immunol. Microbiol. Infect. Dis. 1992;15:221–228. doi: 10.1016/0147-9571(92)90095-9. [DOI] [PubMed] [Google Scholar]
- Greiser-Wilke I., Dreier S., Haas L., Zimmermann B. Genetic typing of classical swine fever viruses—a review. Dtsch. Tierarztl. Wochenschr. 2006;113:134–138. [PubMed] [Google Scholar]
- Haegeman A., Dewulf J., Vrancken R., Tignon M., Ribbens S., Koenen F. Characterisation of the discrepancy between PCR and virus isolation in relation to classical swine fever virus detection. J. Virol. Methods. 2006;136:44–50. doi: 10.1016/j.jviromet.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Hofmann M.A., Brechtbuhl K., Stauber N. Rapid characterization of new pestivirus strains by direct sequencing of PCR-amplified cDNA from the 5′ noncoding region. Arch. Virol. 1994;139:217–229. doi: 10.1007/BF01309467. [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Beer M., Schelp C., Schirrmeier H., Depner K. Validation of a real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J. Virol. Methods. 2005;130:36–44. doi: 10.1016/j.jviromet.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Jong M.H., Chan I.P., Chi C.W., Huang C.C., Chiou T.F., Lee J.T., Hung W.K. Immunoprotection induced by tissue culture-adapted LPC strain of hog cholera virus. Exp. Rep. Taiwan Provincial Res. Inst. Animal Heath. 1989;25:49–58. [Google Scholar]
- Jong M.H., Huang C.C., Chan I.P., Liu T.H., Chi C.W., Chiou T.F., Lee J.T. Development of tissue culture hog cholera vaccine. Exp. Rep. Taiwan Provincial Res. Inst. Animal Heath. 1988;24:33–41. [Google Scholar]
- Kamolsiriprichaiporn S., Morrissy C.J., Westbury H.A. A comparison of the pathogenicity of two strains of hog cholera virus. 2. Virological studies. Aust. Vet. J. 1992;69:245–248. doi: 10.1111/j.1751-0813.1992.tb09871.x. [DOI] [PubMed] [Google Scholar]
- Koenig P., Hoffmann B., Depner K.R., Reimann I., Teifke J.P., Beer M. Detection of classical swine fever vaccine virus in blood and tissue samples of pigs vaccinated either with a conventional C-strain vaccine or a modified live marker vaccine. Vet. Microbiol. 2007;120:343–351. doi: 10.1016/j.vetmic.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Lai Y.L., Chung Y.K., Tan H.C., Yap G., Ooi E.E., Ng L.C. Cost-effective real-time reverse transcriptase PCR (RT-PCR) to screen for Dengue virus followed by rapid single-tube multiplex RT-PCR for serotyping of the virus. J. Clin. Microbiol. 2007;45:935–941. doi: 10.1128/JCM.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophuis R.J., Morrissy C.J., Boyle D.B. Detection and quantitative pathogenesis study of classical swine fever virus using a real time RT-PCR assay. J. Virol. Methods. 2006;131:78–85. doi: 10.1016/j.jviromet.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Office International des Epizooties . Terrestrial Animal Health Code. 16th ed. Office International des Epizooties; Paris, Fance: 2007. World Organisation for Animal Health. Classical swine fever. pp. 252–258 (Chapter 2.6.7) [Google Scholar]
- Pan C.H., Jong M.H., Huang T.S., Liu H.F., Lin S.Y., Lai S.S. Phylogenetic analysis of classical swine fever virus in Taiwan. Arch. Virol. 2005;150:1019–1101. doi: 10.1007/s00705-004-0485-6. [DOI] [PubMed] [Google Scholar]
- Pan C.H., Jong M.H., Huang Y.L., Huang T.S., Chao P.H., Lai S.S. Rapid detection and differentiation of wild-type and three attenuated lapinized vaccine strains of Classical swine fever virus by reverse transcription polymerase chain reaction. J. Vet. Diagn. Invest. 2008;20:448–456. doi: 10.1177/104063870802000406. [DOI] [PubMed] [Google Scholar]
- Paton D.J., McGoldrick A., Greiser-Wilke I., Parchariyanon S., Song J.Y., Liou P.P., Stadejek T., Lowings J.P., Bjorklund H., Belak S. Genetic typing of classical swine fever virus. Vet. Microbiol. 2000;73:137–157. doi: 10.1016/s0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- Payungpom S., Chutinimitkul S., Chaisingh A., Damrongwantanapokin S., Buranathia C., Amonsin A., Theamboonlers A., Poovorawan Y. Single step multiplex real-time RT-PCR for H5N1 influenza A virus detection. J. Virol. Methods. 2006;131:143–147. doi: 10.1016/j.jviromet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Pereda A.J., Greiser-Wilke I., Schmitt B., Rincon M.A., Mogollon J.D., Sabogal Z.Y., Lora A.M., Sanguinetti H., Piccone M.E. Phylogenetic analysis of classical swine fever virus (CSFV) field isolates from outbreaks in South and Central America. Virus Res. 2005;110:111–118. doi: 10.1016/j.virusres.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Sabogal Z.Y., Mogollon J.D., Rincon M.A., Clavijo A. Phylogenetic analysis of recent isolates of classical swine fever virus from Colombia. Virus Res. 2006;115:99–103. doi: 10.1016/j.virusres.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Stegeman A., Elbers A., de Smit H., Moser H., Smak J., Pluimers F. The 1997–1998 epidemic of classical swine fever in the Netherlands. Vet. Microbiol. 2000;73:183–196. doi: 10.1016/s0378-1135(00)00144-9. [DOI] [PubMed] [Google Scholar]
- Suradhat S., Damrongwatanapokin S., Thanawongnuwech R. Factors critical for successful vaccination against classical swine fever in endemic areas. Vet. Microbiol. 2007;119:1–9. doi: 10.1016/j.vetmic.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Thrusfield M. Veterinary Epidemiology. 2nd ed. Blackwell Science Ltd.; Oxford: 1995. Diagnostic testing; pp. 280–282. [Google Scholar]
- Uttenthal A., Le Potier M.F., Romero L., De Mia G.M., Floegel-Niesmann G. Classical swine fever (CSF) marker vaccine. Trial I. Challenge studies in weaner pigs. Vet. Microbiol. 2001;83:85–106. doi: 10.1016/s0378-1135(01)00409-6. [DOI] [PubMed] [Google Scholar]
- van Oirschot J.T. Vaccinology of classical swine fever: from lab to field. Vet. Microbiol. 2003;96:367–384. doi: 10.1016/j.vetmic.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Vilcek S., Nettleton P.F. Pestiviruses in wild animals. Vet. Microbiol. 2006;116:1–12. doi: 10.1016/j.vetmic.2006.06.003. [DOI] [PubMed] [Google Scholar]


