Abstract
PURPOSE
Patients with cancer are predisposed to developing chronic, comorbid conditions that affect prognosis, quality of life, and mortality. While treatment guidelines and care variations for these comorbidities have been described for the general noncancer population, less is known about real-world treatment patterns in patients with cancer. We sought to characterize the prevalence and distribution of initial treatment patterns across a large-scale data network for depression, hypertension, and type II diabetes mellitus (T2DM) among patients with cancer.
METHODS
We used the Observational Health Data Sciences and Informatics network, an international collaborative implementing the Observational Medical Outcomes Partnership Common Data Model to standardize more than 2 billion patient records. For this study, we used 8 databases across 3 countries—the United States, France, and Germany—with 295,529,655 patient records. We identified patients with cancer using SNOMED (Systematized Nomenclature of Medicine) codes validated via manual review. We then characterized the treatment patterns of these patients initiating treatment of depression, hypertension, or T2DM with persistent treatment and at least 365 days of observation.
RESULTS
Across databases, wide variations exist in treatment patterns for depression (n = 1,145,510), hypertension (n = 3,178,944), and T2DM (n = 886,766). When limited to 6-node (6-drug) sequences, we identified 61,052 unique sequences for depression, 346,067 sequences for hypertension, and 40,629 sequences for T2DM. These variations persisted across sites, databases, countries, and conditions, with the exception of metformin (73.8%) being the most common initial T2DM treatment. The most common initial medications were sertraline (17.5%) and escitalopram (17.5%) for depression and hydrochlorothiazide (20.5%) and lisinopril (19.6%) for hypertension.
CONCLUSION
We identified wide variations in the treatment of common comorbidities in patients with cancer, similar to the general population, and demonstrate the feasibility of conducting research on patients with cancer across a large-scale observational data network using a common data model.
INTRODUCTION
Patients with cancer represent a uniquely vulnerable population predisposed to developing a number of comorbid conditions that significantly affect short- and long-term outcomes.1-5 As cancer survivorship continues to increase among an aging population, these comorbidities are often chronic and require long-term treatment that involves multiple therapies, similar to prevalent conditions in the general population, such as hypertension, diabetes, and depression.6-9 Previous studies have associated these comorbidities with decreased survival, quality of life, immune function, or even prognosis and treatment response.8,10-15 In addition, patients with cancer may be particularly vulnerable to developing these three diseases because of the effects of the drugs or treatments they are receiving, or as a sequelae of the natural course of the malignancies themselves.14-18
Numerous treatment guidelines exist for chronic conditions, but it is unclear to what extent these guidelines are followed for patients with cancer in routine, real-world oncology practice.19-21 Despite the known associations of comorbidities, such as depression, hypertension, and diabetes, with cancer outcomes and mortality, the extent of variation in real-world treatment patterns for patients with cancer or the impact of this variation on patient outcomes remains largely unknown.10,13,22 Characterization and a better understanding of practice patterns for chronic, comorbid conditions in cancer represent the first step toward improving care, together with associated outcomes and mortality.
CONTEXT
Key objective
To characterize real-world treatment patterns for the chronic, comorbid conditions of depression, hypertension, and type 2 diabetes mellitus in patients with cancer using the Observational Health Data Sciences and Informatics distributed data network.
Knowledge generated
We found wide variations for the treatment of common comorbidities in patients with cancer across 8 observational databases internationally. We also demonstrate the feasibility of identifying patients with cancer and conducting observational research across a large-scale data network using the Observational Medical Outcomes Partnership common data model.
Relevance
As cancer survivorship continues to increase, optimal management and additional investigation of chronic comorbidities in the population of patients with cancer is becoming increasingly important. Characterizing treatment patterns represents the first step to understanding these variations and their association with outcomes, and improving future management, which investigators can help realize by utilizing the potential of large-scale observational research.
Use of observational health data, particularly through the models of data enclaves or distributed data networks that have been recently endorsed by numerous research and clinical organizations, holds great promise for generating real-world evidence, including this characterization of treatment patterns for chronic comorbidities.23,24 The Observational Health Data Sciences and Informatics (OHDSI) network, which uses the common data model (CDM) developed as part of the Observational Medical Outcomes Partnership (OMOP) to represent data from diverse sources in a standardized format, is one such model with the potential to conduct large-scale network studies on cancer care.25-27 Previous studies have demonstrated the ability of OHDSI to execute a large-scale network study on treatment patterns for chronic diseases across 11 databases in four countries.28 To our knowledge, no previous study has characterized treatment patterns for these comorbidities and chronic diseases in patients with cancer, nor has such a large-scale observational network study been carried out in any cohort of patients with cancer.
Therefore, among patients with cancer, we sought to characterize the prevalence, variation, and distribution of different initial treatment patterns for 3 chronic diseases: hypertension, type 2 diabetes mellitus (T2DM), and depression.
METHODS
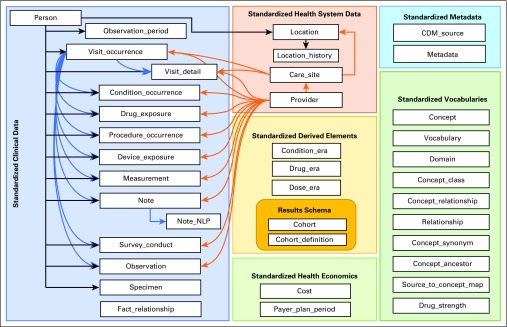
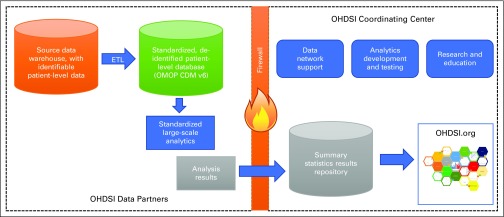
For this study, we used the OHDSI network, a multistakeholder, interdisciplinary, international collaborative implementing the OMOP CDM to standardize observational data from diverse sources.25-27 OMOP CDM is a deep information model (Fig 1) that specifies how to encode and store clinical data at a fine-grained level, including explicitly specified encoding and relationships among concepts using standardized vocabularies.29 Rather than merging databases, source data are locally transformed into the OMOP CDM from which analyses are carried out locally as part of a distributed network. Results in the form of summary statistics are then transmitted in aggregate to the community and the coordinating center (Fig 2). OMOP data represent observational health data, including diagnoses, drug exposures, devices, procedures, visits, measurements, and more, from such sources as electronic health records (EHRs) and claims databases.
FIG 1.
Key domains and tables within the Observational Medical Outcomes Partnership common data model (CDM), version 6. Shaded boxes represent different domains, arrows show linkages between tables within and across domains. Used with permission from Observational Health Data Sciences and Informatics. NLP, natural language processing.
FIG 2.
Observational Health Data Sciences and Informatics (OHDSI) structure and analysis flow diagram. Behind each institution’s firewall, source data are converted to standard Observational Medical Outcomes Partnership (OMOP) format through an extract-transform-load (ETL) process from which large-scale analyses can be executed. Aggregate results and summary statistics are shared across the firewall with the coordinating center, which serves multiple organizational roles including the completion of these analyses Used with permission from OHDSI. CDM, common data model.
Data Sources
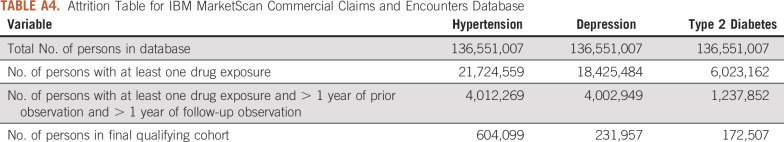
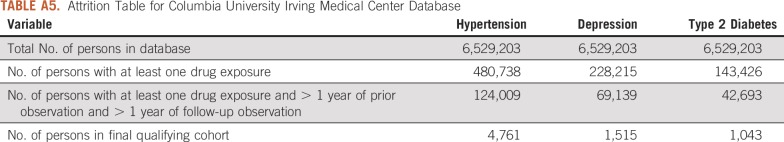
We performed these analyses across a network of observational health care databases, which were standardized into the OMOP Common Data Model, version 5. The complete specification for the OMOP Common Data Model is available on Github.30 A total of 8 databases were included in this analysis, as follows: (1) Columbia University Irving Medical Center (CUIMC), an EHR database from the CUIMC campus of New York-Presbyterian Hospital and its affiliated physician practice; (2) IBM MarketScan Commercial Claims and Encounters, a US employer–based administrative health claims database for active employees, early retirees, COBRA continues, and their dependents insured by employer-sponsored plans (individuals in plans or product lines with fee-for-service plans and fully capitated or partially capitated plans); (3) IBM MarketScan Medicare Supplemental Beneficiaries, an administrative health claims database for Medicare-eligible active and retired employees and their Medicare-eligible dependents from employer-sponsored supplemental plans (predominantly fee-for-service plans); (4) IBM MarketScan Multistate Medicaid, an administrative health claims database for the pooled health care experience of Medicaid enrollees from multiple states; (5) Optum De-Identified Clinformatics Data Mart Database (Optum, Eden Prairie, MN), an administrative health claims database for members of United Healthcare, who enrolled in commercial plans (including Administrative Services Only, 36.31 million), Medicaid (before July 2010, 1.25 million) and Legacy Medicare Choice (before January 2006, 0.36 million) with both medical and prescription drug coverage; (6) IQVIA France, an EHR database from physician practices in France; (7) IQVIA Germany Disease Analyzer, an EHR database from physician practices in Germany; and (8) Stanford University, an EHR database based on clinical data from Stanford University Hospitals.
Study Population
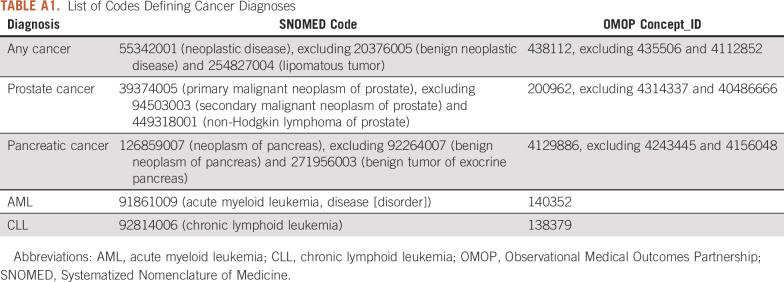
We first created a cohort of patients with cancer defined by cohort entry at the first diagnosis of cancer and cohort exit at the end of observation. Conditions in the OMOP CDM use SNOMED (Systematized Nomenclature of Medicine) as the standard vocabulary for diagnosis codes. We defined our cohort of patients with cancer using a SNOMED diagnosis code for neoplastic disease, excluding benign neoplastic disease or lipomatous tumor. The list of codes used to define cancer are available in Appendix Table A1. We validated this phenotype for cancer using manual chart review of 100 randomly selected patients within the CUIMC database to ensure accurate capture of patients with cancer. Validation showed a positive predictive value of 95.9%, sensitivity of 99%, and specificity of 99.9% for accurately identifying patients with any cancer.
Within that cohort of patients with cancer, we created 3 subcohorts for T2DM, hypertension, and depression. For each of these chronic conditions, entry into the subcohort was determined by the date of first treatment (as defined below) for the chronic disease, with at least 365 days of prior observation before first treatment and at least 365 days of follow-up time post-treatment. We required a diagnosis of cancer and of the chronic disease on or before the first treatment, as well as persistent treatment after initiation of the first treatment, defined as at least one exposure to treatment of the chronic disease during the 121-day to 240-day and the 241-day to 365-day periods postindex. Codes used to define the chronic diseases are included in Appendix Table A2.
Treatment Pattern Analysis
For each patient in the qualifying subcohorts, we identified the sequence of treatments to which they were exposed during the 12 months after first exposure. Treatments were analyzed at the RxNorm—the standard vocabulary for medications—ingredient level, which represents the primary active ingredient in the drug. The sequence was determined by ordering the dates of first exposure to each qualifying ingredient for the disease. An ingredient was determined to be a qualifying ingredient if it met the treatment category for the disease of interest—that is, antihypertensive medications for the disease of hypertension. Codes used to define these treatment categories are included in Appendix Table A3. All patients in the subcohorts were exposed to at least one treatment. If a patient was maintained on the same treatment of the entire 12-month period, meaning all drug exposures were for the same ingredient and no other ingredient for the same disease was observed during that interval, then the person was classified as having a 1-drug treatment sequence. A patient who switched treatments only once during his or her interval would have received a 2-drug treatment sequence. We summarized all sequence combinations that are fewer than 20 drugs. After each patient’s treatment sequence was constructed, we counted the number of unique persons with the same sequence within each data source for each disease. We also stratified the counts by index year of first exposure. Only summary statistics, no patient-level data, were provided by distributed data partners for this analysis. The analysis code also provided each data partner the option to suppress any summary statistics below a minimum cell count number so that all counts < 5 were removed from each data source. We stratified the results by data source to determine if treatment patterns varied by population, region, or data capture process.
To carry out this network study, we developed and posted our study protocol and code on the OHDSI Github in the public domain.31 Summary results were then used to create tabular and graphical summaries of the evidence across the OHDSI data network to characterize the prevalence of treatment sequences by source and year. As a result of the large number of sequences, sequences were truncated at 6 and 3 nodes for the purposes of visualization within a graphical summary. This study was approved by the Columbia University Institutional Review Board under proposal IRB-AAAO7805 and IRB-AAAR9451.
RESULTS
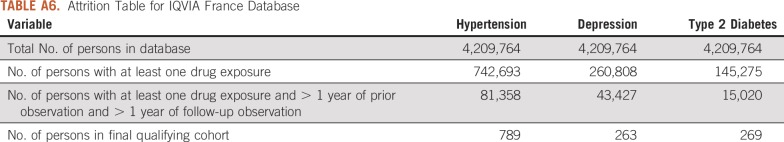
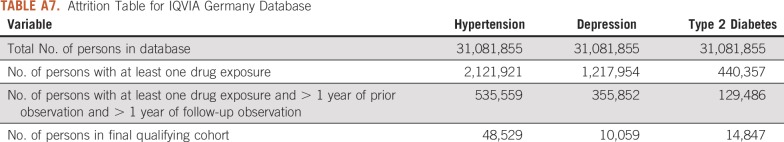
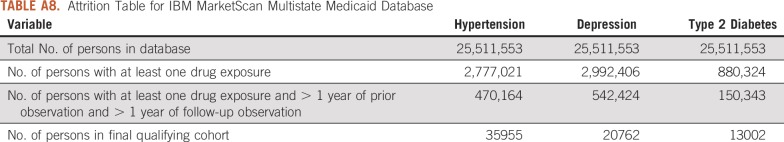
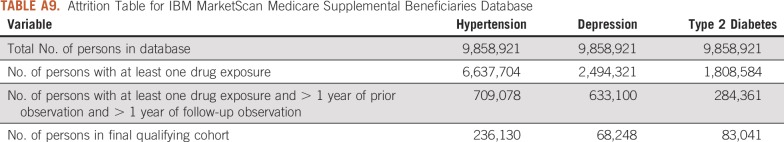
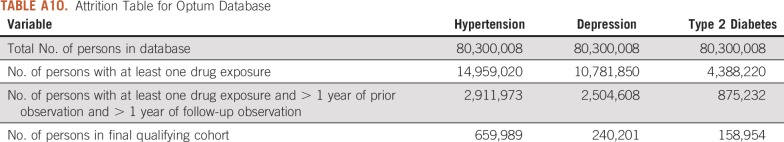
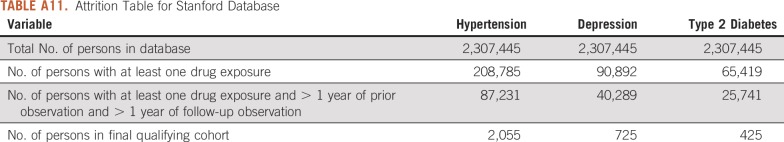
For this study, we used 8 databases, as listed above, across 3 countries that encompassed 295,529,655 patient records. In aggregate, across all databases, there were 1,145,510 patients with cancer initiating treatment for depression, 886,766 patients with cancer initiating treatment for T2DM, and 3,178,944 patients with cancer initiating treatment for hypertension. Across all databases, patient follow-up time ranged from 1 year to 19 years, with the longest treatment sequence being 20 drugs. Attrition tables and counts for each disease and database are available in Appendix Tables A4 to A11.
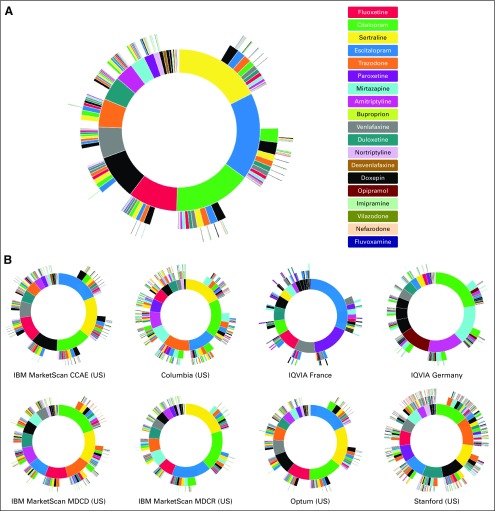
When truncated at 6-node sequences (6 drugs or less), we found 61,052 unique treatment sequences for the treatment of depression in patients with cancer (Fig 3A). Across all databases, the most commonly used initial medications were sertraline (17.5%) and escitalopram (17.5%). Overall, selective serotonin reuptake inhibitors (SSRIs) were by far the most common initial medication, as citalopram (15.5%) and fluoxetine (9.9%) were the third and fourth most frequently prescribed initial medications, respectively. Across databases, SSRIs remained the most common first-line medication, but the specific SSRI used first varied for each database (Fig 3B). Europe seemed to be different in their second most common initial medications, which was paroxetine in France (16.7%) and mirtazapine in Germany (16%), both of which are less commonly used in the United States. Of note, the Stanford database showed greater prescribing of trazadone as the second most common initial medication. Trazadone use seemed to be higher in the EHR databases of Columbia (12.4%) and Stanford (12.7%) compared with other databases.
FIG 3.
Treatment sequences for depression in patients with cancer. (A) Depression results aggregated across all databases, the inner-most circle represents first-line therapy, with each successive surrounding circle representing the next treatments in the sequence. (B) Separate depression results for each database. CCAE, Commercial Claims and Encounters; MDCD, Multistate Medicaid; MDCR, Medicare Supplemental Beneficiaries; US, United States.
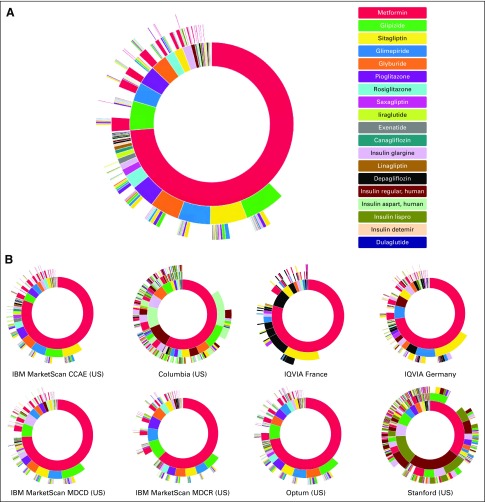
For the treatment of T2DM, when limited to 6-node sequences, we found 40,629 unique treatment sequences in patients with cancer (Fig 4A). In aggregate, the most commonly used initial medication was metformin (73.8%), followed by glipizide (5.83%) and glimepiride (3.78%). Oral medications made up the majority of initial medication choices. Across databases, metformin consistently remained the most common initial medication choice for T2DM (Fig 4B). However, the EHR databases of Columbia and Stanford again stood out for their greater use of regular human insulin (10.7% and 24.9%, respectively), which represented the second most common initial medication at both institutions.
FIG 4.
Treatment sequences for type 2 diabetes mellitus in patients with cancer. (A) Type 2 diabetes mellitus results aggregated across all databases, the inner-most circle represents first-line therapy, with each successive surrounding circle representing the next treatments in the sequence. (B) Separate type 2 diabetes mellitus results for each database. CCAE, Commercial Claims and Encounters; MDCD, Multistate Medicaid; MDCR, Medicare Supplemental Beneficiaries; US, United States.
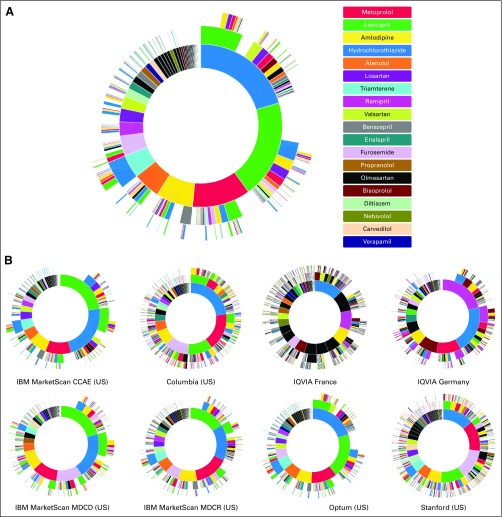
Finally, for the treatment of hypertension, when limited to 6-node sequences, we found 346,067 unique treatment sequences in patients with cancer (Fig 5A). In aggregate, the most commonly used initial drugs for hypertension were hydrochlorothiazide (HCTZ; 20.5%) and lisinopril (19.6%). There was wide variation in the choice of initial agent for hypertension treatment. The five most commonly prescribed initial treatments were thiazide diuretics (HCTZ), angiotensin-converting enzyme–inhibitors (lisinopril), β-blockers (metoprolol, 11.5%; and atenolol, 5.4%), and calcium channel blockers (amlodipine, 7.4%). This variation in the choice of initial agent and number of potential sequences for the treatment of hypertension was also present in individual databases (Fig 5B). One notable difference in Europe was the increased use of ramipril among the top 3 most prescribed initial medications in the IQVIA France (8.6%) and Germany Disease Analyzer (21.0%) databases. Stanford and Columbia showed increased use of furosemide (13.1% and 10.8%, respectively) as an antihypertensive medication compared with other databases.
FIG 5.
Treatment sequences for hypertension in patients with cancer. (A) Hypertension results aggregated across all databases, the inner-most circle represents first-line therapy, with each successive surrounding representing the next treatments in the sequence. (B) Separate hypertension results for each database. CCAE, Commercial Claims and Encounters; MDCD, Multistate Medicaid; MDCR, Medicare Supplemental Beneficiaries; US, United States.
DISCUSSION
This large-scale international network study demonstrates for the first time the wide variation in treatment patterns for the chronic, comorbid conditions of depression, T2DM, and hypertension among millions of patients with cancer. We found more than 61,000 unique treatment sequences for patients with cancer with depression, more than 40,000 unique treatment sequences for patients with cancer with T2DM, and more than 346,000 unique treatment sequences for patients with cancer with hypertension. These extensive variations are similar to practice patterns previously found in the general population.28
Previous research on treatment patterns for these chronic comorbidities in the general population similarly showed significant heterogeneity across and within sources, with 10% of patients with T2DM and depression and 25% of those with hypertension following a unique sequence.28 In addition, both the cancer and general populations shared the most common initial therapies, with SSRIs representing the top four treatments in depression; metformin representing the overwhelming majority for T2DM; and the same top five treatments of HCTZ, lisinopril, amlodipine, atenolol, and metoprolol for hypertension.28 This suggests that therapy considerations, practice patterns, and clinical decision making may not be altered significantly by the concomitant presence of cancer.
With extensive evidence describing the association between comorbidities and cancer outcomes, including mortality, quality of life, prognosis, complications, treatment adherence, and diagnosis delay, this study represents a prerequisite first step toward optimizing the treatment of these conditions.1-5 Studies have previously established the association between T2DM and an increased risk of both cancer mortality and all-cause mortality.9,32-35 Hypertension has also been associated with increased mortality for patients with cancer and portends a better prognosis when observed in response to certain therapies, such as bevacizumab or sunitinib.13-15,17,18 Finally, depression is not only associated with decreased survival time but may also significantly impair quality of life and immune function.8,10-12 It is important to understand which treatment sequences lead to better control of the disease, which, in turn, may affect the outcomes of patients with cancer.
However, despite known disease associations with outcomes, it is unclear whether these variations in treatment patterns or their similarity with the general population represent optimal care. Patients with cancer have different risk profiles, and certain adverse effects of medications may have differential impacts on the outcomes of patients with cancer. For example, metformin has been shown to inhibit cell proliferation and slow cancer growth in laboratory studies, potentially reinforcing its position as first-line therapy for T2DM in patients with cancer, whereas the effects of newer antihyperglycemic agents on cancer risk and progression have yet to be uncovered.36 Although we found lisinopril to be the second most common agent for the treatment of hypertension, it may warrant special consideration or guidelines for more limited use in patients with cancer compared with its first-line status among the general population. This is because of potential concerns for renal toxicity or hyperkalemia, which may be exacerbated by nephrotoxic agents used in chemotherapy or potential preclusion of eligibility for certain cancer treatments if serum creatinine is too elevated, a known adverse effect of this medication. Existing guidelines for the treatment of chronic conditions are largely developed for the general population, and in the limited guidelines that exist for cancer, such as one for depression and anxiety in patients with cancer, they emphasize screening, assessment, and obtaining treatment while making no recommendations about specific antidepressant pharmacologic regimens.10 There have been calls for additional studies to guide therapy for these chronic diseases in patients with cancer while recognizing that this evidence is unlikely to be generated by randomized controlled trials because of cost and time limitations, presenting a key evidence gap that observational research, particularly large-scale studies through data networks, such as OHDSI, can help fill.36
Although our characterization of treatment patterns only represents the first step toward this evidence generation, this study also demonstrates the important feasibility of performing such large-scale observational research on patients with cancer across an international data network using a common data model. To the best of our knowledge, this represents the first international collaborative network study on patients with cancer of this magnitude, identifying millions of patients across different databases with a diagnosis of cancer who were also treated for chronic diseases. The ease of collaboration and interoperability across these disparate databases is made possible by the use of the OMOP CDM for standardization and the extensive suite of open-source tools built by the OHDSI community to decrease barriers to open, collaborative research. This study highlights the potential for high-quality real-world evidence generation using observational data for cancer research, provided that accurate phenotypes—characterizations of patients on the basis of electronic health data—and thorough validation are used to identify the cohorts of patients with cancer of interest.23
Limitations of this study include that our definition of treatment patterns does not characterize patients who switch back to a treatment used previously, as an ingredient will only be listed once within a sequence, nor do these patterns distinguish between switching or augmentation—the addition of a second drug to the treatment regimen—which may underestimate the total number of distinct sequences and their variation; however, this was a rare occurrence and does not affect our primary findings. In addition, data from observational databases, such as EHR, and claims databases may have missing values or incorrect data. Institutions across the OHDSI network attempt to minimize this using standard tools for measuring data quality and constantly developing new metrics to identify deficiencies for improvement. This study does not yet explore associations of the identified variation with clinical outcomes, which we recognize is an important future goal and hope will be the aim of additional OHDSI network studies. Finally, observational data may also be subject to both measured and unmeasured confounding, which is minimized in this high-level descriptive characterization study.
The significant impact that these comorbidities have on mortality, prognosis, and quality of life in patients with cancer suggests that the characterization of treatment patterns shown here should only be considered a first step. Additional studies are needed to understand the cause of these practice pattern variations, uncover their effects on outcomes, and improve our management of these comorbid conditions in this high-risk population of patients with cancer, which, in turn, may help guide future clinical care to improve patient outcomes.
Appendix
TABLE A1.
List of Codes Defining Cancer Diagnoses
TABLE A2.
List of Codes Defining Type 2 Diabetes Mellitus, Hypertension, and Depression Diagnosis
TABLE A3.
List of Codes Defining Treatments
TABLE A4.
Attrition Table for IBM MarketScan Commercial Claims and Encounters Database
TABLE A5.
Attrition Table for Columbia University Irving Medical Center Database
TABLE A6.
Attrition Table for IQVIA France Database
TABLE A7.
Attrition Table for IQVIA Germany Database
TABLE A8.
Attrition Table for IBM MarketScan Multistate Medicaid Database
TABLE A9.
Attrition Table for IBM MarketScan Medicare Supplemental Beneficiaries Database
TABLE A10.
Attrition Table for Optum Database
TABLE A11.
Attrition Table for Stanford Database
Footnotes
Presented at the American Medical Informatics Association 2018 Annual Symposium, San Francisco, CA, November 5, 2018.
Supported by the National Institutes of Health, National Cancer Institute Contract No. HHSN261201700469P and National Library of Medicine Grant No. R01-LM006910.
The views expressed are of the authors and do not necessarily represent the views of the National Institutes of Health or the United States Government.
AUTHOR CONTRIBUTIONS
Conception and design: Ruijun Chen, Patrick Ryan, Gurvaneet Randhawa, George Hripcsak
Financial support: Gurvaneet Randhawa, George Hripcsak
Administrative support: George Hripcsak
Provision of study materials or patients: Ruijun Chen, Patrick Ryan, Christian G. Reich, Rohit Vashisht, Nigam H. Shah, George Hripcsak
Collection and assembly of data: Ruijun Chen, Patrick Ryan, Karthik Natarajan, Thomas Falconer, Christian G. Reich, George Hripcsak
Data analysis and interpretation: Ruijun Chen, Patrick Ryan, Karthik Natarajan, Thomas Falconer, Katherine D. Crew, Rohit Vashisht, Nigam H. Shah, George Hripcsak
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Patrick Ryan
Employment: Janssen Research & Development
Stock and Other Ownership Interests: Johnson & Johnson
Nigam H. Shah
Leadership: Prealize Health
Stock and Other Ownership Interests: Apixio
Consulting or Advisory Role: Apixio
Research Funding: Google (Inst)
Travel, Accommodations, Expenses: Apixio
George Hripcsak
Research Funding: Janssen Research & Development
No other potential conflicts of interest were reported.
REFERENCES
- 1.Edwards BK, Noone A-M, Mariotto AB, et al. Annual Report to the Nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120:1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Søgaard M, Thomsen RW, Bossen KS, et al. The impact of comorbidity on cancer survival: A review. Clin Epidemiol. 2013;5(suppl 1):3–29. doi: 10.2147/CLEP.S47150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satariano WA, Ragland DR. The effect of comorbidity on 3-year survival of women with primary breast cancer. Ann Intern Med. 1994;120:104–110. doi: 10.7326/0003-4819-120-2-199401150-00002. [DOI] [PubMed] [Google Scholar]
- 4.Albertsen PC, Moore DF, Shih W, et al. Impact of comorbidity on survival among men with localized prostate cancer. J Clin Oncol. 2011;29:1335–1341. doi: 10.1200/JCO.2010.31.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tammemagi CM, Neslund-Dudas C, Simoff M, et al. Impact of comorbidity on lung cancer survival. Int J Cancer. 2003;103:792–802. doi: 10.1002/ijc.10882. [DOI] [PubMed] [Google Scholar]
- 6.Bluethmann SM, Mariotto AB, Rowland JH. Anticipating the “silver tsunami”: Prevalence trajectories and comorbidity burden among older cancer survivors in the United States. Cancer Epidemiol Biomarkers Prev. 2016;25:1029–1036. doi: 10.1158/1055-9965.EPI-16-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Extermann M. Measurement and impact of comorbidity in older cancer patients. Crit Rev Oncol Hematol. 2000;35:181–200. doi: 10.1016/s1040-8428(00)00090-1. [DOI] [PubMed] [Google Scholar]
- 8.Chochinov HM. Depression in cancer patients. Lancet Oncol. 2001;2:499–505. doi: 10.1016/S1470-2045(01)00456-9. [DOI] [PubMed] [Google Scholar]
- 9.Bensimon L, Yin H, Suissa S, et al. Type 2 diabetes and the risk of mortality among patients with prostate cancer. Cancer Causes Control. 2014;25:329–338. doi: 10.1007/s10552-013-0334-6. [DOI] [PubMed] [Google Scholar]
- 10.Andersen BL, DeRubeis RJ, Berman BS, et al. Screening, assessment, and care of anxiety and depressive symptoms in adults with cancer: An American Society of Clinical Oncology guideline adaptation. J Clin Oncol. 2014;32:1605–1619. doi: 10.1200/JCO.2013.52.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDaniel JS, Musselman DL, Porter MR, et al. Depression in patients with cancer. Diagnosis, biology, and treatment. Arch Gen Psychiatry. 1995;52:89–99. doi: 10.1001/archpsyc.1995.03950140007002. [DOI] [PubMed] [Google Scholar]
- 12.Pirl WF. Evidence report on the occurrence, assessment, and treatment of depression in cancer patients. J Natl Cancer Inst Monogr. 2004;2004:32–39. doi: 10.1093/jncimonographs/lgh026. [DOI] [PubMed] [Google Scholar]
- 13.Grossman E, Messerli FH, Boyko V, et al. Is there an association between hypertension and cancer mortality? Am J Med. 2002;112:479–486. doi: 10.1016/s0002-9343(02)01049-5. [DOI] [PubMed] [Google Scholar]
- 14.Scartozzi M, Galizia E, Chiorrini S, et al. Arterial hypertension correlates with clinical outcome in colorectal cancer patients treated with first-line bevacizumab. Ann Oncol. 2009;20:227–230. doi: 10.1093/annonc/mdn637. [DOI] [PubMed] [Google Scholar]
- 15.Rini BI, Cohen DP, Lu DR, et al. Hypertension as a biomarker of efficacy in patients with metastatic renal cell carcinoma treated with sunitinib. J Natl Cancer Inst. 2011;103:763–773. doi: 10.1093/jnci/djr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitbart W, Rosenfeld B, Pessin H, et al. Depression, hopelessness, and desire for hastened death in terminally ill patients with cancer. JAMA. 2000;284:2907–2911. doi: 10.1001/jama.284.22.2907. [DOI] [PubMed] [Google Scholar]
- 17.Ranpura V, Pulipati B, Chu D, et al. Increased risk of high-grade hypertension with bevacizumab in cancer patients: A meta-analysis. Am J Hypertens. 2010;23:460–468. doi: 10.1038/ajh.2010.25. [DOI] [PubMed] [Google Scholar]
- 18.Wu S, Chen JJ, Kudelka A, et al. Incidence and risk of hypertension with sorafenib in patients with cancer: A systematic review and meta-analysis. Lancet Oncol. 2008;9:117–123. doi: 10.1016/S1470-2045(08)70003-2. [DOI] [PubMed] [Google Scholar]
- 19. Whelton PK, Carey RM, Aronow WS, et al: 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 71:1269-1324, 2018 [Errata: Hypertension 71:e136-e139, 2018; Hypertension 72:e33, 2018] PubMed.
- 20. Work Group on Major Depressive Disorder. https://psychiatryonline.org/pb/assets/raw/sitewide/practice_guidelines/guidelines/mdd.pdf.
- 21.American Diabetes Association 8. Pharmacologic approaches to glycemic treatment: Standards of medical care in diabetes-2018. Diabetes Care. 2018;41(suppl 1):S73–S85. doi: 10.2337/dc18-S008. [DOI] [PubMed] [Google Scholar]
- 22.Coughlin SS, Calle EE, Teras LR, et al. Diabetes mellitus as a predictor of cancer mortality in a large cohort of US adults. Am J Epidemiol. 2004;159:1160–1167. doi: 10.1093/aje/kwh161. [DOI] [PubMed] [Google Scholar]
- 23.Platt R, Lieu T. Data enclaves for sharing information derived from clinical and administrative data. JAMA. 2018;320:753–754. doi: 10.1001/jama.2018.9342. [DOI] [PubMed] [Google Scholar]
- 24.Randhawa GS, Slutsky JR. Building sustainable multi-functional prospective electronic clinical data systems. Med Care. 2012;50(suppl):S3–S6. doi: 10.1097/MLR.0b013e3182588ed1. [DOI] [PubMed] [Google Scholar]
- 25.Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for observational researchers. Stud Health Technol Inform. 2015;216:574–578. [PMC free article] [PubMed] [Google Scholar]
- 26.Observational Health Data Sciences and Informatics Mission, vision & values. https://www.ohdsi.org/who-we-are/mission-vision-values/
- 27.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hripcsak G, Ryan PB, Duke JD, et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci USA. 2016;113:7329–7336. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reich C, Ryan P, Belenkaya R, et al. OMOP Common Data Model v5.3 Specifications. https://github.com/OHDSI/CommonDataModel
- 30. Observational Health Data Sciences and Informatics: Treatment Pathways in Patients With Cancer. https://www.ohdsi.org/web/wiki/doku.php?id=research:treatment_pathways_in_cancer_12mo.
- 31. Observational Health Data Sciences and Informatics: Common Data Model. https://github.com/OHDSI/CommonDataModel/wiki.
- 32.Hu FB, Manson JE, Liu S, et al. Prospective study of adult onset diabetes mellitus (type 2) and risk of colorectal cancer in women. J Natl Cancer Inst. 1999;91:542–547. doi: 10.1093/jnci/91.6.542. [DOI] [PubMed] [Google Scholar]
- 33.Barone BB, Yeh H-C, Snyder CF, et al. Long-term all-cause mortality in cancer patients with preexisting diabetes mellitus: A systematic review and meta-analysis. JAMA. 2008;300:2754–2764. doi: 10.1001/jama.2008.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Poll-Franse LV, Houterman S, Janssen-Heijnen MLG, et al. Less aggressive treatment and worse overall survival in cancer patients with diabetes: A large population based analysis. Int J Cancer. 2007;120:1986–1992. doi: 10.1002/ijc.22532. [DOI] [PubMed] [Google Scholar]
- 35.Keating NL, O’Malley AJ, Smith MR. Diabetes and cardiovascular disease during androgen deprivation therapy for prostate cancer. J Clin Oncol. 2006;24:4448–4456. doi: 10.1200/JCO.2006.06.2497. [DOI] [PubMed] [Google Scholar]
- 36.Giovannucci E, Harlan DM, Archer MC, et al. Diabetes and cancer: A consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]