Abstract
A transitory, interleukin-25-responsive, group 2 innate lymphoid cell (ILC2) subset induced during type-2 inflammation was recently identified as iILC2s. This study focuses on understanding the significance of this population in relation to tissue-resident nILC2s in the lung and intestine. RNA-sequencing and pathway analysis revealed the AP-1 superfamily transcription factor BATF as a potential modulator of ILC2 cell fate. Infection of BATF-deficient mice with Nippostrongylus brasiliensis showed a selective defect in IL-25-mediated helminth clearance and a corresponding loss of iILC2s in the lung characterized as IL-17RBhigh, KLRG1high, BATFhigh, and Arginase-1low. BATF-deficiency selectively impaired iILC2s as it had no impact on tissue-resident nILC2 frequency or function. Pulmonary-associated iILC2s migrated to the lung after infection where they represented an early source of IL-4 and IL-13. Although the composition of ILC2s in the small intestine were distinct from those in the lung, their frequency and IL-13 expression remained dependent on BATF, which was also required for optimal goblet and tuft cell hyperplasia. Findings support IL-25-responsive ILC2s as early sentinels of mucosal barrier integrity.
One Sentence Summary:
BATF-deficient mice lack iILC2 cells which serve as an important early source of IL-4/IL-13 upon helminth infection.
Introduction
Disruption of mucosal barriers can occur in many ways and often initiates a type-2 immune response (1). Type-2 immunity is the process by which damage to an epithelial barrier is recognized, the pathologic agent is contained or eliminated in a way that minimizes prolonged inflammation, and damage to the underlying tissue is repaired in order to restore barrier integrity. This is orchestrated through the recruitment of both innate and adaptive immune cells to the site of barrier damage. In the case of helminth infection, these cells promote worm expulsion (weep and sweep response) and direct tissue repair (wound healing) (2–4). Innate and adaptive immune cells mediate both of these processes through the production of type-2 cytokines (5).
Tissue-resident group 2 innate lymphoid cells (ILC2s) respond to barrier disruption by sensing epithelial-derived alarmins and serve as early initiators of type-2 responses (6–8). Although enriched at mucosal sites, ILC2s reside in many tissues and respond to IL-25, IL-33, and thymic stromal lymphopoietin (TSLP) released by damaged mucosal epithelium (9, 10). Despite CD4+ T cells being the major producers of IL-4 and IL-13 at the peak of helminth infection, ILC2s expand in number in response to IL-25 and IL-33 and are prolific producers of IL-5 and IL-13 on a per cell basis (6, 11–13). Additionally, ILC2s play an important role in intestinal homeostasis by IL-13-mediated goblet and tuft cell differentiation (14–17) and help to mobilize eosinophils via IL-5 (11). While these tissue-resident ILC2 cells proliferate locally at steady state and localize to adventitia (18–21), they are highly motile within lung tissue after alarmin activation (22). Although tissue-resident ILC2 cells have been the subject of much study, less is known in regard to the significance of circulating ILC2 cells that mobilize in response to type-2 inflammatory cues in the bone marrow and intestine and arrive in the pulmonary vasculature (23, 24).
Until recently, ILC2s were thought to be a homogeneous population when compared to ILC1 and ILC3 subsets (25). However, recent studies have identified two distinct populations of ILC2s characterized by their differential responsiveness to IL-25 and IL-33 (26, 27). The IL-25-responsive subset, called iILC2, expresses high levels of the C-type lectin receptor, KLRG1, and low levels of CD90, and is transiently found in murine lungs early after Nippostrongylus brasiliensis infection or IL-25 administration. This differs from the IL-33-responsive nILC2 subset, which is tissue-resident and displays intermediate levels of KLRG1 and high levels of CD90 (26). Although the relationship between IL-25- and IL-33-responsive ILC2s is not well understood, it has been suggested that tissue-resident nILC2s self-renew at steady state and proliferate locally with some cells likely arriving from the bone marrow in response to IL-33, while migratory iILC2s that arrive in the lung have been suggested to originate from the intestine (19, 20, 23, 24). While the role of tissue-resident nILC2s has been well defined, the significance of this migratory, IL-25-responsive iILC2 subset is less clear and whether there are distinct lineage-determining factors involved in their selective development, fate, or function is not known.
Our previous studies identified the AP-1 superfamily transcription factor basic leucine zipper transcription factor, ATF-like (BATF) as an essential transcription factor involved in helminth clearance (28). BATF is required for the generation of follicular T helper (Tfh) cells and Th2 cells as well as their production of type-2 cytokines (28–31). As a result, the absence of BATF prevents both humoral and cell-mediated aspects of type-2 immunity. Despite its well characterized role in adaptive immunity and type-2 cytokine production, a role for BATF in ILCs has not been described (32). This study reveals that BATF is required by pulmonary iILC2 cells in response to helminth infection, and acts in a cell intrinsic manner. Furthermore, we show that this BATF-dependent migratory iILC2 population is distinct from tissue-resident nILC2s via its low expression of the ILC2 marker arginase-1. Unexpectedly, it is the migratory iILC2 subset and not the tissue-resident subset that acts as the early source of IL-4 and IL-13 in the lung and intestine during infection. In addition, intestinal IL-25-responsive ILC2s serve as key early mediators of goblet and tuft cell hyperplasia as helminths establish residence in the intestine. We propose that BATF-dependent migratory iILC2s act as early responders to damaged mucosal epithelium and orchestrate the initial steps in the re-establishment of barrier integrity at these sites.
Results
RNA sequencing and pathway analysis reveals BATF as a key transcription factor of pulmonary ILC2s
As ILC2s are critical for helminth clearance, we sought to investigate transcriptional differences between KLRG1-positive ILC2s (which include all ILC2 subsets) and KLRG1-negative populations (non-ILC2s; enriched for ILC1 and ILC3 cells) in the lung after IL-33-induced ILC2 expansion. ILC2s were sorted as live, singlet, SSClowFSClowLineage−CD4−CD90+KLRG1pos cells and compared to the KLRG1neg population (fig. S1A–C). Consistent with prior RNA sequencing of ILC2s after IL-33 administration, KLRG1pos and KLRG1neg populations displayed distinct transcriptional profiles, as evident by principal component analysis (fig. S1D) and pairwise hierarchical clustering (fig. S1E) (33, 34). Furthermore, the KLRG1pos cells displayed an enrichment in genes associated with phenotypic markers of ILC2 cells like Il1rl1 (ST2, IL-33 receptor), Il17rb (IL-25 receptor), and Klrg1 (Fig. 1A) as well as increased expression of genes related to ILC2 function such as Arg1, Il4, Il13, and Il5 (Fig. 1B) (33, 35–37). Interestingly, predicting associated transcription factors from annotated affinities (PASTAA) analysis identified the AP-1 transcription factor Atf-1/Batf as among the factors predicted to be regulating significantly enriched genes within KLRG1pos compared to KLRG1neg populations (Fig 1C). PASTAA is a bioinformatic tool used to predict transcription factors that likely regulate gene sets based on how strongly the transcription factor associates with eukaryotic promoters (38). Gene set enrichment analysis was used to calculate normalized enrichment scores for the transcription factors identified by PASTAA in KLRG1pos ILC2s (Fig. 1D).
Figure 1: Batf is expressed by lung ILC2 cells and is predicted to regulate distinct gene sets in KLRG1pos populations.
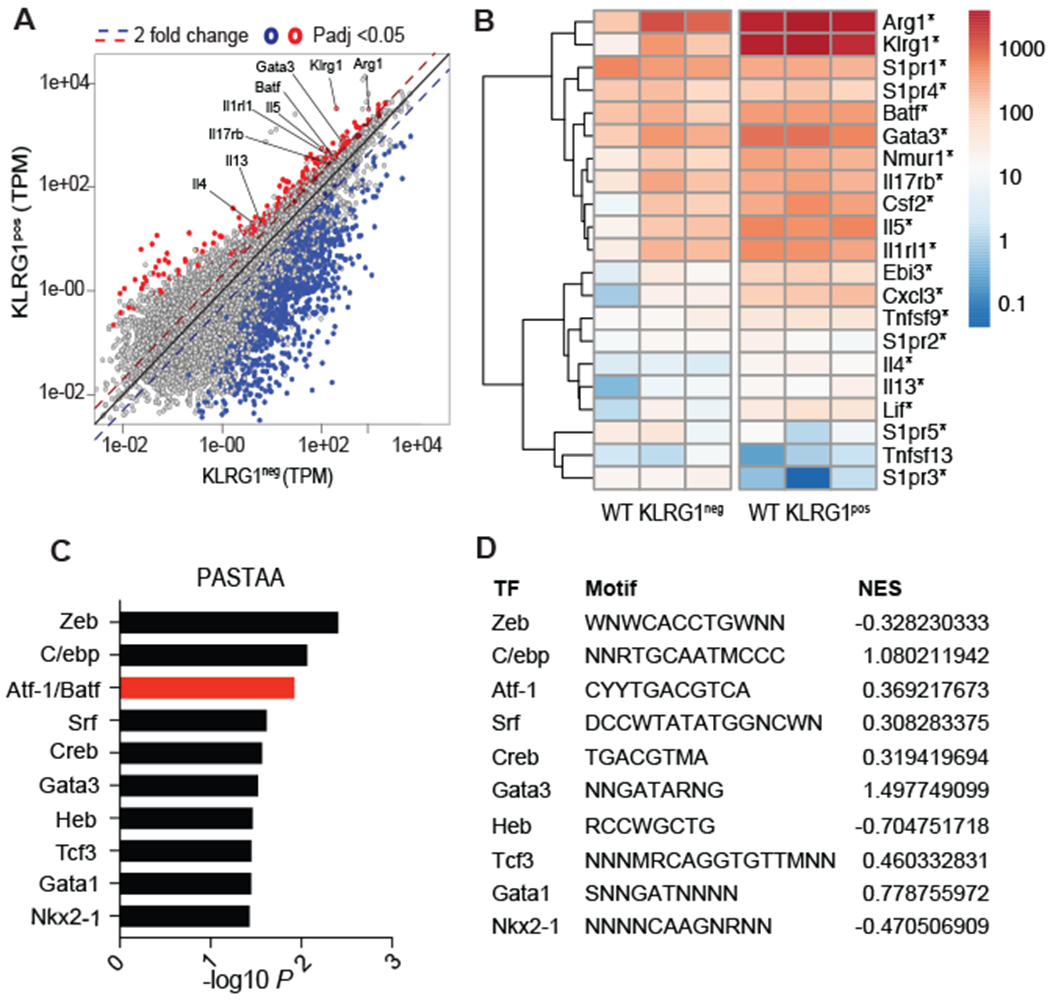
WT C57BL/6 mice were injected intraperitoneally (i.p.) with IL-33 daily for 4 days and pulmonary KLRG1neg (non-ILC2s) and KLRG1pos (ILC2s) were sorted on day 5 and used for RNA-sequencing. (A) Diagonal scatter plot comparing differentially expressed genes (padj <0.05) among KLRG1neg and KLRG1pos cells. (B) Heat map of ILC2-related genes as well as S1P receptor genes upregulated in KLRG1pos ILCs *p≤0.05. (C) Transcription factors predicted by PASTAA analysis to be involved in regulating genes that were enriched among KLRG1pos, compared to KLRG1neg, populations. (D) Normalized enrichment scores of the predicted transcription factors as assessed with gene set enrichment analysis of KLRG1pos ILC2s compared to the KLRG1neg population. Binding motifs as reported by TRANSFAC are also included in the table where S=C or G, W=A or T, R=A or G, Y=C or T, M= A or C, and N=any base. RNA-sequencing analysis was compiled from three separate experiments with n=3-6 mice per group.
BATF selectively promotes IL-25-responsive, KLRG1high iILC2s during helminth infection
ILC2s are identified as live, singlet, SSClowFSClowLin−CD4−CD90+KLRG1+ cells (fig. S2) and are required to clear N. brasiliensis within five days of infection following administration of IL-25 or IL-33 (6, 9). To investigate whether an impairment in ILC2 function exists in BATF-deficient mice, we assessed intestinal worm burden following administration of PBS, IL-25, or IL-33 in wildtype (WT), Rag−/−, Batf−/−, and Rag−/−Batf−/− mice five days after N. brasiliensis infection. While there was no difference in IL-33-dependent helminth expulsion between the groups of mice, Batf−/− and Rag−/−Batf−/− mice were significantly impaired in their ability to clear worms in response to IL-25 (Fig. 2A and B). A closer investigation of pulmonary ILCs revealed that the CD90midKLRG1high IL-25-responsive iILC2 population was absent in Batf−/− mice (Fig. 2C and D) whereas the IL-33-responsive CD90highKLRG1mid nILC2 (Fig. 2C and fig. S3A) and CD90highKLRG1neg (Fig. 2C and fig. S3B) populations remained intact. The N. brasiliensis-induced KLRG1high iILC2s were further phenotyped by low expression of ST2 (IL-33 receptor), high expression of IL-17RB (IL-25 receptor), positive for CD127, and high expression of BATF and GATA3 (Fig. 2E). Together, these data show that BATF deficiency more selectively impairs IL-25-responsive iILC2 cells than IL-33-responsive nILC2 cells.
Figure 2: BATF is required by IL-25-responsive KLRG1high pulmonary ILC2s.
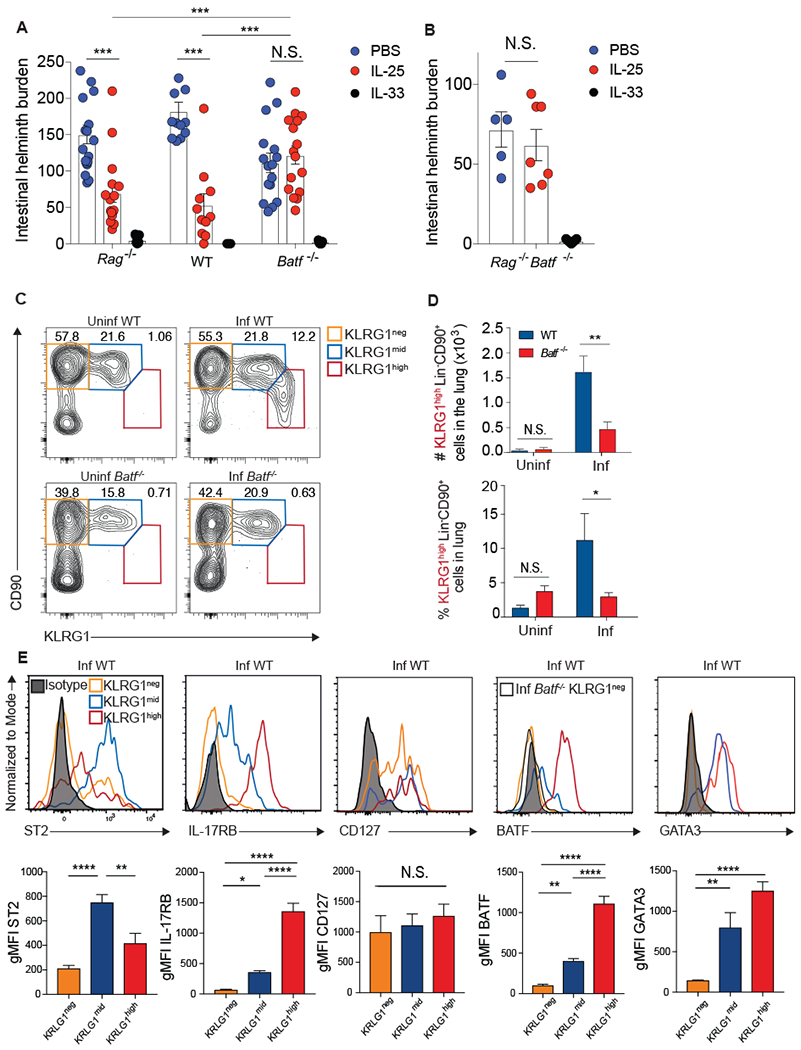
(A and B) Intestinal helminth burden following PBS, IL-25, or IL-33 administration to (A) Rag−/−, WT, or Batf−/− mice or (B) Rag−/−Batf−/− mice five days after infection with N. brasiliensis. Data combined from 6 (A) or 2 (B) independent experiments. (C-E) WT or Batf−/− mice were infected with N. brasiliensis and pulmonary Lin−CD90+ ILC2s were assessed 5 days later by flow cytometry. (C) Flow cytometry plots of different CD90+ ILC subsets based on KLRG1 expression. (D) Number and frequency of KLRG1high ILC2s combined from 7 independent experiments. (E) Representative histograms and compiled gMFI of various surface markers and transcription factors in the indicated ILC populations from infected WT mice. n=6-18 mice pooled from 2-7 /separate experiments. Means ± SEM, *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. Data was analyzed by two-tailed unpaired t test (D), and one-way ANOVA with Tukey post-test (A, B, E).
ILC2s are known to respond to epithelial alarmins such as IL-33, IL-25, and TSLP (6, 39). In the absence of helminth infection, intraperitoneal injections of IL-33 and IL-25, but not TSLP, led to an accumulation of CD90lowKLRG1high ILC2s in the lung (fig. S4A). Further analysis demonstrated that even though IL-33 administration led to accumulation of KLRG1high ILC2s, these cells are not phenotypically distinct from the IL-33-induced KLRG1mid nILC2 subset in regards to ST2 and IL-17RB expression whereas the KLRG1high population induced by IL-25 is unique based on alarmin receptor expression (fig. S4B). Therefore, IL-33 expands nILC2s, which can upregulate KLRG1 expression in response to this alarmin. In contrast, IL-25 leads to the accumulation of iILC2s in the lung, which uniformly display high KLRG1 expression.
Arginase-1 expression differentially defines nILC2s and iILC2s
Arginase-1 (Arg1) has been described as a marker for tissue-resident ILC2s and ILC2 precursors (20, 35, 36), yet its expression among IL-25-responsive relative to IL-33-responsive ILC2s remained uncharacterized. Arg1YFP reporter mice revealed a separate and distinct KLRG1highArglow population that was absent in both uninfected Arg1YFP and N. brasiliensis-infected Batf−/−Arg1YFP mice (Fig. 3A) (40). The majority of KLRG1mid tissue-resident nILC2s showed evidence of high Arg1 (YFP) expression regardless of infection or Batf status. In contrast, KLRG1high iILC2s from infected wildtype Arg1YFP mice expressed little Arg1 (YFP) (Fig. 3B and C). It should be noted that while Arg1 levels in KLRG1high iILC2s are low, expression is not absent as evident by the geometric mean fluorescence intensity (gMFI) of YFP (Arginase) when compared to a WT C57BL/6 reporter-negative mouse (represented by the dashed line in Fig. 3D). Thus, Arg1 expression is a unique identifier of ILC2 subsets in vivo.
Figure 3: KLRG1mid but not KLRG1high pulmonary ILC2s express high levels of Arg-1.
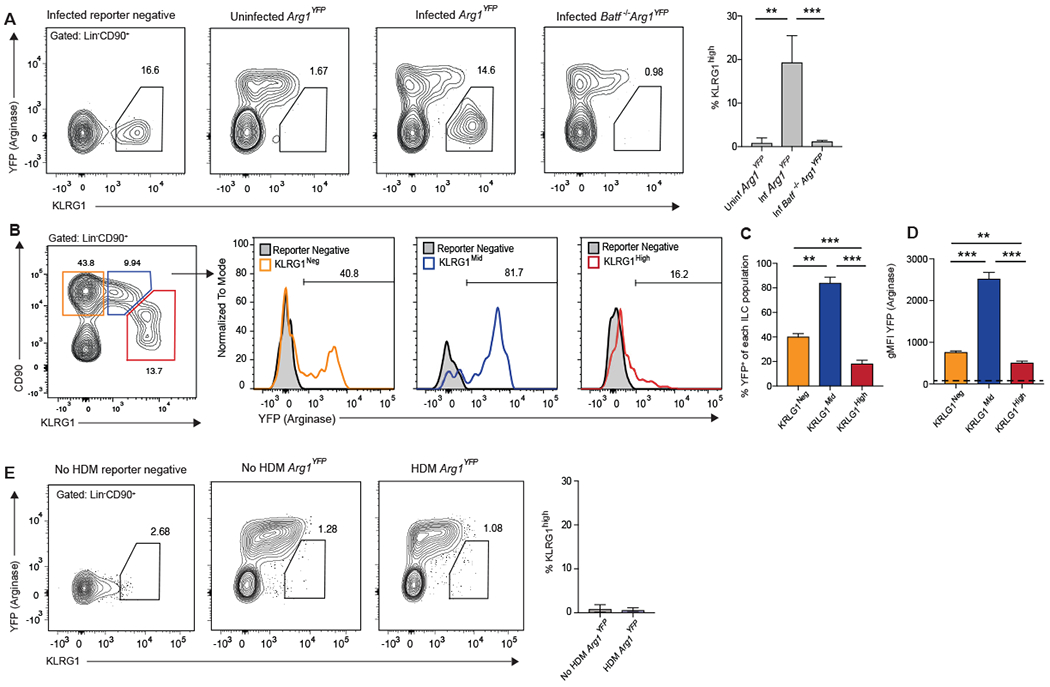
(A-D) Arg1YFP and Batf−/−Arg1YFP mice were infected with N. brasiliensis and pulmonary ILC2s were assessed 5 days later by flow cytometry. (A) Contour plots and combined data of the percentage of KLRG1high iILC2s. (B) Histograms displaying the percentage of YFP+ cells in the gated populations depicted in the contour plot. (C) Compiled data showing the percentage of cells that are YFP+ in (B). (D) Compiled data of the gMFI of YFP of each population in (B). The dashed line represents the gMFI of the reporter negative mouse. Data in (A-D) are from a single experiment with n=2-4 mice per group, representative of 4 individual experiments. (E) Arg1YFP mice were given consecutive daily HDM (10μg) by oropharyngeal aspiration for 4 days, rested on day 5, and pulmonary ILC2s assessed by flow cytometry on day 6. Representative contour plots display the frequency of KLRG1high iILC2s in the lung and the bar graph shows combined data from n=6-7 mice per group compiled from 2 separate experiments. **p≤0.01, ***p≤0.001as determined by one-way ANOVA and Tukey post-hoc analysis (A, C-D) or two-tailed unpaired t test (E).
House dust mite administration does not induce pulmonary iILC2s
We next wanted to understand whether iILC2s could be detected in other forms of acute type-2 pulmonary inflammation. To do this, we treated Arg1YFP mice for four consecutive days with house dust mite extract (HDM) via oropharyngeal aspiration. Unlike N. brasilliensis infection which induced robust iILC2 recruitment to the lung, administration of HDM did not lead to recruitment of iILC2s at the time point tested (Fig 3E). These results indicate that the early induction or recruitment of iILC2 cells to the lung can vary among different settings of type-2 inflammation. The unique events involved in iILC2 induction during helminth infection that are absent in allergen models are likely to reveal important insight into iILC2 biology and function.
BATF works in a cell intrinsic manner to modulate iILC2 cell fate during helminth infection
To determine whether the decreased frequency of pulmonary iILC2s in Batf−/− mice was cell intrinsic, we generated mixed bone marrow chimeric mice. Irradiated WT CD45.1/CD45.2 hosts were reconstituted with equal numbers of WT CD45.2 and Batf−/− CD45.1 bone marrow and infected with N. brasiliensis 8 weeks later (Fig. 4A and fig. S5A). Of the KLRG1high cells, the majority originated from WT bone marrow whereas significantly fewer originated from either residual host cells or Batf−/− bone marrow (Fig. 4B and C). Looking at the total Lin−CD90+ pulmonary ILC compartment, ~23% of the cells were residual CD45.1/CD45.2 host cells indicating their radio-resistant nature, a phenomenon that has already been observed in ILC3s (41) (fig. S5B). Furthermore, there was a major defect in the ability of Batf−/− bone marrow to repopulate the total ILC compartment, as WT bone marrow gave rise to the majority of Lin−CD90+ cells. When gating on ILCs of each donor origin first and then assessing KLRG1 expression, the KLRG1highCD90+ iILC2 subset could only be generated from the WT donor and not Batf−/− bone marrow (fig. S5B and C) further confirming the cell intrinsic requirement for BATF. Surprisingly, we also observed the expansion of pulmonary KLRG1high ILC2s derived from WT bone marrow in uninfected mice (fig. S5D and E). While this expansion required BATF, it does raise the question of whether homeostatic expansion alone can result in increased KLRG1 expression among ILC2s. In support, the apparent lack of nILC2s in the KLRG1mid gate after irradiation indicates that, similar to IL-33 administration (fig. S4A), KLRG1 expression may increase on nILC2s under certain conditions.
Figure 4: Cell intrinsic requirement of BATF for pulmonary KLRG1high ILC2s.
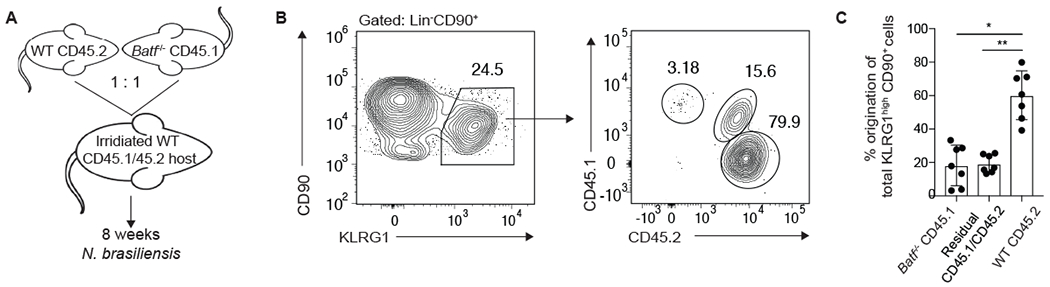
(A) Schematic depicting the generation and infection of bone marrow chimeric mice. Cells were analyzed 5 days after N. brasiliensis infection. (B) Contour plots showing congenic markers delineating Lin−CD90+KLRG1high lung cells that arose from CD45.1 Batf−/− or CD45.2 WT donors or CD45.1/CD45.2 residual host cells. (C) Combined results from 2 separate experiments, n=7. *p≤0.05, **p≤0.01 as determined by one-way ANOVA and Tukey post-hoc analysis.
ILC2s in the small intestine lamina propria (SILP) were also assessed in the bone marrow chimeric mice. Interestingly, in contrast to what was observed in the lung, residual ILC2s from the recipient mice were absent, indicating potential tissue-specific differences in irradiation efficiency. Moreover, while there was a reduction in the frequency of SILP ILC2s in a few Batf−/− mice, this result was not statistically significant across all mice (fig. S5F and G).
KLRG1high iILC2s are the major ILC2-source of IL-4 and IL-13 five days after helminth infection
To further investigate the function of pulmonary ILC2s, we identified immunologically relevant pathways and genes enriched in ILC2s through RNA-sequencing. Utilizing genes that were at least 2-fold enriched in KLRG1pos ILC2s, cytokine activity and cytokine-cytokine receptor interactions were identified as significantly associated pathways by both Gene Ontology Molecular Function and KEGG analysis (Fig. 5A and B). Furthermore, expression of genes for type-2 cytokines such as Il4, Il5, and Il13 was increased in KLRG1pos ILC2s relative to the KLRG1neg population (Fig. 5C). The emphasis on pathways involved in type-2 cytokine expression and ILC2 function was consistent with a key role for BATF in Th2 and Tfh cell function (28, 30, 31).
Figure 5: KLRG1high iILC2s are responsible for type-2 cytokine production in the lung 5 days after N. brasiliensis infection.
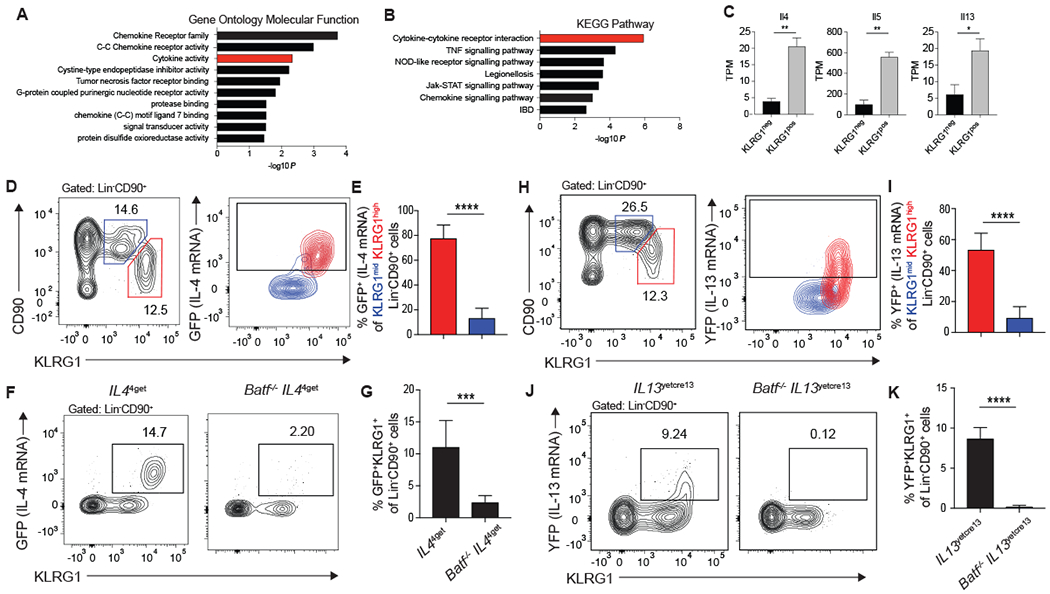
(A-C) RNA sequencing was performed on KLRG1pos and KLRG1neg Lin−CD90+ lung cells from mice treated with IL-33 as described in Figure 1. (A-B) Genes that are significantly different and increased at least two-fold in KLRG1pos relative to KLRG1neg cells were used in (A) Gene Ontology Molecular Function analysis and (B) KEGG Pathway analysis. Results are shown as -log10P values to denote pathways that are significantly associated with genes upregulated in KLRG1high ILC2s. Activity groups or pathways that relate to cytokine signaling are highlighted in red. (C) Quantification of type-2 cytokine expression in KLRG1pos and KLRG1neg populations. (D-G) IL44get and Batf−/−IL44get and (H-K) IL13yetcre13 and Batf−/−IL13yetcre13 mice were infected with N. brasiliensis and pulmonary Lin−CD90+ ILC2s were assessed 5 days later by flow cytometry. (D, H) Contour plot showing IL-4 (GFP) (D) or IL-13 (YFP) (H) cytokine reporter expression of KLRG1mid (blue) and KLRG1high (red) ILC2s. Gates were set according to a reporter negative control. (E, I) Bar graphs comparing the percent of KLRG1mid and KLRG1high ILC2s that express either IL-4 (GFP) with n=8 mice combined from 3 separate experiments (E) or IL-13 (YFP) with n=9-10 mice combined from 4 experiments (I). (F, J) Representative contour plots of IL-4 (GFP) (F) or IL-13 (YFP) (J) and KLRG1 expression on Lin−CD90+ lung ILC2s. (G, K) Bar graphs comparing the percent of KLRG1+ ILCs expressing IL-4 (GFP) with n=6-8 mice combined from 3 separate experiments (G) or IL-13 (YFP) with n=6-7mice combined from 3 separate experiments (K). *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 as determined by two-tailed unpaired t test.
To expand on these findings and extend the analysis of type-2 cytokine production to nILC2 and iILC2 subsets, C57BL/6 Il44get-GFP reporter mice were infected with N. brasiliensis and 5 days later IL-4 mRNA transcription, as detected by green fluorescent protein (GFP) expression, was assessed by flow cytometry in the KLRG1high and KLRG1mid ILC2 subsets. IL-4 expression during the early response to helminths is restricted to KLRG1high iILC2s with very little contribution from KLRG1mid nILC2s (Fig. 5D and E). The role of BATF in this process is clear as there is very little IL-4 mRNA being transcribed in the total Lin−CD90+ ILC compartment in Batf−/− IL44get-GFP mice (Fig. 5F and G). A similar pattern is seen in IL13Yetcre13-YFP reporter mice where the KLRG1high iILC2 subset is responsible for significantly more IL-13, as detected by yellow fluorescent protein (YFP) expression, than the KLRG1mid nILC2s (Fig. 5H and I). Again, BATF is critical for IL-13 transcription by ILCs as there is essentially no reporter expression in Batf−/− IL13Yetcre13-YFP mice (Fig. 5J and K). Defects in IL-4 and IL-13 production by Batf−/− ILC2s was confirmed using IL-4 and IL-13 protein reporter mice (12, 42) (fig. S6). These data highlight the importance of KLRG1high iILC2s in initiating the type-2 cytokine response in the lung during helminth infection and identify BATF as a key modulator of iILC2 function in type-2 immunity.
Expression of IL-13 five days after helminth infection is confined to migratory KLRG1high iILC2s
Since pulmonary KLRG1high iILC2s are a transient population which may migrate to the lung from peripheral sites (26), we investigated the chemokine gene signature of ILC2s from our initial RNAseq study. Utilizing genes that were enriched at least 2-fold in KLRG1pos ILC2s, Gene Ontology Molecular Function and KEGG pathway analysis indicated that overall chemokine receptor family and C-C chemokine receptor activity were significantly more likely to be involved in the recruitment of KLRG1pos ILC2s to the lung than expected by chance (Fig. 6A and B). In support, an array of chemokine receptors were shown to be differentially regulated between KLRG1pos relative to KLRG1neg ILCs (Fig. 6C). Specifically, enhanced expression of Ccr2, Ccr3, and Ccr1, all of which have been shown to mediate leukocyte migration to the lung during allergic inflammation (reviewed in (43)) as well as Cxcr6, which directs ILC2s to the lung (44), were detected in KLRG1pos ILC2s relative to the KLRG1neg population (Fig. 6C). To investigate whether the IL-13-producing KLRG1high iILC2 population is indeed migratory, we administered the sphingosine-1-phosphate receptor agonist, FTY720, to IL13Yetcre13-YFP mice upon infection with N. brasiliensis and found that mice treated with FTY720 exhibited a significantly reduced frequency and number of KRLG1high iILC2s in the lung compared to mice given saline (Fig. 6D and E). Similarly, blockade of lymphocyte migration led to a near complete absence of IL-13-producing Lin−CD90+KLRG1+ ILC2s in the lung (Fig. 6F and G) indicating that at this early stage of helminth infection, the majority of IL-13 is produced by migratory KLRG1high iILC2s rather than tissue-resident nILC2.
Figure 6: Appearance of KLRG1high pulmonary iILC2s in the lung is blocked by FTY720.
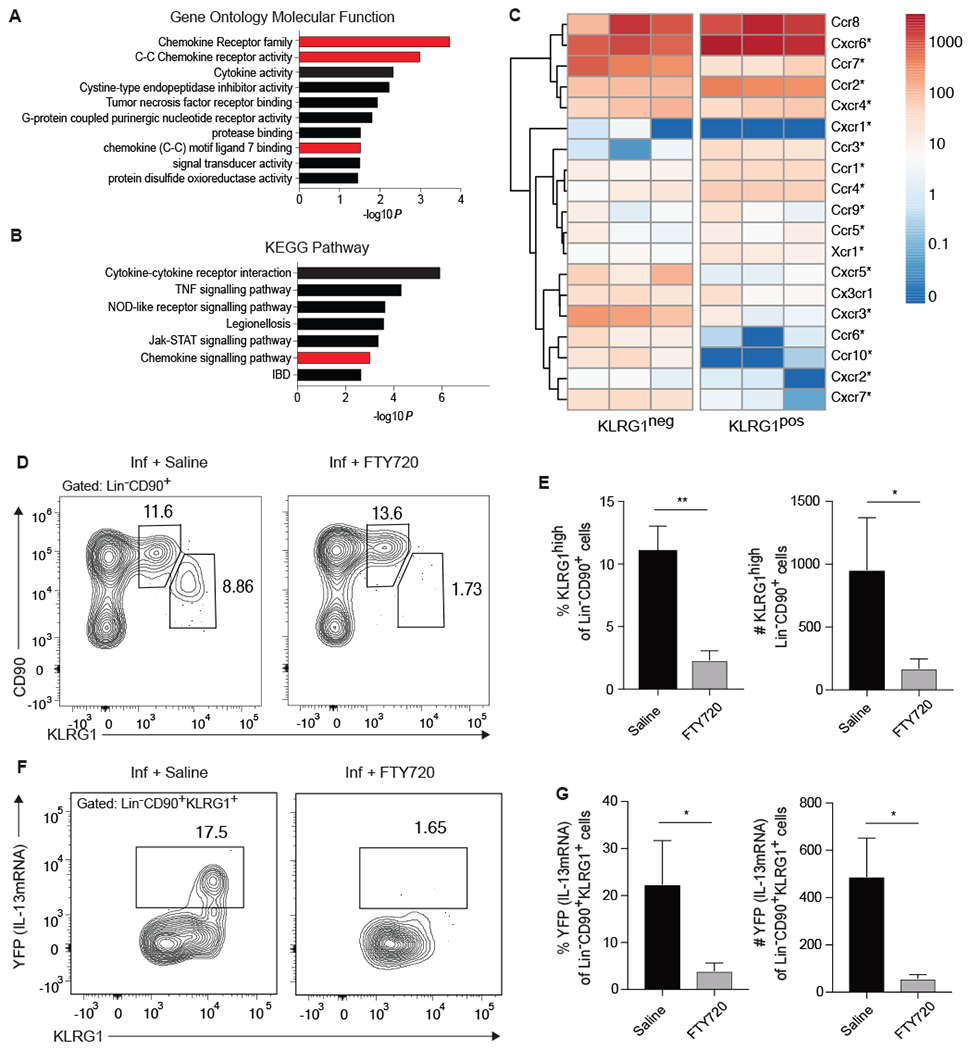
(A-C) RNA sequencing was performed on KLRG1pos and KLRG1neg Lin−CD90+ lung cells from mice treated with IL-33 as described in Figure 1. (A) Gene Ontology Molecular Function and (B) KEGG Pathway analyses highlighting gene groups and pathways that are significantly associated with genes that are at increased at least two-fold in KLRG1pos relative to KLRG1neg populations, as performed in Figure 5. Gene groups and pathways associated with cell trafficking are highlighted in red. (C) Heatmap of chemokine receptor expression in indicated populations from three separate RNA-sequencing experiments. Significant differential expression is denoted by an asterisk. (D-G) IL13yetcre13 mice were infected with N. brasiliensis on day 0 and given saline control or FTY720 on days 0, 2, and 4 after infection. Pulmonary Lin− CD90+ ILC2s were assessed on day 5 by flow cytometry. (D) Representative contour plots of the frequency of KLRG1mid and KLRG1high lung ILC2s. (E) Combined frequency and number of KLRG1high cells, n=3 from a single experiment representative of 3 independent experiments. (F) Contour plots of YFP (IL-13mRNA) expression within the KLRG1+ subset of Lin−CD90+ ILC2s. (G) Combined frequency and number of YFP+ Lin−CD90+KLRG1high cells, n=3 from a single experiment representative of 3 independent experiments. *p≤0.05, **p≤0.01 as determined by two-tailed unpaired t test.
Because intraperitoneal injection of IL-33 led to an expansion of KRLG1high ILC2s that resembled IL-25-induced KLRG1high iILC2s, we investigated whether any of these KLRG1high IL-33-responsive ILC2s might be migrating from the periphery. Mice given IL-33 and FTY720 displayed similar frequencies and numbers of CD90+ KLRG1high ILC2s as mice given only IL-33 (fig. S7A and B). Thus, unlike IL-25 which promotes the appearance of circulating iILC2 cells in the lung, IL-33 increased KLRG1 expression on nILC2 but did not lead to the mobilization of iILC2 cells.
ILC2s are present at a reduced frequency in the intestine of BATF-deficient mice but fail to produce IL-13
Intestinal homeostasis and helminth expulsion are modulated by ILC2 responsiveness to IL-25 (14–16). This prompted us to investigate whether BATF influenced ILC2s in the small intestine lamina propria (SILP). Like those in the lung, SILP ILC2s were gated on live, singlet, SSC-AlowFSC-AlowLin−CD90+CD4− cells (fig. S2). Unlike the lung where intermediate and high levels of KLRG1 identify unique subsets of nILC2 and iILC2s, only KLRG1high ILC2s were found in the intestine of naïve or N. brasiliensis-infected mice (Fig. 7A). This KLRG1-expressing population represents the only ILC2 population in this tissue. For this reason, we refer to SILP ILC2s as “KLRG1pos”. Moreover, this KLRG1pos population was also observed in the SILP regardless of Batf status, although infected Batf−/− mice displayed a significantly reduced frequency compared to infected WT mice (Fig. 7A and B). To determine whether BATF controlled type-2 cytokine expression in SILP ILC2s similar to what was observed for lung ILC2s, IL-13 production was assessed after helminth challenge in IL13Yetcre13-YFP or Batf−/−IL-13Yetcre13-YFP mice. The results again indicate a critical role for BATF in IL-13 transcription by ILC2, as significantly reduced frequencies of YFP+ cells were detected in the SILP of Batf−/− mice compared to their Batf-sufficient counterparts (Fig. 7C and D).
Figure 7: BATF is required for ILC2 function in the small intestine after helminth infection.
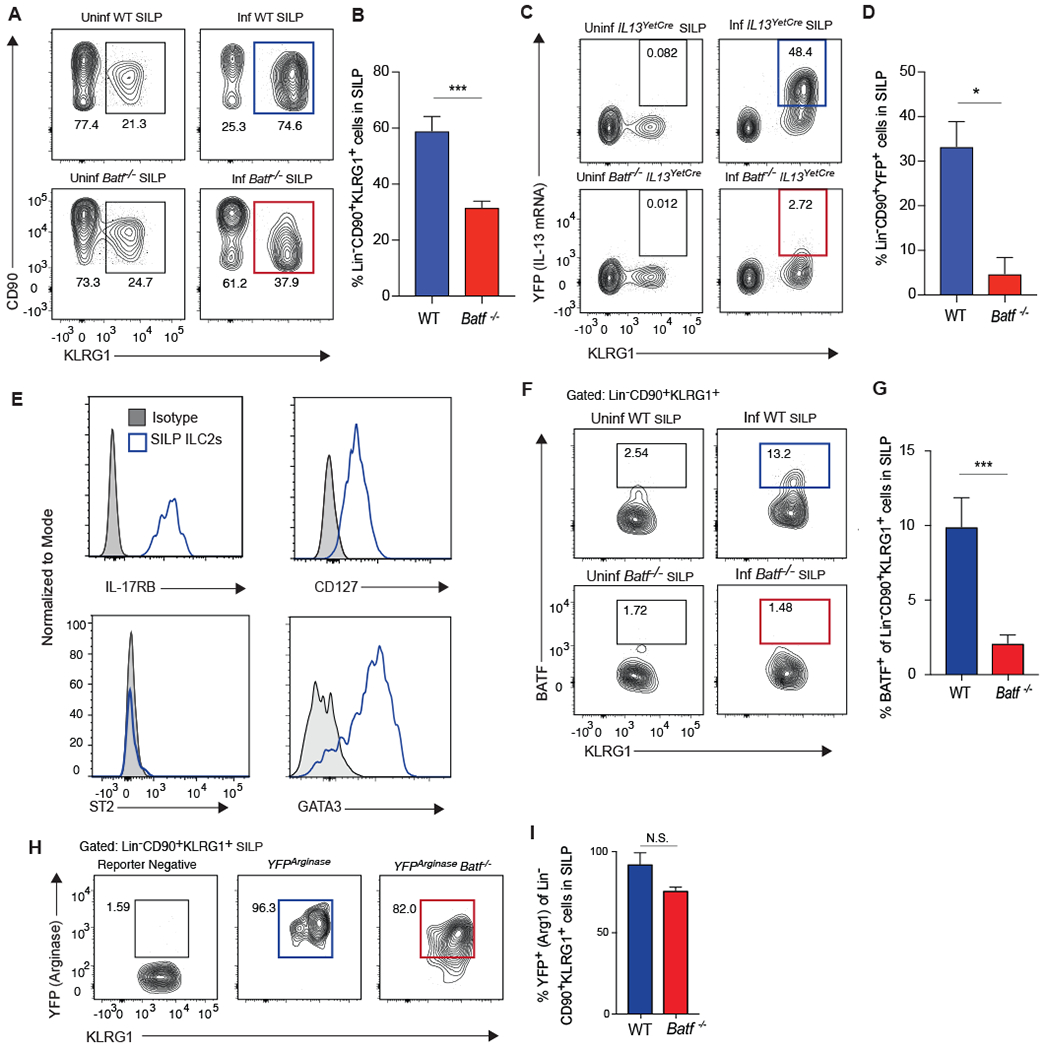
(A, B) WT or Batf−/− mice were infected with N. brasiliensis or remained uninfected. Lin− CD90+ KLRG1+ ILC2s in the small intestine lamina propria (SILP) were assessed 5 days after infection by flow cytometry. (A) Contour plots of SILP ILC2s in indicated mice pre- and post-infection. (B) Compiled data comparing the frequency of Lin− CD90+ KLRG1+ cells in the SILP between infected WT (blue) and Batf−/− (red) mice, n=7-9 mice, combined from 3 separate experiments. (C, D) IL13yetcre13 and Batf−/−IL13yetcre13 were infected with N. brasiliensis or remained uninfected. Lin− CD90+ KLRG1+ ILC2s in the SILP were assessed 5 days later by flow cytometry. (C) Contour plots of YFP (IL-13) expression versus KLRG1 expression. (D) Compiled data comparing the frequency of Lin− CD90+ KLRG1+ cells that express YFP (IL-13) in the SILP between infected WT (blue) and Batf−/− (red) mice, n=5 mice, combined from 2 separate experiments. (E) Representative histograms of various surface markers and transcription factors in SILP ILC2s from infected WT mice. (F, G) WT and Batf−/− mice were infected with N. brasiliensis or remained uninfected and Lin−CD90+KLRG1+ ILC2s in the SILP were assessed 5 days later by flow cytometry. (F) Representative contour plots showing the percentage of Lin− CD90+ KLRG1+ ILC2s expressing BATF. Gates were set according to Batf−/− mice. (G) Bar graph showing the percentage of Lin− CD90+ KLRG1+ ILC2s expressing BATF, n=6-8 mice, combined from 3 separate experiments. (H, I) Arg1YFP and Batf−/−Arg1YFP mice were infected with N. brasiliensis and SILP ILC2s were assessed 5 days later by flow cytometry. (H) Representative contour plots showing the percentage of Lin−CD90+KLRG1+ ILC2s that express YFP (Arg1 mRNA). Gates were set according to reporter negative mice. (I) Bar graph showing the percentage of Lin− CD90+ KLRG1+ ILC2s that express YFP (Arg1) between infected WT (blue) and Batf−/− (red) mice, n=6 mice, combined from 2 separate experiments. **p≤0.01, ***p≤0.001 as determined by two-tailed unpaired t test.
These results show that ILC2 populations may differ between the lung and SILP. This is interesting since pulmonary iILC2s have been reported to originate from the SILP (24). To explore this further, we phenotypically profiled the KLRG1pos SILP ILC2s and found that they resemble KLRG1high pulmonary iILC2s in their high expression of IL-17RB, CD127, and GATA3 (Fig. 7E). Also consistent with the iILC2 phenotype, KLRG1+ SILP ILC2s lacked significant ST2 expression (Fig. 7E). The lack of ST2 expression by SILP ILC2s reveals differential alarmin responsiveness among ILC2 cells in different mucosal tissues. Despite these similarities, several major differences were evident between lung KLRG1high and SILP ILC2 populations. First, while BATF expression was detected among all KLRG1high iILC2s in the lung, only a subset of ILC2s in the SILP expressed this transcription factor (Fig. 7F and G). Interestingly, helminth infection expands the BATF-expressing SILP ILC2s relative to uninfected mice. Second, Arginase-1 expression was elevated among all KLRG1pos ILC2s in the SILP but not KLRG1high iILC2s in the lung (Fig. 7H and I). Moreover, the presence of ILC2s in the SILP is not significantly affected by FTY720 administration, indicating that the majority of KLRG1pos SILP ILC2s are resident to the intestine (fig. S7C and D).
BATF-deficient mice have impaired tuft and goblet cell hyperplasia after helminth infection
In the SILP, ILC2-derived IL-13 is critical for worm expulsion via goblet cell hyperplasia, smooth muscle contractility, and increased tuft cell generation. Thus, we investigated whether BATF-deficiency would ultimately affect tuft cells and mucin-producing epithelial cells after N. brasiliensis infection. Immunohistochemistry of IL-25-producing tuft cells, marked by expression of doublecortin-like kinase 1 (DCLK-1), revealed a significant decrease in cell number/mm of villus in Batf−/− mice compared to WT mice 8 days after N. brasiliensis infection (Fig. 8A and B). Furthermore, BATF-deficiency led to a significant reduction of mucin-producing (MUC2) epithelial cells (Fig. 8C and D). These results are consistent with Batf−/− mice having decreased frequencies of IL-13-producing ILC2s relative to WT mice in their small intestine during N. brasiliensis infection (Fig 7C and D).
Figure 8: BATF-deficient mice have reduced tuft cells and mucin-producing epithelial cells after helminth infection.
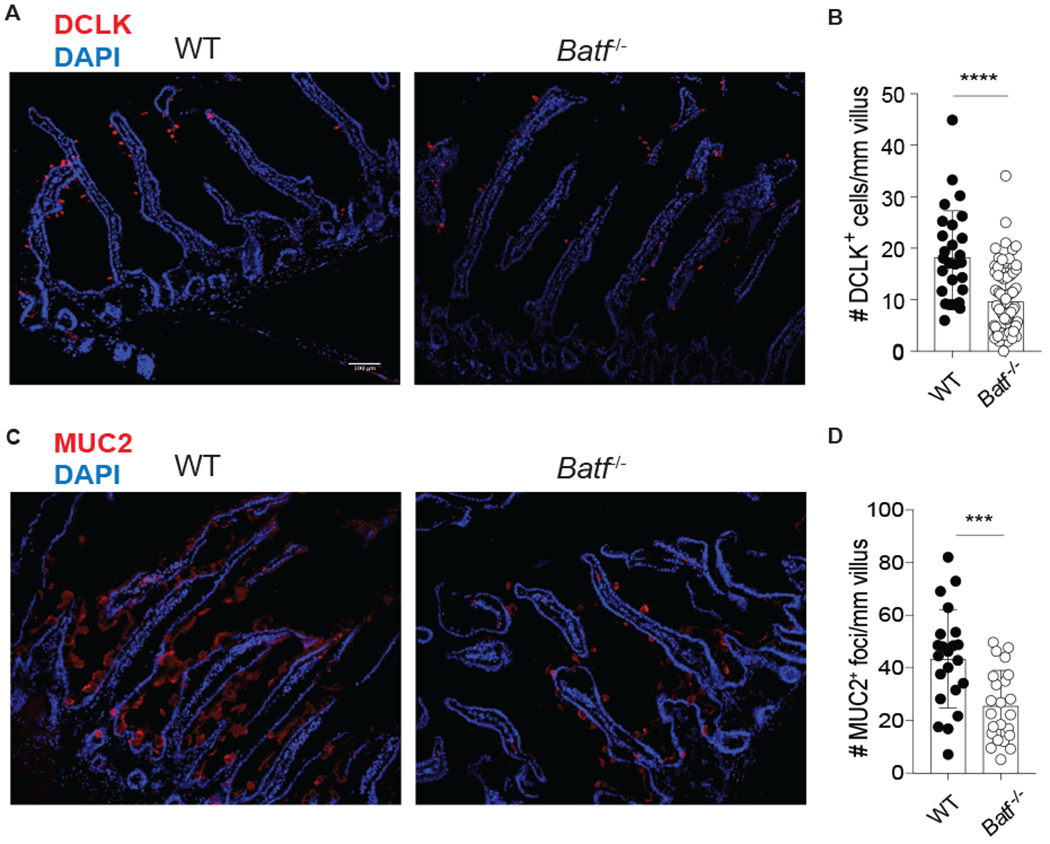
WT and Batf−/− mice were infected with N. brasiliensis and immunohistochemistry was performed on the small intestine 8 days later. (A) Representative image of DAPI (blue) and DCLK (red) staining. (B) Bar graphs depicting the number of DCLK+ cells/mm villus, n=28-77 individual villi counted from 4 individual mice per group, combined from 2 separate experiments. (C) Representative image of DAPI (blue) and MUC2 (red) staining. (D) bar graphs depicting the number of MUC2+ foci/mm villus, n=22-23 individual villi from 2 mice per group from one experiment. Scale bar represents 100μm. ****p≤0.0001 as determined by two-tailed unpaired t test.
Discussion
The role of tissue-resident ILC2s during helminth infection and allergic disease is well defined (45). These cells are prominent producers of IL-5 and IL-13 and likely work with canonical Th2 cells to promote eosinophil mobilization, wound healing, and worm clearance. In addition to their role in anti-helminth immunity and allergic disease, tissue-resident ILC2s modulate mucosal barrier homeostasis by responding to tuft cell-derived IL-25. Together, their positioning in tissues and their responsiveness to tissue-derived signals supports the long-held idea that these tissue-resident nILC2 cells act as early sensors of mucosal barrier integrity. The data provided herein shows that migratory iILC2s also contribute to barrier immunity, and serve as an important, early source of IL-4 and IL-13 in the lung after helminth infection (fig. S8). Thus, while tissue-resident nILC2s are prominent type-2 cytokine producers in the lung at the peak of the response, circulating iILC2s appear to be transient, early initiators of type-2 immune hallmarks at mucosal sites.
The selective dependence on the AP-1 superfamily transcription factor BATF in the fate and function of iILC2s relative to tissue-resident nILC2s provides further support for the unique nature of these populations in settings of type-2 immunity and helps to explain some of the heterogeneity observed among ILC2 populations in response to different tissue alarmins. Prior single cell RNA-sequencing revealed that both IL-25- and IL-33-responsive ILC2s expressed BATF, but no significance of its expression in ILCs was assigned (32, 33). Herein, the data indicates that BATF, although expressed in nILC2s, is highly expressed in iILC2s. BATF works in a cell intrinsic manner among iILC2s and appears critical in both the mobilization of iILC2s into circulation as well as the expression of IL-4 and IL-13. While we show that expression of GATA3 and IL-13 is common to both iILC2s and tissue-resident nILC2s, IL-4 expression is not commonly observed in mature nILC2s (6, 8, 9). As such, the observation that migratory iILC2s express both IL-4 and IL-13 is intriguing as this is reminiscent of Th2 cells which make both cytokines. IL-4 production has been reported among some intestinal ILC2s in response to colonization by the helminth Heligmosomoides polygyrus (46). The factors involved in migratory iILC2 expression of IL-4 are not clear, but the high expression of BATF in this population indicates one potential mechanism as we and others have shown that BATF plays an important role in IL-4 production by T cells (28–30). Future studies specifically investigating the regulome of migratory iILC2 cells and their relationship to other ILC and type-2 cytokine producing immune cell populations will be important in order to better understand these functional differences (47).
Another unexpected difference between migratory and tissue-resident ILC2s is their expression of Arginase-1 (Arg1) which has been used to define ILC2s in mice and humans (20, 35, 36). Arg1 plays a key role in ILC2 metabolism and allergic inflammation. Here we show that while tissue-resident nILC2s in the lung and intestine all express Arg1, the majority of migratory lung iILC2 cells express very little if any of this enzyme. This may indicate either that iILC2 are a unique Arg1low ILC2 subset independent from nILC2 or ILC2 precursors (18, 20, 35, 36), or that Arg1 expression is tied to tissue residency. In this context, circulating iILC2s may be Arg1-negative while they transit to the lung via the blood. Indeed, the lack of Arg1 in iILC2 cells is revealing when placed in the context of prior publications suggesting that these cells arise from intestinal emigrants (24). Given that all ILC2s in the intestine express high levels of Arg1 and pulmonary iILC2s lack significant expression of this enzyme, if pulmonary iILC2s originate in the intestine, then these intestinal emigrants would have to extinguish expression of Arg1 prior to arrival in the lung. If this is the case, the time it takes for an intestinal ILC2 to make its way to the lung as an iILC2 would have to be longer than 16-24 hours given the half-life of YFP in the Arg1YFP reporter mouse. This may indicate that Arg1 expression is tied to tissue-specific signals received by ILC2s as these cells establish a more permanent tissue-residency. Recent evidence that Th2 cells become “licensed” to become tissue-resident effectors only after they enter the lung and sense alarmins supports this idea (48). Indeed, the tissue in which an ILC resides can impart unique identities (49).
Alternatively, migratory iILC2 cells may be replaced by IL-33-responsive populations emigrating from the bone marrow or are outcompeted by proliferating tissue-resident populations already established at mucosal sites (23, 50). Although it was recently argued that migratory iILC2s do not contribute to the expansion of pulmonary ILC2 numbers after N. brasiliensis infection, a clear decrease (10%) of neonatally-derived tissue-resident ILC2 cells was observed in the lung after helminth infection compared to controls indicating de novo replacement of the local tissue-resident population does occur (20). Further studies are required to delineate between these models. In any event, Arg1 expression serves as a unique identifier for iILC2 from nILC2 cells and helps to stratify tissue resident and circulating ILC2 populations.
One limitation of this study is that it remains unclear whether human ILC2s are similarly composed of distinct ILC2 subsets that differentially respond to tissue alarmins. Human ILC2s that lack IL-33 receptor expression also lack Arginase-1 expression (36). This is phenotypically consistent with the migratory, IL-25-responsive population described here in mice and whether this represents a BATF-dependent migratory iILC2 equivalent in humans is intriguing. Another consideration lies within the model of ILC2 induction. The differences in response to parasitic helminth infection, which mobilizes iILC2s to the lung, compared to allergen exposure, which does not, suggests a more nuanced role for ILC2 cells at mucosal barriers than previously appreciated. How these different ILC2 subsets are influenced by environmental factors, disease states, and lifestyle choices will be an important next step in our understanding of ILC2 biology and their orchestration of type-2 immunity.
Materials and Methods
Study Design:
When possible, a sample size of four to six mice was chosen for mouse studies involving N. brasiliensis infection and cytokine reporters. This sample size was chosen to maximize significance and minimize use of animals (based on prior publications). All such studies were repeated at least twice. All studies involving N. brasiliensis infection and analysis in the lung had a defined endpoint of 5 days (Figs. 2–7) given pulmonary iILC2s peak at day 5 in this setting of type-2 inflammation. Worm clearance was assessed on day 5 when alarmins were provided (Fig. 2). Tuft cells and mucus production were assessed on day 8 corresponding to when worm expulsion occurs in the N. brasiliensis model of helminth infection (Fig. 8). All data including outliers were included in the data analysis and figures with the exception of one mouse that did not receive a productive infection determined by a lack of worms in the intestine at day 5 and lung eosinophilia similar to uninfected controls. The specific number of mice used and experimental repeats performed are included in each figure legend.
The major research objective outlined at the start of the study was to identify whether BATF played a role in ILC2 development and/or function. The initial hypothesis that BATF would influence ILC2s was based on the requirement of this transcription factor for the function of Th2 and Tfh cells in helminth clearance. Once a role for BATF was confirmed specifically in IL-25-mediated helminth clearance in both WT and Rag−/− mice, we hypothesized that BATF played a specific role in IL-25-responsive ILC2 subsets as compared to tissue-resident IL-33-responsive ILC2 populations.
In these studies, mouse tissues were used as research subjects. For experimental design, the models of type-2 immunity used were helminth infection and house dust mite administration. The helminth N. brasiliensis was used and all infections and methods were performed under controlled laboratory settings. The study was performed so that Batf−/− mice were compared to WT counterparts. For scoring of tuft cells and mucus-producing epithelial cells in intestinal sections, slides were blinded prior to analysis and scoring.
Mice:
C57BL/6 Batf−/− (51), Rag1−/−, Arg1YFP (40), and wildtype CD45.2 mice were originally purchased from Jackson Laboratories. C57BL/6 Il44get/KN2 and Il13YetCre/YetCre and BALB/c Il44get and Il13Smart were provided by Richard Locksley (University of California, San Francisco). Batf−/− mice were crossed onto the following mRNA and/or protein reporter mice: IL-4 (Il44get or Il4KN2), IL-13 (Il13YetCre) and Arginase-1 (Arg1YFP). C57BL/6 Batf−/− mice were crossed to Rag1−/− mice to generate Rag−/−Batf−/− mice. Additionally, C57BL/6 Batf−/− mice were bred to BALB/c mice for >10 generations and crossed onto the following reporter mice: IL-4 mRNA (Il44get) and IL-13 protein (Il13Smart). Both male and female mice were used in all experiments except for the bone marrow chimera studies in which only female donors were used to avoid potential rejection. All mice were maintained in specific pathogen free conditions and in accordance with guidelines established by the Institutional Animal Care and Use Committee at National Jewish Health. Mice used in these experiments were aged 6 to 12 weeks.
Preparation of single cell suspensions for flow cytometry and sorting:
Murine lungs were harvested, rinsed with PBS, finely minced using a razor blade and digested with 250μg/ml collagenase, 250μg/ml liberase, 1mg/ml hyaluronidase, and 200μg/ml DNAse in RPMI1640 for 30 minutes. In some experiments, murine lungs were dissociated using the 37c_m_LDK_1 program on a gentleMACS Octo Dissociator with Heaters (Miltenyi) with the enzyme cocktail listed above. Cells were then subjected to erythrocyte lysis and filtered prior to staining.
Murine small intestine was prepared according to a previous report (14). Briefly, 10cm of small intestine distal to the stomach was isolated, flushed with PBS, opened longitudinally, and the Peyer’s patches were removed. Tissue was incubated at room temperature, rocking in HBSS with 5% FBS, 10mM HEPES, 10mM DTT, and 5mM EDTA for 15 minutes. This was repeated in fresh solution after vigorous vortexing. Next, intestines were incubated with rocking in HBSS with 5% FBS and 10mM HEPES for 20 minutes and then vortexed vigorously. Each intestine was then cut into 3-5mm pieces and incubated in HBSS with 5% FBS, 10mM HEPES, 200μg/ml DNAse, and 40μg/ml liberase. Tissue was further dissociated in GentleMACS C tubes, quenched in 10% FBS, filtered through 80μm nylon mesh, and then lymphocytes were separated by centrifugation with lymphocyte separation medium (Corning 25-072).
Staining for flow cytometry:
Single-cell suspensions of mouse lung and small intestine were incubated with Fc block (TruStain fcX; Biolegend). Cells were then stained with antibodies to mouse: CD3 (145-2C11), CD4 (RM4-5), CD8 (53-6.7), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD45.1 (A20), CD45.2 (104), CD49b (DX5), CD90.2 (30-H12), CD127 (SB/199), B220 (RA3-6B2), FcεR1α (MAR-1), Gr-1 (RB6-8C5), IL-17RB (9B10), KLRG1 (2F1), NK1.1 (PK136), ST2 (DIH9), TCRγδ (GL3), TER119 or human: CD2 (S5.5), or CD4 (S3.5). Cells were resuspended in 2% FCS containing DAPI. For intracellular transcription factor staining, Fc receptors were blocked and cells were stained with live/dead fixable violet dye (ThermoFisher) prior to fixation and permeabilization (Foxp3 transcription factor staining kit; eBioscience) according to manufacturer’s instructions. Cells were then incubated with anti-mouse/human BATF (MBM7C7) or anti-mouse GATA3 (TWAJ). Data were collected on a LSRII (BD) or a LSRFortessa (BD) and analyzed with FlowJo software v10 (Treestar). For analysis of ILCs, cells were gated as lineage-negative which excludes cells labelled with the following antibodies: CD3, CD4, CD8, TCRγδ, CD11b, CD11c, CD19, B220, CD49b, FcεR1α, Gr-1, NK1.1, and Ter119.
RNA Sequencing and Bioinformatics Analysis:
Following IL-33 treatment, Linneg, CD90+ KLRG1neg or KLRG1pos populations from WT Il44get mice were sorted to >85% purity into a solution of 50% FCS, 50% PBS, and 20 U RNAase inhibitor on a BD FACSAria Fusion. RNA was isolated (RNeasy Micro Kit; Qiagen) and libraries were constructed by SMARTer 1st strand cDNA synthesis (SMARTer® Ultra™ Low Input RNA Kit– v4; Clontech) and full-length dscDNA amplification by LD-PCR, followed by purification and validation. Samples were then fragmented, tagged, and indexed (Nextera XT) and sequenced on a HiSeq 2500 as routinely performed by the NJH Genomics Facility. FASTQ files were generated using the Illumina bcl2fastq converter (version 2.17) and read quality was assessed using FastQC (version 0.11.5). Nextera TruSight adapters were trimmed with skewer (version 0.2.2) (52) and mapped with STAR aligner (version 2.4.1d) (53) to the GRCm38 assembly of the mouse genome using gene = annotations from Ensembl version 90 (http://aug2017.archive.ensemble.org/index.html). Gene reads were counted with featureCounts from Subread software package (v1.5.2) (54). Pairwise comparisons were conducted using the Wald test in the DESeq2 package (version 1.81) (55) for the R statistical software (version 3.2.0). p-values were adjusted for multiple testing using the method by Benjamini & Hochberg (56). For Principal components analysis (PCA), heatmaps, and expression plots, raw read counts per sample and gene were normalized to transcripts per million (TPM). PCA was performed using the prcomp function in R version 3.3.2. Heatmaps were generated using the pheatmap function (version 1.0.8) (https://CRAN.R-project.org/package=pheatmap) in R version 3.3.2. Gene ontology (molecular function), KEGG pathway, and prediction of transcription factors was conducted using DAVID (Database for Annotation, Visualization and Integrated Discovery) version 6.8 functional annotation tool (https://david.ncifcrf.gov/tools.jsp). PASTAA (Predicting Associated Transcription Factors from Annotated Affinities) was perfomed using the webtool at http://trap.molgen.mpg.de/cgi-bin/pastaa.cgi) and validated by utilizing gene sets within collection 3-Transcription factor targets in MSigDB. Gene Set Enrichment Analysis was performed with the WebGestalt 2019 webtool assessing Transcription factor targets within the network functional database (http://www.webgestalt.org). Transciption factor binding motifs were reported by TRANSFAC v7.4. All pathway analyses utilized expression data from the KLRG1pos ILCs that were significantly different from KLRG1neg population and at least 2-fold enriched.
N. brasiliensis infection:
N. brasiliensis were prepared as previously described (57), and 500 L3 larvae in 0.9% saline were injected subcutaneously in the center rear flank of mice. Mice were euthanized 5 or 8 days post-infection.
Administration of epithelial alarmins:
Mice were given i.p. recombinant IL-33 (Biolegend, 300ng), IL-25 (R&D, 300ng), or TSLP (R&D, 800ng) daily for three days, rested one day, and then lung ILC2 populations were assessed by flow cytometry.
Epithelial alarmin-induced helminth expulsion:
Mice were infected with 500 L3 N. brasiliensis larvae. On the day of infection and for an additional 3 days, mice were administered sterile PBS or 500ng recombinant IL-33 (Biolegend) or IL-25 (R&D Systems) i.p. Mice were euthanized 5 days post-infection and the small intestine was opened longitudinally, placed in HBSS at 37°C for 1 hour, and worms were manually enumerated.
Administration of HDM:
Mice were given 10μg HDM extract in 50μl PBS daily for 4 days by oropharyngeal aspiration, rested one day, and euthanized the following day for analysis of pulmonary ILC2 populations by flow cytometry as described.
Generation of bone marrow chimeras:
CD45.1/CD45.2 wildtype (WT) mice were irradiated with 1 dose of 900 rads and reconstituted with a 1:1 ratio of 2x106 CD45.2 WT and 2x106 CD45.1 Batf−/− bone marrow cells by tail-vein injection. Mice were maintained on a diet containing trimethoprim and sufametoxazole (SEPTRA) for 4 weeks before being switched to normal chow and were rested for a total of 8 weeks prior to N. brasiliensis infection.
Treatment with FTY720:
FTY720 was purchased from Sigma Aldrich and administered i.p. at ~1μg/g mouse weight in sterile 0.9% saline. FTY720 was given on the day of N. brasiliensis infection or IL-33 administration and re-administered 2 and 4 days post-infection/treatment.
Immunohistochemistry and quantification of DCLK+ and MUC2+ cells:
Mice were euthanized 8 days following infection and the first 10 cm of the duodenum and jejunum were flushed with PBS, fixed in 4% paraformaldehyde for 4 hours, washed in PBS overnight, and placed in 30% sucrose for 6 hours before embedding in OCT medium. Eight micron cryosections for both DCLK and MUC2 staining were cut and placed on slides for histology. For detection of DCLK+ cells, slides were incubated with: 1% H2O2 in PBS for 45 minutes, TNB and Fc block for 30 minutes, and biotin/avidin for 30 minutes each. Tissues were stained with 1:1000 rabbit anti-DCLK (Abcam 31704) in TNB followed by 1:500 biotinylated donkey anti-rabbit antibody (Jackson ImmunoResearch), streptavidin-HRP (Perkin Elmer), 1:200 555-tyramide (Perkin Elmer), and DAPI and mounted with a coverslip (Vectashield). For detection of MUC2+ cells, tissues were blocked with 5% BSA and Fc block and stained with 1:200 anti-mouse MUC2 (Santa Cruz H-300), followed by goat anti-rabbit 647 (LifeTechnologies), and DAPI. Images were obtained on a Marianas microscope. Images were blinded and the number of DCLK+ cells/mm villus were quantified using Fiji (ImageJ) where only intact villi were assessed. A similar blinded quantification was used for MUC2 staining but since mucin is released from the cell, the number of MUC2+ foci/mm villus was assessed by enumerating only the regions where discrete MUC2 staining originates.
Statistical Analyses:
Data are the mean ± SEM from the specified number of mice combined from multiple experiments unless otherwise noted. Statistical calculations were performed with Prism 7.0 software (GraphPad). Comparison of three or more groups was performed using a one-way ANOVA test, followed by Tukey post hoc analysis. Comparison of data between two groups was analyzed using a two-tailed unpaired t test to determine statistical significance. In the figures indicated, N.S. designates non-significant statistical differences. In all figures, only statistical differences between WT and Batf−/− mice in either naïve, infected, or treated groups are displayed. Statistically significant differences are indicated by *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
RNA-seq data availability:
The data and software code used to generate and support this study will be available from corresponding authors upon request. RNA-sequencing data is freely available in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE125816).
Supplementary Material
Table S1. Raw Data Spreadsheet
Fig. S1. Gating strategy for sorting cells for RNAseq
Fig. S2. Gating strategy for murine pulmonary and small intestine lamina propria ILCs.
Fig. S3. Quantification of KLRG1neg and KLRG1mid pulmonary ILC2s.
Fig. S4. Induction of pulmonary ILC2s by alarmins.
Fig. S5. Bone marrow chimeric mice reveal KLRG1high ILC2s require BATF and expand even in the absence of infection.
Fig. S6. Quantification of type-2 cytokine protein reporter production by lung ILC2s.
Fig. S7. FTY720 treatment does not affect nILC2s in the lung or any ILC2 population in the SILP.
Fig. S8. Graphical Summary.
Acknowledgments:
We thank Dr. Richard Locksley for mouse reporter strains; Shirley Sobus and Josh Loomis for assistance with flow cytometry, sorting, and microscopy; Mary Chapman and Kendra Walton for assistance and generation of RNA sequencing libraries; Dr. Erwin Gelfand for access to gentleMACS dissociator; Dr. Claudia Jakubzick, Sophie Gibbings, Dr. Mark Dell’Aringa, Ivy Brown, Mary Schoenbach, Ann Miller, and Julio Barerra for technical and laboratory support; and Dr. Matt Rosenbaum, Samantha Crow, Michael Crumrine, and the NJH Biological Resource Center for excellent animal care and support.
Funding: This work was supported by NIH grant AI119004 (R.L.R.) and Windsweep Farm Fellowship (M.M.M).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The data used to generate and support this study will be available from corresponding authors upon request. RNA-sequencing data is freely available in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE125816. All mice are either commercially available or available under a material transfer agreement. For IL44get C57BL/6 mice which are under a current MTA, please contact the corresponding authors to initiate a request for access to these mice.
References and Notes
- 1.Pulendran B, Artis D, New paradigms in type 2 immunity. Science 337, 431–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen JE, Sutherland TE, Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Seminars in immunology 26, 329–340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locksley RM, Asthma and allergic inflammation. Cell 140, 777–783 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gieseck RL 3rd, Wilson MS, Wynn TA, Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol 18, 62–76 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Bao K, Reinhardt RL, The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, McKenzie AN, Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fallon PG, Ballantyne SJ, Mangan NE, Barlow JL, Dasvarma A, Hewett DR, McIlgorm A, Jolin HE, McKenzie AN, Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J Exp Med 203, 1105–1116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S, Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 463, 540–544 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Price AE, Liang HE, Sullivan BM, Reinhardt RL, Eisley CJ, Erle DJ, Locksley RM, Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proceedings of the National Academy of Sciences of the United States of America 107, 11489–11494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halim TY, Krauss RH, Sun AC, Takei F, Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 36, 451–463 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, Locksley RM, Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 502, 245–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM, Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology 13, 58–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien TF, Bao K, Dell’Aringa M, Ang WX, Abraham S, Reinhardt RL, Cytokine expression by invariant natural killer T cells is tightly regulated throughout development and settings of type-2 inflammation. Mucosal Immunol 9, 597–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Moltke J, Ji M, Liang HE, Locksley RM, Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS, Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P, Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gracz AD, Samsa LA, Fordham MJ, Trotier DC, Zwarycz B, Lo YH, Bao K, Starmer J, Raab JR, Shroyer NF, Reinhardt RL, Magness ST, Sox4 Promotes Atoh1-Independent Intestinal Secretory Differentiation Toward Tuft and Enteroendocrine Fates. Gastroenterology 155, 1508–1523 e1510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bando JK, Liang HE, Locksley RM, Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol 16, 153–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY, Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, Locksley RM, Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AV, Tward AD, Krummel MF, Peng T, Molofsky AB, Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity 50, 707–722 e706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puttur F, Denney L, Gregory LG, Vuononvirta J, Oliver R, Entwistle LJ, Walker SA, Headley MB, McGhee EJ, Pease JE, Krummel MF, Carlin LM, Lloyd CM, Pulmonary environmental cues drive group 2 innate lymphoid cell dynamics in mice and humans. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, Peebles RS Jr., IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J Exp Med 215, 263–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF Jr., Paul WE, Germain RN, S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E, Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. Immunology 13, 145–149 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U, Williamson PR, Urban JF Jr., Paul WE, IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat Immunol 16, 161–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, Fokkens WJ, Cupedo T, Spits H, Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol 12, 1055–1062 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Bao K, Carr T, Wu J, Barclay W, Jin J, Ciofani M, Reinhardt RL, BATF Modulates the Th2 Locus Control Region and Regulates CD4+ T Cell Fate during Antihelminth Immunity. J Immunol 197, 4371–4381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahoo A, Alekseev A, Tanaka K, Obertas L, Lerman B, Haymaker C, Clise-Dwyer K, McMurray JS, Nurieva R, Batf is important for IL-4 expression in T follicular helper cells. Nature communications 6, 7997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, Taparowsky EJ, Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. The Journal of experimental medicine 207, 933–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM, The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature immunology 12, 536–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy TL, Tussiwand R, Murphy KM, Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nature reviews. Immunology 13, 499–509 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, Haas BJ, Tickle TL, Trombetta JJ, Baral P, Klose CSN, Mahlakoiv T, Artis D, Rozenblatt-Rosen O, Chiu IM, Levy BD, Kowalczyk MS, Regev A, Kuchroo VK, The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saenz SA, Siracusa MC, Monticelli LA, Ziegler CG, Kim BS, Brestoff JR, Peterson LW, Wherry EJ, Goldrath AW, Bhandoola A, Artis D, IL-25 simultaneously elicits distinct populations of innate lymphoid cells and multipotent progenitor type 2 (MPPtype2) cells. J Exp Med 210, 1823–1837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bando JK, Nussbaum JC, Liang HE, Locksley RM, Type 2 innate lymphoid cells constitutively express arginase-I in the naive and inflamed lung. J Leukoc Biol 94, 877–884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monticelli LA, Buck MD, Flamar AL, Saenz SA, Tait Wojno ED, Yudanin NA, Osborne LC, Hepworth MR, Tran SV, Rodewald HR, Shah H, Cross JR, Diamond JM, Cantu E, Christie JD, Pearce EL, Artis D, Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol 17, 656–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klose CSN, Mahlakoiv T, Moeller JB, Rankin LC, Flamar AL, Kabata H, Monticelli LA, Moriyama S, Putzel GG, Rakhilin N, Shen X, Kostenis E, Konig GM, Senda T, Carpenter D, Farber DL, Artis D, The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roider HG, Manke T, O’Keeffe S, Vingron M, Haas SA, PASTAA: identifying transcription factors associated with sets of co-regulated genes. Bioinformatics 25, 435–442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohapatra A, Van Dyken SJ, Schneider C, Nussbaum JC, Liang HE, Locksley RM, Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis. Mucosal Immunol 9, 275–286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, Locksley RM, Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC, Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med 211, 1723–1731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M, A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 23, 419–429 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lloyd C, Chemokines in allergic lung inflammation. Immunology 105, 144–154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Chen S, Chi Y, Yang Y, Chen X, Wang H, Lv Z, Wang J, Yuan L, Huang P, Huang K, Corrigan CJ, Wang W, Ying S, Kinetics of the accumulation of group 2 innate lymphoid cells in IL-33-induced and IL-25-induced murine models of asthma: a potential role for the chemokine CXCL16. Cellular & molecular immunology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambrecht BN, Hammad H, The immunology of asthma. Nat Immunol 16, 45–56 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Pelly VS, Kannan Y, Coomes SM, Entwistle LJ, Ruckerl D, Seddon B, MacDonald AS, McKenzie A, Wilson MS, IL-4-producing ILC2s are required for the differentiation of TH2 cells following Heligmosomoides polygyrus infection. Mucosal Immunol 9, 1407–1417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF Jr., Davis FP, Kanno Y, O’Shea JJ, Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang HE, Pollack JL, Gate RE, Haliburton GE, Ye CJ, Marson A, Erle DJ, Locksley RM, A tissue checkpoint regulates type 2 immunity. Nat Immunol 17, 1381–1387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, Locksley RM, Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol 19, 1093–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Sullivan TE, Rapp M, Fan X, Weizman OE, Bhardwaj P, Adams NM, Walzer T, Dannenberg AJ, Sun JC, Adipose-Resident Group 1 Innate Lymphoid Cells Promote Obesity-Associated Insulin Resistance. Immunity 45, 428–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM, The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang H, Lei R, Ding SW, Zhu S, Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y. a. H., Yosef, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 57.Voehringer D, Reese TA, Huang X, Shinkai K, Locksley RM, Type 2 immunity is controlled by IL-4/IL-13 expression in hematopoietic non-eosinophil cells of the innate immune system. J Exp Med 203, 1435–1446 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw Data Spreadsheet
Fig. S1. Gating strategy for sorting cells for RNAseq
Fig. S2. Gating strategy for murine pulmonary and small intestine lamina propria ILCs.
Fig. S3. Quantification of KLRG1neg and KLRG1mid pulmonary ILC2s.
Fig. S4. Induction of pulmonary ILC2s by alarmins.
Fig. S5. Bone marrow chimeric mice reveal KLRG1high ILC2s require BATF and expand even in the absence of infection.
Fig. S6. Quantification of type-2 cytokine protein reporter production by lung ILC2s.
Fig. S7. FTY720 treatment does not affect nILC2s in the lung or any ILC2 population in the SILP.
Fig. S8. Graphical Summary.


