SUMMARY
Bovine calf scours reported to be caused by multiple aetiologies resulting in heavy mortality in unweaned calves and huge economic loss to the dairy farmers. Among these, cryptosporidiosis is an emerging waterborne zoonoses and one of the important causes of neonatal calf diarrhoea. Poor immune response coupled with primary cryptosporidial infections predispose neonatal calves to multiple secondary infections resulting in their deaths. In the present study, faecal samples from 100 diarrhoeic calves randomly picked up out of 17 outbreaks of bovine calf diarrhoea in periurban Ludhiana, Punjab in Northern India were subjected to conventional (microscopy, modified Zeihl–Neelsen (mZN) staining) and immunological and molecular techniques (faecal antigen capture ELISA and PCR) for detection of primary Cryptosporidium parvum infection as well as other frequently reported concurrent pathogens, viz. rotavirus and coronavirus, Salmonella spp., Escherichia coli, Clostridium perfringens and Eimeria spp. The faecal antigen capture ELISA and PCR revealed 35% prevalence of C. parvum in contrast to 25% by mZN staining with a relatively higher prevalence (66·7%) in younger (8–14-day-old) calves. The detection rate of the other enteropathogens associated with C. parvum was 45·71% for C. perfringens followed by Salmonella spp (40·0%), rotavirus (36·0%), coronavirus (16·0%), E. coli (12·0%) and Eimeria spp (4·0%) The sensitivity for detection of C. parvum by ELISA and mZN staining in comparison to PCR was 97·14% and 72·72%, respectively. An important finding of the study was that C. parvum alone was found in only 10% of the diarrhoeic faecal samples, whereas, majority of the samples (90%) showed mixed infections ranging from a combination of two to five agents. This is the first documentary proof of C. parvum and associated pathogens responsible for severe periurban outbreaks of bovine calf diarrhoea culminating in heavy mortality from Northern India.
Key words: Bovine calf diarrhoea, concurrent infections, Cryptosporidium parvum, ELISA, histopathology, PCR, periurban outbreaks
INTRODUCTION
Diarrhoea, a common clinical presentation in bovine calves has a multifactorial aetiology [1], including Cryptosporidium and Eimeria spp., rotavirus, coronavirus, enteropathogenic Escherichia coli and Salmonella spp. [2]. The neonatal diarrhoeic syndrome caused by Cryptosporidium parvum is usually observed in 5–35-day-old claves with maximum incidence in the second week of life [3], resulting in enormous direct and indirect economic loss to dairy industry [4–6]. In addition to bovine calves, it is known to infect humans, domestic and wild animals, and birds [7–11]. The cryptosporidia, highly resistant to harsh environmental conditions, transmitted by faeco-oral route, have attained an increased zoonotic importance posing a great risk to dairy workers, milk handlers and general public through water contamination. The disease has particularly generated a great public health interest after a large human waterborne outbreak in Milwaukee in 1993 [11, 12].
Difficulties in the clinical diagnosis of infectious diarrhoea arise from frequent non-specific clinical signs and lesions, the presence of asymptomatic infections, the involvement of multiple agents, and the interplay of intrinsic and extrinsic factors that predispose the host to enteric dysfuction and pathology [13]. Traditionally, the infectious agents of diarrhoea have been identified using conventional techniques [14, 15]. However, these techniques are labour intensive, time consuming and less sensitive; therefore, advanced diagnostic methods may prove more efficacious in determining the precise aetiology of calf diarrhoea complex and thereby helpful in planning strategies for better herd management, disease surveillance and control programmes.
In India, extensive research work has been conducted on bovine calf diarrhoea with majority of infectious aetiological agents incriminated using conventional methods. Studies on Cryptosporidium in the country have mainly focused on the morphological identification of oocysts in faeces [5, 7, 8, 11, 16–20] with little emphasis on immunomolecular methods [21–24]. These previous investigations were concentrated on isolated reports of cryptosporidia alone without an insight into multiple aetiologies. Even globally, very few studies have been conducted on Cryptosporidium-associated complex aetiology of bovine calf diarrhoea outbreaks [6, 25]. In fact very few comprehensive investigations ellucidating the most recognised agents, viz. protozoa (C. parvum, Eimeria spp.) viruses (rota and corona) and bacteria (E. coli, Salmonella spp., Clostridium perfringens) have been attempted [26–30]. Moreover, a comparative evaluation of multiple aetiological factors by traditional, immunological and molecular methods with a pursuit to determine their relative sensitivity using faecal samples is lacking. The present study deals with 17 periurban outbreaks of bovine calf diarrhoea investigated thoroughly comparing faecal examination, faecal ELISA and faecal PCR (most trusted methods) from 100 cases with 43% mortality.
MATERIALS AND METHODS
Study area and sample collection
The study was conducted in Ludhiana, located between north latitude 30°34′ and 31°01′ and east longitude 75°18′ and 76°20′. It is most populated and centrally located district of Punjab (India) with a huge periurban dairy animal population. The representative faecal samples were collected in sterile plastic bags directly from the rectum of 100 calves covering 17 outbreaks (four to six animals each) of diarrhoea from the organised dairy farms located in the periurban areas consisting of about 300 organised dairy farms in a radius of 25 km. To determine the exact age-wise distribution of aetiological factors of neonatal bovine diarrhoea, the animals were divided into five sub-groups, i.e. 1–7 days (group I), 8–14 days (group II), 15–21 days (group III), 22–30 days (group IV) and 31–60 days (group V) of age. Faecal samples were stored at −20 °C for ELISA and PCR.
The study was conducted after approval of the Institutional Animal Ethical Committee and consent of the dairy farmers was taken for collection of samples.
Detection of Cryptosporidium
Microscopic examination of faecal smears was conducted for bacteria and cryptosporidia by employing Leishman's and modified Zeihl–Neelsen's (mZN) stains [31].
Detection of infectious agents by ELISA
All the faecal samples collected from 100 neonatal bovine calves during scour outbreaks were subjected to faecal ELISA by using the protocol of commercial ELISA kits for C. parvum, rotavirus, coronavirus and E. coli (K99) (Bio-X Easy-Digest, Bio K 151; Bio-X Diagnostics, Belgique) and C. perfringens (type C) (BIO K 269; Bio-X Diagnostics, Belgique).
DNA extraction
DNA of target agents C. parvum, E. coli, Salmonella, Clostridium were extracted from faecal specimens using Nucleo-poreR Stool DNA Mini kit (HiMedia, Mumbai, India) as per the manufacturer's instructions.
RNA extraction
RNA from all the faecal samples for detection of coronaviruses was extracted using RNASure® Mini kit (Genetix, India; Table 1).
Table 1.
Primers used for PCR
| Sr. no. | Agent | Primer | Sequence (5′−3′) | PCR conditions | Product size (base pair) | Reference |
|---|---|---|---|---|---|---|
| 1 | Bovine coronavirus (N) | BCoVF | CCGATCAGTCCGACCAATC | Initial denaturation 95 °C (5 min), 40 cycles of denaturation 94 °C (30 s), annealing 55 °C (1 min), elongation 72 °C (1 min) and final elongation at 72 °C (7 min) | 406 | Tsunemitsu et al. (1999) [60] |
| BCoVR | AGAATGTCAGCCGGGGTAT | |||||
| 2 | Escherichia coli K99+ (K99) | K99F | GCGACTACCAATGCTTCTGCGAATAC | 230 | Cho et al. (2010) [25] | |
| K99R | GAACCAGACCAGTCAATACGAGCA | |||||
| 3 | Salmonella (16SrDNA) | 16 SF | TGTTGTGGTTAATAACCGCA | 575 | Lin et al. (1996) [61] | |
| 16SR | CACAAATCCATCTCTGGA | |||||
| 4 | Cryptosporidium (COWP) | BB-3F | GCGAAGATGACCTTTTGATTTG | 194 | Balatbat et al. (1996) [48] | |
| BB-4R | AGGATTTCTTCTTCTGAGGTTCC | |||||
| 5 | Clostridium perfringens | Clostri. F | AAAGATGGCATCATCATTCAAC | Initial denaturation 94 °C (5 min), 35 cycles of denaturation 94 °C (1 min), annealing 53 °C (1 min), elongation 72 °C (1 min) and final elongation at 72 °C (7 min) | 279 | Yoo et al. (1997) [62] |
| Clostri. R | TACCGTCATTATCTTCCCCAAA |
Confirmation of infectious agent by PCR
PCR was employed for Cryptosporidium, coronavirus, Salmonella, E coli and C. perfringens individually. The details of primers used and amplification conditions are given in Table 1. PCR reaction mixture 25 µl was constituted by adding 2·5 µl of 10× PCR buffer (HiMedia, Mumbai, India), 0·75 µl of 50 mM MgCl2, 2μl of template DNA, 1 µl of 20 pmol/μl of forward and reverse primer each, 0·2 µl of Taq DNA polymerase 5 U/μl and rest with nuclease-free water. Amplified PCR products were separated by gel electrophoresis using 1% agarose and visualisation of the product was carried out using UV transilluminator (Alpha Imager, San Jose, California, USA).
First strand cDNA synthesis
For the first strand cDNA synthesis from the RNA sample, the kit (Fermantas, Thermo Fisher Scientific, Waltham, MA, USA) was used, and the reaction was set up as per the manufacturer's instructions. In brief, the following reagents were added into a sterile nuclease-free tube on ice total RNA (0·1–5 µg), random hexamer primer (1 µl), nuclease-free water to make volume 11 µl. Subsequently, 5× reaction buffer (4 µl), Ribolock RNAase inhibitor (20 units/μl) 1 µl, 10 mM dNTP mix 2 µl, M-MuLV Reverse Transcriptase (20 units/μl), 2 µl was added and volume was made upto 20 µl with nuclease-free water. The above contents were mixed gently and centrifuged and incubated for 5 min at 25 °C, followed by 60 min at 37 °C, and finally the reaction was terminated by heating at 70 °C for 5 min. The reverse transcription reaction product was directly used in PCR.
Mortality analysis
During the investigation of 17 outbreaks in question, 43 claves died within 1 weak after sample collection, and the tissues from representative animals were collected for histopathology.
Statistical analysis
The data were statistically analysed by χ2 test using SPSS 16.0 software.
Sensitivity of PCR in relation to ELISA, mZ–N stain and Leishman stain for detection of Cryptosporidium was done following the formulae of Perrin and Sureau [32].
RESULTS AND DISCUSSION
Detection of Cryptosporidium by conventional and immunological and molecular methods
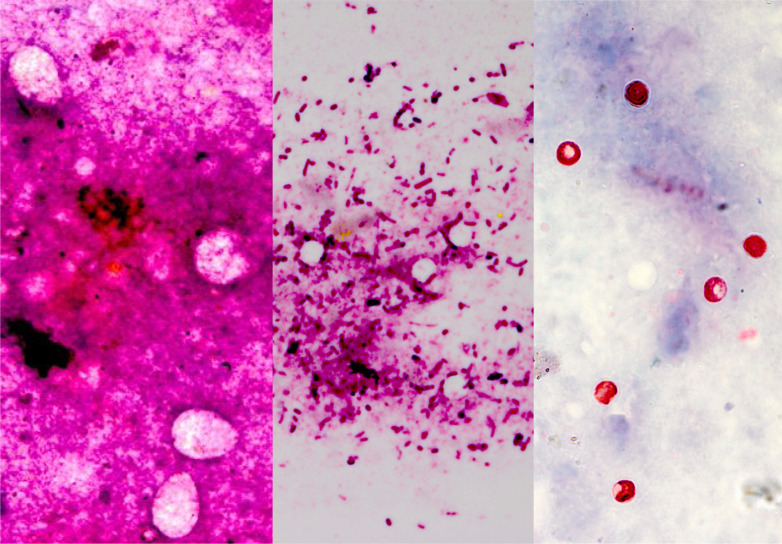
Microscopic examination of the faecal smears stained with Leishman's stain showed cryptosporidial and eimerial oocysts, besides other faecal inclusions, viz. cocci, thick and thin bacilli, stumpy bacilli/coccobacilli, squamous epithelial cells, intestinal epithelial cells, cell debris, undigested plant fibres(s), red blood cells (RBCs), fungus/yeast, bile pigment, leucocytes and undigested (Table 3). The Leishman-stained smears proved highly useful in preliminary screening of faecal smears for the presence or absence of coocidia, particularly cryptosporidia. Morphometric analysis of oocysts of Cryptosporidium (n = 10; 10 field each sample) in Leishman (4·24 ± 0·28 µm) and mZN-stained (4·26 ± 0·29 µm) faecal smears revealed non-significant (0·16 NS) variation in the size. In the Romanowsky-stained faecal smears, the cryptosporidial oocysts were seen as hollow round bodies and with a size compatible with that recorded in this study and documented earlier by mZN staining [33] and the coocidian oocysts were larger hollows and oval (Fig.1a, b); but on mZN staining, cryptosporidial oocysts were seen as bright red, oval to spherical bodies in 25% cases (Fig. 1c; Table 2). Both the faecal antigen-ELISA and PCR showed an improved 35% prevalence of Cryptosporidium in diarrhoeic calves. In fact, direct visualisation by light microscopy of cryptosporidia in faeces or intestinal contents as well as the detection of its antigens (e.g. Ag-ELISA) or nucleic acids (e.g. PCR) in specimens have been widely accepted as alternative diagnostic methods [9, 34].
Table 3.
Microscopic findings in Leishman's-stained faecal smears
| Microscopic findings | Age | Total | Rank | r-value | ||||
|---|---|---|---|---|---|---|---|---|
| 1–7 days (n = 9) | 8–14 days (n = 3) | 15–21 days (n = 30) | 22–30 days (n = 19) | 31–60 days (n = 39) | ||||
| Cocci | 6 (66·67) | 2 (66·67) | 23 (76·67) | 15 (78·95) | 26 (66·67 | 72 (72·00) | 1 | −0·011 |
| Thick bacilli | 2 (22·22) | 2 (66·67) | 17 (56·67) | 14 (73·68) | 28 (71·79) | 63 (63·00) | 2 | 0·598 |
| Thin bacilli | 5 (55·56) | 3 (100·00) | 21 (70·00) | 14 (73·68) | 14 (35·90) | 57 (57·00) | 4 | −0·669 |
| Stumpy bacilli/coccobacilli | – | – | 5 (16·67) | 4 (21·05) | 7 (17·95) | 16 (16·00) | 7 | 0·699 |
| Squamous epithelial cells | – | 1 (33·33) | 4 (13·33) | – | 8 (20·51) | 13 (13·00) | 8 | 0·092 |
| Other cells and cell debris | 4 (44·44) | 3 (100·00) | 17 (56·67) | 13 (68·42) | 25 (64·10) | 62 (62·00) | 3 | −0·061 |
| Undigested plant fibre(s) | 2 (22·22) | 1 (33·33) | 4 (13·33) | 1 (5·26) | 3 (7·69) | 11 (11·00) | 10 | −0·710 |
| RBCs | 2 (22·22) | 3 (100·00) | 13 (43·33) | 6 (31·58) | 14 (35·90) | 38 (38·00) | 5 | −0·303 |
| Fungus/yeast | 1 (11·11) | – | 8 (26·67) | 5 (26·32) | 4 (10·26) | 18 (18·00) | 6 | 0·108 |
| Eimeria spp. | – | 1 (33·33) | 5 (16·67) | – | 1 (2·56) | 7 (7·00) | 12 | −0·431 |
| Cryptosporidium spp. | 1 (11·11) | 1 (33·33) | 1 (33·33) | 5 (26·32) | 2 (5·13) | 10 (10·00) | 11 | −0·349 |
| Thin filamentous granulated bacilli (Actinomycetes) | 2 (22·22) | 1 (33·33) | 8 (26·67) | – | – | 11 (11·00) | 10 | −0·824 |
| Pigment | – | – | 8 (26·67) | – | 4 (10·26) | 12 (12·00) | 9 | 0·157 |
| Leucocytes | – | – | 1 (3·33) | – | 2 (5·13) | 3 (3·00) | 13 | 0·744 |
| Undigested fat | – | – | – | 1 (5·26) | – | 1 (1·00) | 14 | 0·146 |
Fig. 1.
Leishman-stained faecal smears showing large oval bodies resembling Eimeria spp. oocysts (left frame; a), small hollow round bodies resembling cryptosporidial oocysts (middle frame; b) and modified Zeihl–Neelson-stained faecal smear conforming the presence of cryptosporidial oocysts (right frame; c) besides several other faecal inclusions (original magnification × 1000×).
Table 2.
Detection of Cryptosporidium parvum by conventional and modern techniques in different age groups of diarrhoeic bovine calf faeces (n = 100)
| Age group (days) | Number of animals | mZn stain positive (%) | ELISA positive (%) | PCR positive (%) |
|---|---|---|---|---|
| 1–7 | 9 | 3 (33·3) | 3 (33·3) | 3 (33·3) |
| 8–14 | 3 | 1 (33·3) | 2 (66·7) | 2 (66·6) |
| 15–21 | 30 | 9 (30·0) | 10 (30·0) | 10 (30·0) |
| 22–30 | 19 | 5 (26·3) | 7 (36·8) | 7 (36·8) |
| 31–60 | 39 | 7 (17·9) | 13 (30·8) | 13 (30·8) |
| Overall | 100 | 25 (25·0) | 35 (33·0) | 35 (33·0) |
| χ2 | 1·446 |
The age-wise prevalence of Cryptosporidium by PCR and antigen detection ELISA showed non-significant variation with highest rate of 66·6% in 8–14 days group calves (Table 2). The maximal prevalence of Cryptosporidium was recoded in the 2-week-old diarrhoeic bovine calves. Similar prevalence of cryptosporidial infection in neonatal diarrhoeic calves with a gradual reduction with age has been reported previously from various regions of India [5, 21, 22, 35] and across the globe [27, 36–39]. In the present study, all calves positive by faecal ELISA were also positive for the Cryptosporidium by PCR, except one. The sensitivity of ELISA and mZN staining with respect to PCR was 97·14% and 72·72%, respectively (Table 4). Comparable results regarding sensitivity between ELISA and PCR, matching those of ours, were previously recorded by other workers [25, 40] irrespective of the variations in the type of reagents and kits used. Settawy and Fathy [41] observed high detection rate by PCR (24·4%) and low by microscopy (18·6%), while it was 20·9% by ELISA. Their data showing higher sensitivity of PCR compared with ELISA are contradictory to our results showing equivalent positivity. In the differentiation of the aetiology of the diarrhoea in neonatal calves, PCR is being increasingly employed for the detection of Cryptosporidium during the last two decades mainly because of its accuracy, specificity and sensitivity and it has made significant contributions in understanding the epidemiology of Cryptosporidium infection [42–46]. Moreover PCR has added advantages such as ease of use, ability to analyse large numbers of samples simultaneously, relatively low cost and ability to speciate (eliminating false positives and cross-reactions of antibodies to non-pathogenic species) [45]. Though nested PCR involving a second round of amplifications has been applied to Cryptosporidium to increase specificity and sensitivity [35, 47, 48], yet it has not been generally recommended for use in diagnostic laboratories owing to a major risk of contamination with PCR products [45, 49].
Table 4.
Sensitivity of ELISA, mZN and Leishman's staining of faecal smears with PCR as gold standard for determination of Cryptosporidium parvum in diarrhoeic bovine calf faeces (n = 100)
| Tests | PCR | Total | Per cent sensitivity | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| ELISA | Positive | 34 | 1 | 33 | 97·14 |
| Negative | 1 | 64 | 67 | ||
| Total | 35 | 65 | 100 | ||
| mZN stain | Positive | 24 | 1 | 25 | 72·72 |
| Negative | 9 | 66 | 75 | ||
| Total | 33 | 67 | 100 | ||
Detection of Cryptosporidium-associated enteropathogens
The progression of diarrhoea with multifactorial aetiology is rapid. Hence, a quick diagnosis is critical for not only rapidly confirming the causes but also helping clinicians and cattle producers to implement appropriate interventions in a timely manner. Antigen capture ELISAs are well known for rapid turnaround, high-throughput testing and portability [50]; whereas, PCR is especially useful for detecting viruses that are difficult to isolate in cell culture or bacteria that require a long time to grow [51]. Therefore, all the samples tested positive for other enteropathogens either/or by faecal antigen capture ELISA or PCR were considered positive as Cryptosporidium-associated enteropathogens.
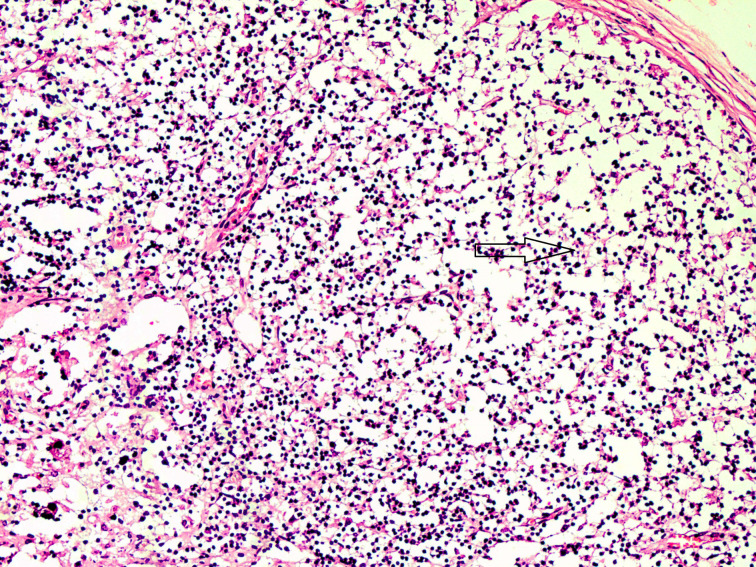
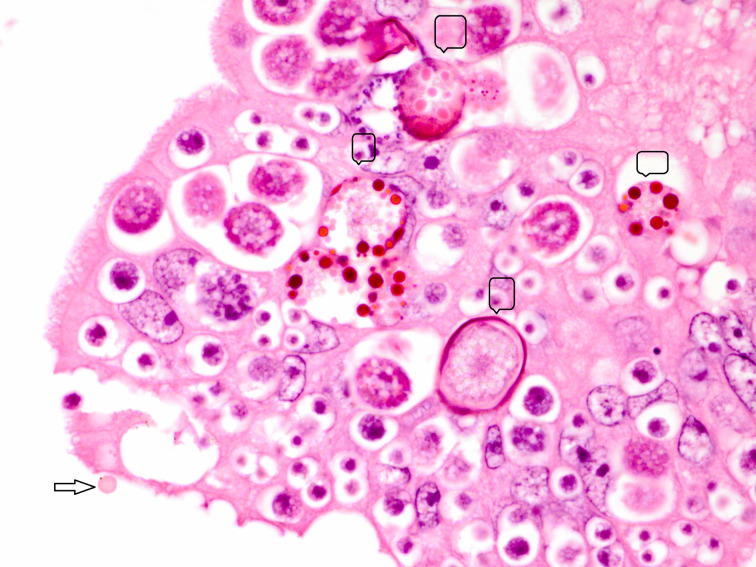
Electrophoretic analysis of PCR products of Salmonella spp., C. perfringens and bovine coronavirus in association with C. parvum are shown in Figure 2. Majority of the cases, in this study, showed as many as 13 combinations of Cryptosporidium-associated infections (71·40%) with Cryptosporidium alone in 28·60% diarrhoeic calves only (Table 5). Moreover, the age-wise faecal prevalence of Cryptosporidium as a single agent and combination aetiologies with it, showed a decreasing and increasing trend (χ2 13·28, P < 0·01), respectively (Table 5). This indicates that initial primary infection with Cryptosporidium led to significant enterocytic damage favouring flare up of other secondary infections with the passage of time. In addition, the findings of severe intestinal epithelial damage (Fig. 3), lymphoid depletion in mesenteric lymph nodes (Fig. 4) and demonstration of Cryptosporidium along with coccidiosis (eimeriosis) (Fig. 5) were amply augmented by histopathology of the representative tissue samples collected from calf mortality during the outbreaks. Histopathology is considered as a gold standard for demonstrating the intestinal involvement and cryptosporidia in one go in the animals dying of diarrhoeal disease [6]. Furthermore, the haematobiochemical findings of significant neutrophilic leucocytosis, lymphopaenia and anaemia along with significant increase in serum albumin and blood urea nitrogen with corresponding significant decrease in total globulins hinted at secondary suppurative infection, dehydration and immunosuppression, respectively, as have been reported by the authors earlier [52, 53]. Contrary to our findings, de la Fuente et al. [27] reported a significant age-associated decrease in the detection rate of mixed infections. In fact, the epidemiological studies have proven that diarrhoea is more severe in mixed infection(s), especially in immunocompromised individuals [54]. In this study, the Cryptosporidium-associated pathogens responsible for bovine calf scour outbreaks and mortality were C. perfringens 45·71% (16/25), followed by Salmonella spp 40·0% (10/25), rotavirus 36·0% (9/25), coronavirus 16·0% (4/25), E. coli 12·0% (3/25) and Eimeria spp. 4·0% (1/4), respectively, which fell nearly in line with that for C. perfringens (54·0%) and in contrast to rotavirus (87%), coronavirus (11·1%), E. coli (27·8%) and Salmonella spp. (1·8%) in central Spain [55]. Almost similar infection rate of Cryptosporidium (21·28%) with Blastocystis 19·15%, Giardia (51·06%) and Enterocytozoon 36·17%, although in a higher 3–5-month age group of diarrhoeic calves was reported [30]. The severity of Cryptosporidium in combination with other agents, or whether other agents extend the risk period for clinical cryptosporidiosis remains to be proved further experimentally [27]. The prevalence of each of pathogen and disease incidence can vary due to geographical location of the farms, farm managemental practices and herd size. A heavy mortality of 43% was recorded in the present study. A similar high mortality has been reported due to diarrhoea in the USA (57%) and Korea (53·4%) in unweaned dairy calves [56–57].
Fig. 2.
Agarose gel electrophoretic analysis showing PCR amplified products of multiple agents of diarrhoea. Lane 1: Salmonella sp(s) positive (575 bp), lane 2: Salmonella sp negative, lane 3: Clostridium perfringens (Cl) positive (279 bp), lane 4: C. perfringens (Cl) negative, lane M: DNA marker (100 bp plus, SRL), lane 5: bovine coronavirus (Co) positive (406 bp), lane 6: bovine coronavirus (Co) negative, lane 7: Cryptosporidium parvum (Cr) positive (194 bp), lane 8: C. parvum (Cr) negative control.
Table 5.
Detection of Cryptosporidium and its combination with other enteropathogens in different age groups of diarrhoeic calves
| Enteropathogens detected | 1–7 days (N = 9) (%) | 8–14 days (N = 3) | 15–21 days (N = 30) | 22–30 days (N = 19) | 31–60 days (N = 39) | Total (N = 100) |
|---|---|---|---|---|---|---|
| Cryptosporidium pavrum | – | 2 (66·67) | 6 (20·00) | 1 (5·26) | 1 (2·56) | 10 (10·00) |
| C. pavrum + rotavirus | – | – | – | – | 2 (5·13) | 2 (2·00) |
| C. pavrum + Clostridium perfringens | 1 (11·11) | – | 1 (3·33) | 2 (10·53) | 1 (2·56) | 5 (5·00) |
| C. pavrum + Salmonella spp. | – | – | 1 (3·33) | 1 (5·26) | 2 (5·13) | 4 (4·00) |
| C. pavrum + Escherichia coli | – | – | – | – | 1 (2·56) | 1 (1·00) |
| C. pavrum + rotavirus + Salmonella spp. | – | – | – | – | – | 1 (1·00) |
| C. pavrum + Eimeria + C. perfringens | – | – | – | 1 (5·26) | – | 1 (1·00) |
| C. pavrum + rotavirus + C. perfringens | 1 (11·11) | – | – | – | 1 (2·56) | 2 (2·00) |
| C. pavrum + coronavirus + C. perfringens | – | – | 1 (3·33) | 1 (5·26) | – | 2 (2·00) |
| C. pavrum + E. coli + Salmonella spp. | – | – | – | – | 1 (2·56) | 1 (1·00) |
| C. pavrum + rotavirus + C. perfringens + Salmonella spp. | – | – | – | – | 3 (7·69) | 3 (7·69) |
| C. pavrum + rotavirus + coronavirus + C. perfringens | 1 (11·11) | – | – | – | – | 1 (1·00) |
| C. pavrum + coronavirus + C. perfringens + Salmonella spp. | – | – | – | 1 (5·26) | – | 1 (1·00) |
| C. pavrum + E. coli + C. perfringens + Salmonella spp. | – | – | – | – | 1 (2·56) | 1 (1·00) |
| Total | 3 (33·33) | 2 (66·67) | 10 (33·33) | 7 (36·8) | 13 (33·3) | 35 (35·00) |
| Per cent concurrent infection | 33·33 | – | 40·00 | 85·71 | 92·30 | 71·40 |
Figures in parentheses indicate percentage.
All the agents shown in the table were detected by both ELISA and PCR except Salmonella spp., for which only PCR was performed and for rotavirus only ELISA was performed.
Fig. 3.
Severe intestinal damage due to multiple aetiological agents characterised by massive superficial and deep necrosis extending into crypts with formation of marked debris in the lumen. H&E × 4× original magnification.
Fig. 4.
Section of a mesenteric lymph node of the small intestine segment affected with severe diarrhoea showing massive lymphoid cell depletion. H&E × 10× original magnification.
Fig. 5.
Section of small intestine showing various developmental stages of coccidia and an oocyst (arrow) of Cryptosporidium present superficially. H&E × 100× original magnification.
Among the various risk factors analysed during the study, poor hygiene, overcrowding, bad calf nutrition, including deprivation of colostrum and inadequate milk feeding, and sudden weather changes were found to be the major epidemiological factors. Poor managemental practices, including the method of cleaning, the type of flooring and the frequency of cleaning and deprivation of colostrum in suckling calves were also found to be the main risk factors associated with cryptosporium infections [58, 59].
From this first comprehensive report of 17 cryptosporidial outbreaks from Punjab State of North India, diagnosed based on a combination of techniques, we conclude that when C. parvun occurs in association with other concurrent enteropathogens, it may result into severe infection leading to outbreaks of neonatal bovine calf diarrhoea, resulting in huge mortality perpetuated by poor managemental practices and immunosuppression as augmented by histopathology of lymph nodes. In addition to C. parvum, the other aetiological agents, viz. C. perfringens, Salmonella spp and E. coli detected in the present study also carry marked zoonotic potential in children and immunocompromised people.
ACKNOWLEDGEMENTS
Thanks are due to the Director of Research, Guru Angad Dev Veterinary and Animal Sciences University for providing financial support to carry out the research work under RKVY project entitled, ‘Rapid and precise diagnostics of important gastrointestinal and respiratory diseases of bovines’.
REFERENCES
- 1.Otter A, Cranwell M. Differential diagnosis of diarrhoea in adult cattle. In Practice 2007; 29: 9–19. [Google Scholar]
- 2.Bangoura B, Daugschies A. Influence of experimental Eimeria zuernii infection in calves on electrolyte concentrations, acid–base balance and blood gases. Parasitology Research 2007; 101: 1637–1645. [DOI] [PubMed] [Google Scholar]
- 3.Gay CC. Intestinal diseases in ruminants. In: Kahn CM, ed. Merck Veterinary Manual. U.S.A, Merck and Co. inc, N.J.: Whitehouse Station, 2005, pp. 220–233. [Google Scholar]
- 4.Haschek B, et al. Detection of bovine torovirus in neonatal calf diarrhoea in Lower Austria en Styria (Austria). Journal of Veterinary Medicine. B, Infectious Diseases and Veterinary Public Health 2006; 53: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh BB, et al. Prevalence of Cryptosporidium parvum infection in Punjab (India) and its association with diarrhoea in neonatal dairy calves. Veterinary Parasitology 2006; 140: 162–165. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y, Yoon KJ. An overview of calf diarrhoea – infectious etiology, diagnosis, and intervention. Journal of Veterinary Science 2014; 15(1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay UKD, et al. Prevalence of cryptosporidiosis in man and animals in and around Calcutta. Journal of Veterinary Parasitology 2000; 14: 167–168. [Google Scholar]
- 8.Kumar D, Sreekrishnan R, Das SS. Cryptosporidiosis in man and animals in Pondicherry. Indian Journal of Animal Sciences 2004; 74: 261–263. [Google Scholar]
- 9.Bhat SA, Juyal PD, Singla LD. Bovine cryptosporidiosis: brief review of its distribution in India. Trends in Parasitology Research 2013; 2(2): 5–13. [Google Scholar]
- 10.Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clinical Microbiology Reviews 2004; 17: 72–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randhawa SS, Randhawa SS, Zahid UN, Singla LD, Juyal PD. Drug combination therapy in control of cryptosporidiosis in Ludhiana district of Punjab. Journal of Parasitic Diseases 2012; 36: 269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacKenzie WR, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. The New England Journal of Medicine 1994; 331: 161–167. [DOI] [PubMed] [Google Scholar]
- 13.Athanassious R, et al. Detection of bovine coronavirus and type A rotavirus in neonatal calf diarrhoea and winter dysentery of cattle in Quebec: evaluation of three diagnostic methods. Canadian Veterinary Journal 1994; 35: 163–169. [PMC free article] [PubMed] [Google Scholar]
- 14.Ruest N, Faubert GM, Couture Y. Prevalence and geographical distribution of Giardia spp. and Cryptosporidium spp. in dairy farms in Quebec. Canadian Veterinary Journal 1998; 39: 697–700. [PMC free article] [PubMed] [Google Scholar]
- 15.Nasir A, et al. Prevalence of Cryptosporidium parvum infection in Lahore (Pakistan) and its association with diarrhoea in dairy calves. International Journal of Agriculture & Biology 2009; 11(2): 221–224. [Google Scholar]
- 16.Nooruddin M, Sarma DK. Role of Cryptosporidium in calf diarrhoea: in annual report. Livestock Advisor 1987; 12: 49. [Google Scholar]
- 17.Dubey JP, Fayer R, Rao JR. Cryptosporidial oocysts in faeces of water buffalo and zebu calves in India. Journal of Veterinary Parasitology 1992; 6: 55–56. [Google Scholar]
- 18.Das G, et al. Prevalence of Cryptosporidium infection in cattle. Journal of Veterinary Public Health 2005; 2: 15–17. [Google Scholar]
- 19.Jeyabal L, Ray DD. 2005. Cryptosporidial infection in cattle and buffaloes. Journal of Veterinary Parasitology 2005; 19: 165–166. [Google Scholar]
- 20.Randhawa SS, et al. Therapeutic management of cryptosporidiosis in cross bred dairy calves. Indian Veterinary Journal 2012; 89: 17–19. [Google Scholar]
- 21.Roy SS, et al. Observations on the epidemiology of bovine cryptosporidiosis in India. Veterinary Parasitology 2006; 141: 330–333. [DOI] [PubMed] [Google Scholar]
- 22.Paul S, et al. Comparative evaluation and economic assessment of coprological diagnostic methods and PCR for detection of Cryptosporidium spp. in bovines. Veterinary Parasitology 2009; 164: 291–295. [DOI] [PubMed] [Google Scholar]
- 23.Venu R, et al. Factors influencing on prevalence of Cryptosporidium infection in south Indian dairy calves. Journal of Parasitic Diseases 2013; 37(2): 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat SA, et al. Comparison of nested PCR and microscopy for the detection of cryptosporidiosis in bovine calves. Journal of Parasitic Diseases 2014; 38: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho YI, et al. Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhoea in feces. Journal of Veterinary Diagnostic Investigation 2010; 22(4): 509–517. [DOI] [PubMed] [Google Scholar]
- 26.Xiao L, Herd RP, Rings DM. Concurrent infections of Giardia and Cryptosporidium on two Ohio farms with calf diarrhoea. Veterinary Parasitology 1993; 51(1–2): 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Fuente R, et al. Cryptosporidium and concurrent infections with other major enteropathogens in 1–30-day-old diarrhoeic dairy calves in central Spain. Veterinary Parasitology 1999; 80: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García A, et al. Rotavirus and concurrent infections with other enteropathogens in neonatal diarrhoeic dairy calves in Spain. Comparative Immunology, Microbiology and Infectious Diseases 2000; 23(3): 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becher KA, et al. Molecular epidemiology of Giardia and Cryptosporidium infections in dairy calves originating from three sources in Western Australia. Veterinary Parasitology 2004; 123: 1–9. [DOI] [PubMed] [Google Scholar]
- 30.Fayer R, Santin M, Dumitru M. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitology Research 2012; 111: 1349–1355. [DOI] [PubMed] [Google Scholar]
- 31.O.I.E. Cryptosporidiosis. In: Linnane S, Pearson JE, eds. Manual of Standards for Diagnostic Tests and Vaccines for Terrestrial Animals, 6th edn. OIE Biological Standards Commission; adopted by the International Committee of the OIE. Paris, France: OFFICE INTERNATIONAL DES EPIZOOTIES, 2008, pp. 1197–1200. [Google Scholar]
- 32.Perrin P, Sureau P. A collaborative study of an experimental kit for rapid rabies enzyme immunodiagnosis. Bulletin World Health Organization 1987; 65: 489–493. [PMC free article] [PubMed] [Google Scholar]
- 33.Brar APS, et al. Validation of Romanowsky staining as a novel screening test for the detection of faecal cryptosporidial oocysts. Journal of Parasitic Diseases 2017; 41(1): 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singla LD, et al. Antigen based diagnosis of Cryptosporidium parvum infection in cattle and buffalo faeces. Indian Journal of Animal Science 2013; 83(1): 37–39. [Google Scholar]
- 35.Bhat SA, Juyal PD, Singla LD. Prevalence of cryptosporidiosis in neonatal buffalo calves in Ludhiana district of Punjab, India. Asian Journal of Animal and Veterinary Advances 2012; 7: 512–520. [Google Scholar]
- 36.Castro-Hermida JA, González-Losada YA, Ares-Mazás E. Prevalence of and risk factors involved in the spread of neonatal bovine cryptosporidiosis in Galicia (NW Spain). Veterinary Parasitology 2002; 106(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trotz-Williams LA, et al. Prevalence of Cryptosporidium parvum infection in southwestern Ontario and its association with diarrhoea in neonatal dairy calves. Canadian Veterinary Journal 2005; 46: 349–351. [PMC free article] [PubMed] [Google Scholar]
- 38.Kvac M, Kouba M, Vitovec J. Age-related and housing dependence of Cryptosporidium infection of calves from dairy and beef herds in South Bohemia, Czech Republic. Veterinary Parasitology 2006; 137: 202–209. [DOI] [PubMed] [Google Scholar]
- 39.Fayer R, Santin M, Dargatz D. Species of Cryptosporidium detected in weaned cattle on cow-calf operations in the United States. Veterinary Parasitology 2010; 170: 187–192. [DOI] [PubMed] [Google Scholar]
- 40.Van den BD, et al. Comparison of four rapid diagnostic tests, ELISA, microscopy and PCR for the detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in feces. Journal of Microbiological Methods 2015; 110: 78–84. [DOI] [PubMed] [Google Scholar]
- 41.Settawy MA EL, Fathy GM. Evaluation and comparison of PCR, coproantigen ELISA and microscopy for diagnosis of Cryptosporidium. Journal of American Science 2012; 8(12): 1378–1385. [Google Scholar]
- 42.Xiao L, Ryan UM. Molecular epidemiology. In: Fayer R, Xiao L, eds. Cryptosporidium and Cryptosporidiosis. Boca Raton: CRC Press IWA Publishing, 2008. [Google Scholar]
- 43.Webster KA, et al. Detection of Cryptosporidium parvum oocysts in faeces: comparison of conventional coproscopical methods and the polymerase chain reaction. Veterinary Parasitology 1996; 61: 5–13. [DOI] [PubMed] [Google Scholar]
- 44.Morgan UM, et al. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. Journal of Parasitololgy 1997; 83: 825–830. [PubMed] [Google Scholar]
- 45.Morgan UM, Thompson RCA. PCR detection of Cryptosporidium: the way forward? Parasitology Today 1998; 14(6): 241–245. [DOI] [PubMed] [Google Scholar]
- 46.Morgan UM, et al. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. Journal of Clinical Microbiology 1998; 36(4): 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer CL, Palmer CJ. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in waste water. Applied Environmental Microbiology 1996; 62: 2081–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balatbat AB, et al. Detection of Cryptosporidium parvum DNA in human faeces by nested PCR. Journal of Clinical Microbiology 1996; 34: 1769–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persing DH. Diagnostic Molecular Microbiology: Principles and Applications. Washington, DC: Published by American Society for Microbiology, 1993, pp. 51–87. [Google Scholar]
- 50.Fenner F, MacLachlan NJ, Dubovi EJ, eds. Fenner's Veterinary Virology, 4th edn. Burlington: Academic Press, 2011, pp. 288–290. [Google Scholar]
- 51.Espy MJ, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clinical Microbiology Reviews 2006; 19: 165–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brar APS, Sood NK, Ahuja CS, Sandhu BS, Gupta K and Singh CK. Serum biochemical changes in neonatal diarrhoeic claves of different age groups. Indian Journal of Veterinary Pathology 2014; 38: 14–17. [Google Scholar]
- 53.Brar APS, Ahuja CS, Sood NK, Sandhu BS and Gupta K. Haematological changes in neonatal diarrhoeic claves of different age groups. Indian Journal of Veterinary Pathology 2015; 39: 73–77. [Google Scholar]
- 54.Morin M, et al. Diarrhoea of newborn calves. II. Agents responsible for the disease on Quebec dairy farms. Med. VeÂt. Quebec 1980; 10: 60–65. [Google Scholar]
- 55.Bartels CJM, et al. Prevalence, prediction and risk factors of enteropathogens in normal and non-normal faeces of young Dutch dairy calves. Preventive Veterinary Medicine 2010; 93: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.USDA. Dairy 2007 Part II: Changes in the U.S. Dairy Cattle industry, 1991–2007. Fort Collins: USDA-APHIS-VS, CEAH, 2008, pp. 57–61. [Google Scholar]
- 57.Hur TY, et al. The dairy calf mortality: the causes of calf death during ten years at a large dairy farm in Korea. Korean Journal of Veterinary Research 2013; 53: 103–108. [Google Scholar]
- 58.Mohammed HO, Wade SE, Schaaf S. Risk factors associated with Cryptosporidium parvum infection in dairy cattle in southeastern New York State. Veterinary Parasitology 1999; 83: 1–13. [DOI] [PubMed] [Google Scholar]
- 59.José A, et al. Prevalence of and risk factors involved in the spread of neonatal bovine cryptosporidiosis in Galicia (NW Spain). Veterinary Parasitology 2002; 106: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsunemitsu H, Smith DR, Saif LJ. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Archives of Virology 1999; 144(1): 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin CK, Tsen HY. Use of two 16S DNA targeted oligonucleotides as PCR primers for the specific detection of Salmonella in foods. Journal of Applied Bacteriology 1996; 80: 659–666. [DOI] [PubMed] [Google Scholar]
- 62.Yoo HS, et al. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. Journal of Clinical Microbiology 1997; 35: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]