Abstract
Purpose
To report the clinical features and genetic basis of three previously unreported families with punctiform and polychromatic pre-Descemet corneal dystrophy (PPPCD).
Design
Observational case series.
Methods
Full ophthalmic assessment was performed for members of three unreported families with PPPCD. Structural and biomechanical alterations of the cornea were screened. Whole-exome-sequencing (WES) was performed on the first family. Novel or rare variants that segregated with the affected status were screened for in the other two families with Sanger sequencing. Identified variants that segregated with the affected status in all families were characterized using in silico prediction tools and/or in vitro splice assays. Additionally, two previously reported PPPCD families were screened for variants identified in the three unreported PPPCD families.
Results
Twelve of 21 examined members of the three unreported families were diagnosed with PPPCD. The only refractive, topographic or biomechanical abnormality associated with PPPCD was a significantly increased corneal stiffness. WES and Sanger sequencing identified two variants that segregated with the affected status in the all three families: a rare intronic PDZD8 c.872+10A>T variant and a novel missense PRDX3 c.568G>C (p.Asp190His) variant. The same PRDX3 variant was identified in the previously reported PPPCD family expressing the common PPPCD phenotype, and is predicted by in silico prediction tools to be damaging to protein function.
Conclusions
PPPCD is associated with an alteration of corneal biomechanics and a novel missense variant in PRDX3. Screening of additional families will determine whether all families demonstrate a PRDX3 variant, or whether locus heterogeneity may exist for PPPCD.
Keywords: Corneal dystrophy, punctiform, polychromatic, genetics, cornea
INTRODUCTION
Punctiform and polychromatic pre-Descemet corneal dystrophy (PPPCD) is a rare corneal dystrophy first described by Fernandez-Sasso et al in 1979.1 Typically asymptomatic without any reported visual disturbance, PPPCD is characterized by the presence of punctiform, multicolored opacities in the posterior stroma, immediately anterior to Descemet membrane.1–7 According to the second edition of the IC3D classification of corneal dystrophies, PPPCD is currently considered a subtype of pre-Descemet corneal dystrophy (PDCD), which is classified as a “Category 4” dystrophy (suspected, new, or previously documented corneal dystrophies, where the evidence for it being a distinct entity is not yet convincing).8
To our knowledge, only ten families with PPPCD have been reported in the literature as case reports (Table 1).1–7 While an autosomal dominant inheritance has been suggested, the inheritance pattern and genetic basis have yet to be elucidated. Additionally, it is also unknown whether PPPCD is associated with any other corneal abnormalities as a complete ophthalmic assessment of individuals with PPPCD has not been reported.
Table 1.
Description of reported families with punctiform and polychromatic pre-Descemet corneal dystrophy in the literature.
| Family | Authors | Country | Publication Year | Affected members | Proposed Inheritance | Family Origin |
|---|---|---|---|---|---|---|
| 1 | Fernandez-Sasso et al1 | Argentina | 1979 | 8 | Autosomal dominant | French/Spanish Pyrenees |
| 2 | Lisch et al6 | Germany | 1984 | 2 | Autosomal dominant | Non reported |
| 3 | Tzelikis et al5 | Brazil | 2007 | 3 | Autosomal dominant | Non reported |
| 4 | Brazil | 2007 | 1 (family members not analysed) | Non reported | Non reported | |
| 5 | Dolz-Marco et al3 | Spain | 2013 | 2 | Autosomal dominant | East Spain |
| 6 | Coelho et al4 | Brazil | 2015 | 1 (family members not analysed) | Non reported | Non reported |
| 7 | Brazil | 2015 | 1 (family members not analysed) | Non reported | Non reported | |
| 8 | Lagrou et al2 | Canada | 2016 | 3 | Autosomal dominant | Columbia (USA) & Northern Spain (ancestral family) |
| 9 | Henriquez Recine et al7 | Spain | 2018 | 8 | Autosomal dominant | Central Spain |
| 10 | Spain | 2018 | 3 | Autosomal dominant | Central Spain | |
| 11 | Alió del Barrio et al | Spain | 2019 | 9 | Autosomal dominant | East Spain |
| 12 | Spain | 2019 | 3 | Autosomal dominant | East Spain | |
| 13 | Spain | 2019 | 1 | Insufficient Sample | East Spain |
We present three previously unreported PPPCD pedigrees, in which we performed a comprehensive ophthalmic assessment of affected and unaffected family members, as well as the results of whole exome and Sanger sequencing in these and two previously reported pedigrees to identify the genetic basis of PPPCD.
MATERIALS AND METHODS
Three previously unreported (PPPCD family 1, family 2 and family 3) and two previously reported (PPPCD family 4 and family 5)7 PPPCD pedigrees were identified and family members enrolled in this observational case series and in the authors’ ongoing study of inherited ocular disorders. Informed written consent was obtained from all subjects in this study according to the tenets of the Declaration of Helsinki and approval for this study was obtained from the Institutional Review Board at the University of California at Los Angeles (UCLA IRB#11–000020) and the ethical committee from Vissum Corporación.
Clinical Evaluation
All affected and unaffected individuals from three previously unreported families (PPPCD family 1–3) that agreed to participate in the study received a full ophthalmic examination including: slit lamp biomicroscopy, fundoscopy, corneal endothelial specular microscopy (Noncom Robo, Japan), corneal topography (including anterior keratometry, pachymetry and corneal aberrometry with 6-mm pupil) (Sirius, CSO, Italy), ocular aberrometry (Osiris, CSO, Italy), anterior segment optical coherence tomography (AS-OCT) (MS-39, CSO, Italy), corneal biomechanics (Ocular Response Analyser, OftalTech, Spain) and ocular scattering index (OSI) (HD Analyzer, Visiometrics, Spain). Affected individuals also underwent corneal confocal biomicroscopy (Confoscan 4, Nidek, Japan). Two individuals under 5 years of age were included in the study, but due to the expected lack of cooperation, only a clinical examination (slit lamp examination and fundoscopy) was performed. The diagnosis of PPPCD was based on the presence of polychromatic crystals located into the posterior corneal stroma (in a pre-Descemet membrane location) on slit lamp examination that appeared as hyperreflective pre-Descemetic opacities with confocal and specular microscopy (Figures 1 and 2).
Figure 1.
Pedigrees of three previously unreported Spanish PPPCD families: family 1 (A), family 2 (B), and family 3 (C) with punctiform and polychromatic pre-Descemet corneal dystrophy. Question marks (?) indicate unexamined individuals. “WES” indicates individuals in whom whole exome sequencing was performed. Individuals heterozygous for the PRDX3 c.568G>C variant are indicated by +/− and individuals who lack the variant are indicated by −/−. (D) The heterozygous PRDX3 c.568G>C variant (Refseq: NM_006793.4) was confirmed by Sanger sequencing in all affected individuals.
Figure 2.
Slit lamp photomicrographs of an individual with punctiform and polychromatic pre-Descemet corneal dystrophy (Figure 1A, III–15) demonstrating multiple polychromatic posterior stromal opacities in each eye.
Although a comprehensive past medical history, including a complete medication history, was taken from each of the individuals recruited into the study, a physical examination to identify systemic or metabolic associations was not performed given the last of evidence of extraocular manifestations of PPPCD in previous reports.1–7
Statistical Analysis
SPSS software for Windows was used for statistical analysis (IBM SPSS Statistics, version 22.0). Analysis was performed for all variables with non-parametric tests due to the small sample size (n<30). Thus, the Mann-Whitney test was applied to assess differences between groups (affected and unaffected), except for the nominal variable “sex”, for which the Pearson’s chi-squared test (X2 test) was used. Differences were considered statistically significant when p<0.05.
DNA isolation
After informed consent was obtained, saliva samples were collected from members of each of the three unreported families and the two previously reported families7 with the Oragene Saliva Collection Kit (DNA Genotek, Inc., Ottawa, Canada) and genomic DNA was isolated using the Oragene prepIT-L2P Kit (DNA Genotek, Inc.) according to the manufacturer’s instructions.
WES and variant calling
WES was performed on genomic DNA derived from affected and unaffected members of PPPCD family 1. DNA libraries were prepared using the TruSeq DNA Sample Preparation Kit v2 (Illumina Inc., San Diego, CA) and exome capture was performed with the SeqCap EZ Exome Library v3.0 (Roche NimbleGen, Inc., Madison, WI). Paired-end sequencing (2×150 bp) was performed with Illumina’s HiSeq 4000 by Genewiz (South Plainfield, NJ). Using the Qiagen’s Biomedical Genomics Workbench 5.0 (Qiagen, Redwood City, CA), the generated sequence reads were aligned to the Hg38 human genome reference, and aligned reads were annotated with the Ensembl 88 transcript database. Called variants were annotated with the dbSNP 150 database.
Filtering of WES variants
Using Qiagen’s Ingenuity Variant analysis software, variants identified by WES analysis in the affected and unaffected members of PPPCD family 1 were filtered to exclude any variant with: a quality score < 20, read count < 5, or minor allele frequency (MAF) > 0.5% in either the ExAC, 1000genomes, or genomeAD databases; present in a homozygous state; absent in any of the affected individuals; and present in any of the unaffected individuals. To make allowances for a potential false positive/negative call by WES for any particular variant in one individual, additional filtering and analyses were performed to exclude variants: absent in 2 or more of 6 affected and present in any unaffected individuals; or absent in any of the affected individuals and present in 2 or more of 4 unaffected individuals.
PCR amplification and Sanger sequencing
Primers were designed to amplify the genomic regions containing: filtered variants identified by WES; exons and/or introns of PDZD8 (Refseq Gene ID: 118987), PRDX3 (Refseq Gene ID: 10935), and OR2M5 (Refseq gene ID: 80000) (see Supplemental Table 1 for primer sequences); and variants used for mini-haplotype analysis. DNA amplification by polymerase chain reaction (PCR) was performed in 25μL reaction volumes containing 25–40 ng of genomic DNA, 2.5 pico-moles of each primer, and GoTaq® Green Master mix (Promega, Madison, WI) according to the manufacturer’s recommendations. The PCR protocol consisted of a denaturizing step at 95°C for 3 minutes, followed by a 35x cycle of: a denaturing step at 95°C for 30 seconds, annealing step at 60°C for 30 seconds, and elongation step at 72°C for 30–60 seconds. Sanger sequencing was performed by Laragen, Inc. (Culver City, CA).
In silico variant prediction and scoring
Filtered variants identified by WES were analysed by online tools SIFT9, Polyphen-210, CADD11, 12, Provean13, and/or Human Splicing Finder14 to predict each variant’s impact on protein function or splicing.
Mini-haplotype analyses
To determine the haplotype of the genomic region encompassing the PRDX3 c.568G>C variant on chromosome 10, rare proximal variants were identified in the WES data from PPPCD family 1 and screened in affected individuals from families 2–4 who harboured the PRDX3 c.568G>C variant (see Supplemental Table 1 for primer sequences).
In vitro splice assay
A 1444 bp region of PDZD8, containing either the wild-type sequence or the c.872+10A>T variant, was amplified from genomic DNA obtained from either an affected or unaffected individual of PPPCD family 1. The amplified PDZD8 fragment contained exon 1, along with 264 bp of the 5’ UTR and 308 bp of intron 1 that flank exon 1, and was amplified using the following primer sequences: Forward- 5’-GAATT CCCATATGGAGTGGAGGCCTGAGGGA-3’ and Reverse- 5’-GAATTCCCATATGCCTGGGGATTAGGGTAGGCT-3’. Both primers were designed with a Ndel restriction site at their 5’ ends to be used for cloning the amplified PDZD8 fragment into the pTBNde(min) plasmid (plasmid# 15125, a gift from Franco Pagani, Addgene, Cambridge, MA) that contains a modified version of the α-globin-fibronectin-EDB minigene.15, 16
The splicing assay was performed by transfecting HEK293T cells with each minigene plasmid using Lipofectamine® LTX (Life Technologies, Grand Island, NY) according to the manufacturer’s recommended protocols. Total RNA from transfected HEK293T was extracted using TRI Reagent (Sigma-Aldrich Corp., St. Louis, MO) and complementary DNA (cDNA) was synthesized using the Superscript Ill First-Strand kit (Life Technologies) according to the manufacturer’s recommendations. Reverse transcription PCR (RT-PCR) was performed using previously published RT-PCR protocols with primers targeting the flanking fibronectin exons (see Supplemental Table 1 for primer sequences).17
RESULTS
Clinical evaluation of PPPCD families
Seventeen members from family 1, six members from family 2 and two members from family 3 were enrolled in the clinical study (Figure 1). Slit lamp examination demonstrated bilateral, symmetric, punctiform and polychromatic opacities in the deep stroma immediately anterior to Descemet membrane in eight individuals from family 1, three individuals from family 2, and one individual from family 3 (Figures 1 and 2). There was no significant difference in the mean age between the affected (42.9 years; range 8–79) and unaffected (33.3 years; range 1–69) individuals (p= 0.25) or in the percentage of men in the affected (41.6%; 5/12) and unaffected groups (53.8%; 7/13; p= 0.38).
Comprehensive clinical examination failed to reveal associated ophthalmic disorders in the affected individuals. In addition, no ophthalmic surgical interventions were documented in any of the study patients’ medical records, with an exception of bilateral cataract surgery in an affected 79-year-old individual. Corneal imaging could not be performed due to limited cooperation in two patients: a 4-year-old child (unaffected based on slit lamp examination) and a 1-year-old infant (unaffected based on a portable slit lamp examination).
Anterior Segment Imaging
Anterior segment OCT imaging of the corneas of affected individuals demonstrated faint, hyperreflective, pre-Descemetic opacities (Figure 3-top), which were more easily visualized with Scheimpflug imaging (Figure 3-bottom). These opacities were absent in the unaffected individuals.
Figure 3.
Anterior segment OCT (top) and Scheimpflug (bottom) imaging of an individual with punctiform and polychromatic pre-Descemet corneal dystrophy (Figure 1A, III–11) demonstrating hyper-reflective posterior stromal opacities (indicated by yellow arrows) that are more easily identified with Scheimpflug imaging.
Specular and confocal microscopic imaging
Specular microscopic imaging of the corneal endothelium revealed a normal endothelial cell mosaic in both groups, with similar cellular density, coefficient of variation and percentage of hexagonality (Table 2). Affected individuals demonstrated multiple, round, hyperreflective opacities at the pre-Descemetic level, immediately anterior to an unremarkable endothelial cell layer (Figure 4-right). Confocal microscopic imaging of affected individuals demonstrated an unremarkable corneal stroma other than for the extracellular, pre-Descemetic opacities that measured approximately 10μm in diameter (Figure 4-left).
Table 2.
Clinical evaluation of PPPCD families.
| PPPCD Affected | Unaffected | P Value | |||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Refractive | |||||
| Ref Sphere (D) | −0.42 | 2.54 | −2.08 | 3.26 | 0.12 |
| Ref Cylinder (D) | −0.79 | 0.66 | −0.48 | 0.45 | 0.1 |
| CDVA (decimal) | 0.1 | 0.05 | 1.02 | 0.06 | 0.43 |
| Tomographic | |||||
| Anterior Km (D) | 44.33 | 1.93 | 43. 94 | 1.35 | 0.67 |
| Topo Cylinder (D) | −0.98 | 0.5 | −1.13 | 0.44 | 0.2 |
| CCT (μm) | 528.65 | 30.69 | 539.4 | 25.48 | 0.24 |
| Thinnest (μm) | 517.75 | 32.81 | 530.15 | 28.27 | 0.21 |
| Kmax (D) | 44.83 | 1.96 | 44.51 | 1.43 | 0.74 |
| Wavefont | |||||
| Corneal Total HOA (μm) | 0.59 | 0.37 | 0.46 | 0. 1 | 0.35 |
| Corneal Coma (μm) | 0.33 | 0.18 | 0.31 | 0.1 | 0.95 |
| Corneal Sph (μm) | 0.22 | 0.18 | 0.22 | 0.07 | 0.11 |
| Endothelial specular microscopy | |||||
| CD (cells/mm2) | 2563 | 708.7 | 2572 | 344.83 | 0.84 |
| CV | 43 | 40.3 | 33 | 5.56 | 0.12 |
| Hexagonality (%) | 61 | 10.13 | 61 | 9.26 | 0.48 |
| Ocular abberometry | |||||
| Strehl Ratio (PSF) | 0.37 | 0.15 | 0.39 | 0.17 | 0.85 |
| OSI | 1.24 | 1.19 | 0.82 | 0.53 | 0.62 |
| Biomechanical | |||||
| CH (mm Hg) | 10.32 | 1.44 | 9.85 | 1.52 | 0.26 |
| CRF (mm Hg) | 10.77 | 1.32 | 9.93 | 1.24 | 0.02* |
| Intraocular Pressure | |||||
| IOP (mm Hg) | 14.6 | 3.5 | 13.25 | 2.15 | 0,22 |
Ref: refractive; D: diopters; CDVA: corrected distance visual acuity; dec: decimal scale; Km: mean keratometry; Topo: topographic; CCT: central corneal thickness; Thinnest: thinnest pachymetric point; Kmax: maximum keratometry; HOA: higher order aberrations; Sph: spherical aberration; CD: cell density; CV: coefficient of variation; 6A: hexagonality; PSF: point spread function; OSI: ocular scatter index; CH: corneal hysteresis; CRF: corneal resistance factor].
statistically significant differences.
Figure 4.
Confocal microscopy (left) and specular microscopy (right) imaging of the posterior corneal stroma of an individual with punctiform and polychromatic pre-Descemet corneal dystrophy (Figure 1B, II–4) demonstrating hyper-reflective opacities distributed at the level of Descemet membrane.
Cornea biomechanics
Corneal biomechanical evaluation demonstrated increased corneal hysteresis (CH; p=0.26) and significantly increased corneal resistance factor (CRF; p=0.02) in individuals with PPPCD compared with unaffected individuals (Table 2).
Vision, refraction and corneal topography
There were no statistically significant differences in any of the other analysed visual, refractive, keratometric, pachymetric or corneal aberrometric parameters (Table 2).
WES analysis of PPPCD family 1
DNA samples were collected from 13 members (8 affected and 5 unaffected) of PPPCD family 1 (Figure 1A). WES was performed on DNA samples from six affected (II:6, III:7, III:11, III:13, IV:4, IV:8) and four unaffected individuals (III:5, III:8, III:9, III:14). After excluding variants with low quality (quality score < 20) and low read counts (read counts < 5), 281,004 unique variants (SNV and indels) were collectively identified in the 10 individuals. After excluding homozygous variants (given the observed autosomal dominant inheritance pattern in PPPCD family 1), 108,844 heterozygous variants were evaluated for allele frequency, revealing that 29,336 were novel or rare (MAF < 0.5%). After filtering variants based on segregation with the affected phenotype, no novel or rare heterozygous coding region variants were present in all 6 affected individuals and not present in any of the 4 unaffected individuals. While WES primarily targets the coding regions of the genome, the non-coding regions of the genome that are close to intron-exon junctions are also typically captured and sequenced. As such, screening of the 29,336 novel or rare heterozygous variants revealed two intronic variants, PDZD8 c.872+10A>T (based on transcript NM_173791.4) and GREB1L c.4229–25T>C (based on transcript NM_001142966.2), that segregated with the affected phenotype in the 10 members of PPPCD family 1 who underwent WES (Table 3). Sanger sequencing performed to validate the WES results for these two intronic variants confirmed the results of WES in each of the 10 individuals. Sanger sequencing of PDZD8 and GREB1L in the remaining three individuals of PPPCD family 1 who did not undergo WES (III:15, IV:6, IV:9) demonstrated that PDZD8 c.872+10A>T continued to segregate with the affected status while GREB1L c.4229–25T>C did not.
Table 3.
Variants identified by WES
| # of affecteds containing variant by WES | # of unaffecteds containing variant by WES | Chr | Position | Gene | Transcript ID | Transcript Variant | Amino acid change | dbSNP ID | gnom AD MAF (%) | SIFT/PolyPhen-2 Function Prediction | Confirmed by Sanger sequencing to be present in 6 of 6 affected and 0 of 4 unaffecteds? | If confirmed by Sanger sequencing, does variant segregate in 3 additional family members? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 of 6 | 0 of 4 | 10 | 117374346 | PDZD8 | NM_17379 1.4 | c.872+10A>T | - | rs201808439 | 0.006 | - | Yes | Yes |
| 6 of 6 | 0 of 4 | 18 | 21505785 | GREB1L | NM_00114 2966.2 | c.4229-25T>C | - | rs775835476 | 0.004 | - | Yes | No |
| 5 of 6 | 0 of 4 | 1 | 248145920 | OR2M5 | NM_00100 4690.1 | c.773T>C | p.M258T | rs146014040 | 0.056 | Activating/Benign | Yes | Yes |
| 5 of 6 | 0 of 4 | 5 | 194925 | LRRC14B | NM_00108 0478.2 | c.1117 G>A | p.V373I | rs200063605 | 0.077 | Tolerated/Benign | No | - |
| 5 of 6 | 0 of 4 | 5 | 10280458 | CMBL | NM_13880 9.3 | c.733A>C | p.M245L | rs182773279 | 0.086 | Tolerated/Benign | No | - |
| 5 of 6 | 0 of 4 | 7 | 150692529 | GIMAP2 | NM_01566 0.2 | c.243G>A | p.M81I | rs927523889 | - | Activating/Benign | No | - |
| 5 of 6 | 0 of 4 | 8 | 8702651 | CLDN23 | NM_19428 4.2 | c.253G>A | p.V85I | rs372410779 | 0.005 | Tolerated/Benign | No | - |
| 5 of 6 | 0 of 4 | 10 | 17796225 | TMEM236 | NM_00109 8844.2 | c.777C>A | p.N259K | rs1001234857 | 0.037 | Tolerated/Benign | No | - |
| 5 of 6 | 0 of 4 | 10 | 119169326 | PRDX3 | NM_00679 3.4 | c.568G>C | p.D190H | None | - | Damaging/Prob. Dama. | Yes | Yes |
| 5 of 6 | 0 of 4 | 18 | 23784125 | LAMA3 | NM_00112 7717.2 | c.1571 G>A | p.R524H | rs201845068 | 0.036 | None/Poss. Damaging | Yes | No |
| 6 of 6 | 1 of 4 | 1 | 226736078 | ITPKB | NM_00222 1.3 | c.1381C>T | p.P461S | rs35823273 | 0.396 | Tolerated/Benign | No | - |
| 6 of 6 | 1 of 4 | 7 | 74120997 | LIMK1 | NM_00231 4.3 | c.1729C>T | p.P577S | rs147218553 | 0.018 | Tolerated/Benign | No | - |
| 6 of 6 | 1 of 4 | 7 | 76482982 | DTX2 | NM_00110 2596.1 | c.743A>G | p.N248S | rs145151450 | 0.314 | Tolerated/Benign | No | - |
To allow for a false positive/negative call by WES for any particular variant in one individual, re-analysis of the WES data was performed using less stringent criteria for filtering variants (see Methods), which led to the identification of: 8 heterozygous novel or rare coding region variants present in 5 of 6 affected individuals and not present in any of the 4 unaffected individuals; and 3 heterozygous rare coding region variants present in 6 of 6 affected individuals and not present in more than 1 unaffected individual (Table 3). Sanger sequencing validation of these 11 total coding region variants did not identify any false positives; however, Sanger sequencing revealed 3 variants, OR2M5 c.773T>C, PRDX3 c.568G>C and LAMA3 c.1571G>A, to be false negatives in one of the 6 affected individuals, thereby confirming each of these three variants to be present in all 6 affected individuals and not present in any of the 4 unaffected individuals. Sanger sequencing screening in the three additional family members who did not undergo WES revealed OR2M5 c.773T>C and PRDX3 c.568G>C continued to segregate with the affected status while LAMA3 c.1571G>A did not (Table 3). The OR2M5 c.773T>C variant was predicted by SIFT to have an activating impact on protein function but was predicted to be benign or neutral by PolyPhen and Provean (Table 3). In contrast, PRDX3 c.568G>C variant was predicted by SIFT, PolyPhen and Provean to deleteriously impact protein function, and also obtained a CADD score of 31, which places this variant in the top 0.1% of deleterious substitutions in the human genome (Table 3).12
Screening of PDZD8, PRDX3, and OR2M5 in PPPCD families 2 and 3
Given that PDZD8 c.872+10A>T, OR2M5 c.773T>C, and PRDX3 c.568.G>C segregated with the affected status in PPPCD family 1, we performed Sanger sequencing of all three genes in three affected (I:1, II:1, II:4) and two unaffected (I:2, II:2) members of PPPCD family 2 and in one affected individual (II:1) and one unaffected individual (II:3) in PPPCD family 3 (Figure 1B and 1C). Sanger sequencing of the OR2M5 coding region did not reveal a novel or rare variant in either family 2 or family 3. Sanger sequencing of PDZD8 and PRDX3 revealed the same PDZD8 c.872+10A>T and PRDX3 c.568G>C variants identified in PPPCD family 1 in the heterozygous state in the affected individuals and absent in the unaffected individuals of family 2 and family 3. The PDZD8 and PRDX3 variants are both located on chromosome 10 within ~2Mb from each other. Based on a previously performed RNA- seq analyses of adult human corneal gene expression, both PDZD8 and PRDX3 are expressed in ex vivo keratocytes and endothelial cells, with RPKM values of: 4.25 and 8.67 for PDZD8, respectively; and 8.43 and 33.91 for PRDX3, respectively.18
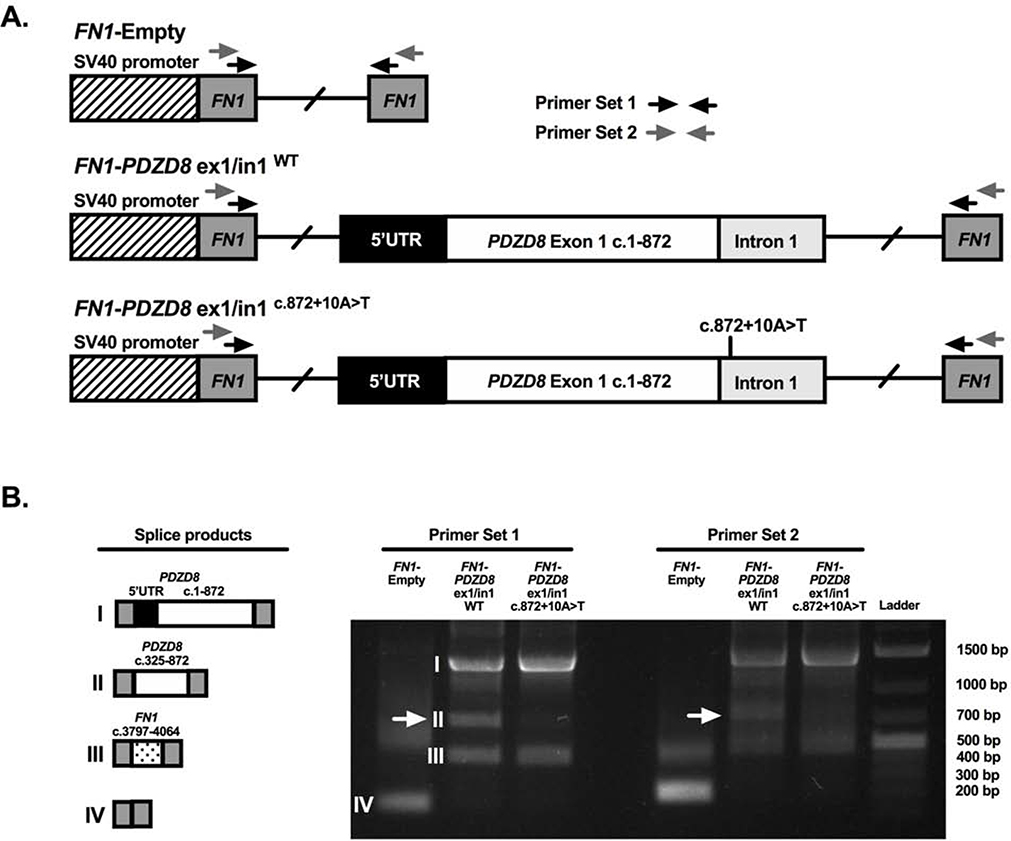
Impact of PDZD8 c.872+10A>T on splicing
In silico analysis performed using Human Splicing Finder v3.1 predicted that the PDZD8 c.872+10A>T variant activates an intronic cryptic donor splice site, potentially altering splicing. To determine whether the PDZD8 c.872+10A>T variant does in fact alter splicing, an in vitro splice assay was performed by inserting a genomic sequence containing the PDZD8 exon 1 and partial intron 1 (either with the c.872+10A>T variant or the wild-type sequence) in between two flanking fibronectin 1 (FN1) exons residing within a FN1 minigene plasmid, which was subsequently transfected into HEK293T cells. Using cDNA generated from the transfected HEK293T cells, RT-PCR demonstrated the c.872+10A>T variant caused the loss of a transcript-splice product (denoted by a ~700bp band) that was detected in the FN1/PDZD8 minigene with the wild-type sequence (Supplemental Figure 1). Sequencing of the ~700bp band revealed the c.325 to c.872 region of PDZD8 exon 1 spliced in between the two flanking FN1 exons. Sequencing of the ~1400bp and ~400bp bands revealed transcript products, flanked by the two minigene FN1 exons, containing either: the PDZD8 5’UTR and exon 1 regions; or a FN1 exonic region (c.3797–4064, NM_212482), respectively (Supplemental Figure 1).
Screening of PRDX3, PDZD8, and OR2M5 in two previously published PPPCD families
Genomic DNA samples were obtained from members of two previously reported PPPCD families: 3 affected individuals from PPPCD family 4 and 7 affected individuals from PPPCD family 5.7 Screening of the PDZD8 exon1/intron1 region in each of the affected individuals from both families did not reveal the c.872+10A>T variant. Screening of the PDZD8 promoter and coding regions and the OR2M5 coding region in two affected individuals from both families failed to reveal a novel or rare variant. Screening of the PRDX3 coding region in each of the affected individuals from both families identified the same c.568G>C variant in all three of the affected individuals of PPPCD family 4 but did not identify a novel or rare variant in PPPCD family 5.
Identification of ancestral mini-haplotype in families 1–3
Rare variants that are adjacent to the PRDX3 c.568G>C variant were genotyped in the four PPPCD families that demonstrated the PRDX3 c.568G>C variant to determine whether this variant likely arose from a common ancestor (Supplemental Figure 2). The same mini-haplotype was identified in the three previously unreported families (PPPCD families 1, 2, and 3) but not in PPPCD family 4, suggesting that the PRDX3 c.568G>C variant likely arose from a common ancestor in PPPCD families 1–3 and independently in PPPCD family 4.
DISCUSSION
The aim of this study was to perform a through corneal phenotypic analysis and to elucidate the genetic basis of punctiform and polychromatic pre-Descemet corneal dystrophy after the identification of three unreported families. In order to corroborate the results of genetic analysis in these three families, we recruited members of two recently reported families for additional screening.7 One of these families (Family 4) demonstrated the typical PPPCD phenotype, with localization of the opacities to the pre-Descemetic posterior stroma, as was observed in the three unreported families. However, affected members of family 5 presented an atypical PPPCD phenotype in that the opacities were distributed throughout all levels of the corneal stroma, indicating that this family may have a dystrophy that is clinically and genetically distinct from PRDX3-associated PPPCD.7 What is common to each of these five families, and indeed to eight of the 13 families reported to date is location in Spain or Spanish ancestry (Table 1). In addition, four of the five other families reported to date have been in Brazil (family origin non reported). Considering that Brazil and all South America have received strong immigration from the Iberian peninsula since the XVI century after the discovery of America, it is likely that the causative mutation(s) originated in the Iberian peninsula centuries ago.
Our investigation identified two variants, PRDX3 c.568G>C and PDZD8 c.872+10A>T, that segregated with the affected status in multiple PPPCD pedigrees. PRDX3 c.568G>C (p.Asp190His) is novel (not reported in the dbSNP database) and was identified in 4 of 5 pedigrees affected with PPPCD, with haplotype analysis indicating that this variant likely derived from an independent event in family 4. However, given the ~2 Mb distance between the two variants, we cannot rule out the possibility the PRDX3 c.568G>C variant arose from the same founder in all four families. The PDZD8 c.872+10A>T variant, identified in 3 of 5 PPPCD families, is not novel but is rare, and was demonstrated to impact splicing in vitro; whether or not splicing is altered in vivo by this variant, and is not simply an artifact of the in vitro splice assay, is yet to be determined. However, each of these 3 families in which it was identified also demonstrated the PRDX3 c.568G>C variant, which is only ~2Mb away from PDZD8 c.872+10A>T variant. Therefore, it is likely that these two variants on chromosome 10, along with the other rare variants within the shared mini-haplotype, were inherited from a common ancestor in PPPCD families 1–3. Given that both PRDX3 c.568G>C and PDZD8 c.872+10A>T were identified in PPCD families 1–3 that likely share a common ancestor but the novel PRDX3 c.568G>C variant was also found in a fourth unrelated PPPCD pedigree, PRDX3 is likely the causative gene for PPPCD.
The PRDX3 gene belongs to the thioredoxin family of peroxidases and encodes a mitochondrial antioxidant peroxidase that is responsible for regulating mitochondrial reactive oxygen species (mROS).19–21 Overexpression of PRDX3 has been reported in various cancers, while knockdown of PRDX3 was demonstrated to increase mtDNA oxidation and silencing of PRDX3 promoted enhanced invasive properties in HepG2 cells.22–26 Interestingly, a significant decrease of PRDX3 protein expression was reported in corneal endothelium derived from patients affected with Fuchs endothelial corneal dystrophy (FECD) compared to healthy controls, suggesting that corneal endothelial cells affected with FECD are less able to withstand oxidant-induced damage, possibly contributing to the pathogenesis of the disease.27 In the same report, while PRDX3 protein was shown to be expressed in normal corneal endothelium, PRDX3 protein was demonstrated to not be expressed in either normal corneal stroma or epithelium.27 While our identification of PRDX3 expression in ex vivo human corneal endothelial cells using RNA-seq corroborates this report, we found PRDX3 to be expressed in corneal stromal keratocytes, again using RNA-seq data.18 Functional studies will elucidate how the c.568G>C variant impacts the expression, localization, and function of PRDX3 in the cornea and whether or not the polychromatic crystalline-like opacities located in the stromal extra-cellular matrix are byproducts of aberrant PRDX3 proteins. To date, histopathologic examination of only one corneal button obtained post-mortem from an affected individual has been performed, which indicated that the opacities may represent focal lipid accumulations.28
According to the second edition of the IC3D classification of corneal dystrophies, PPPCD is currently classified as a subtype of PDCD as a category 4 dystrophy, indicating that “…the evidence for it, being a distinct entity is not yet convincing”.8 While the presence of punctate opacities anterior to Descemet membrane is a common feature of each subtype of PDCD, the PDCD subtypes differ in terms of inheritance, age of onset and morphology of the deposits. Herein, we present a comprehensive clinical characterization of PPPCD and report the association of a segregating PRDX3 missense variant in four PPPCD pedigrees with an autosomal dominant inheritance pattern. Therefore, we suggest that PPPCD might be considered a distinct inherited disorder and reclassified as a category 1 dystrophy, defined as a “…well-defined corneal dystrophy in which the gene has been mapped and identified and the specific mutations are known.” 8
Supplementary Material
ACKNOWLEDGMENTS
This publication has been carried out in the framework of the Red Temática de Investigación Cooperativa en Salud (RETICS), reference number RD16/0008/0012, financed by the Instituto Carlos III - General Subdirection of Networks and Cooperative Investigation Centers (R&D&I National Plan 2008–2011) and the European Regional Development Fund (Fondo Europeo de Desarrollo Regional FEDER). Support also provided by National Eye Institute Grants R01 EY022082 (A.J.A.), P30 EY000331 (core grant), the Walton Li Chair in Cornea and Uveitis (A.J.A.), the Stotter Revocable Trust and an unrestricted grant to Stein Eye Institute from Research to Prevent Blindness.
The authors thank Dr. Carmen Cardona (Vissum Corporación, Alicante, Spain) for her support on the clinical assessment of the families included in the study.
Biography
Jorge L. Alió del Barrio MD, PhD, FEBOS-CR specialised in Ophthalmology at the Ramón y Cajal University Hospital (Madrid, Spain) and he has completed Cornea Fellowships at Guy’s and St Thomas’ Hospital and Moorfields Eye Hospital (London, UK). He is currently working as a surgeon in the Cornea, Cataract and Refractive Surgery Service of Vissum Corporación (Alicante, Spain). He is also Associate Professor at Universidad Miguel Hernández (Alicante, Spain) School of Medicine.
Footnotes
Conflict of interest: No financial disclosures. The authors, their families, their employers and their business associates have no financial or proprietary interest in any product or company associated with any device, instrument or drug mentioned in this article. The authors have not received any payment as consultants, reviewers or evaluators of any of the devices, instruments or drugs mentioned in this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fernandez-Sasso D, Acosta JE, Malbran E. Punctiform and polychromatic pre-Descemet’s dominant corneal dystrophy. Br J Ophthalmol 1979;63(5):336–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagrou L, Midgley J, Romanchuk KG. Punctiform and Polychromatophilic Dominant Pre-Descemet Corneal Dystrophy. Cornea 2016;35(4):572–575. [DOI] [PubMed] [Google Scholar]
- 3.Dolz-Marco R, Gallego-Pinazo R, Pinazo-Duran MD, Diaz-Llopis M. Crystalline subtype of pre-descemetic corneal dystrophy. J Ophthalmic Vis Res 2014;9(2):269–271. [PMC free article] [PubMed] [Google Scholar]
- 4.Coelho LM, Muinhos GK, Tanure MAG, Almeida HGd, Sieiro RdO. Distrofia policromática posterior da córnea. Revista Brasileira de Oftalmología 2015;74:186–188. [Google Scholar]
- 5.Tzelikis PFdM Santos URd, Tanure MAG Trindade FC. Distrofia corneana policromática posterior. Revista Brasileira de Oftalmologia 2007;66(4):262–266. [Google Scholar]
- 6.Lisch W, Weidle EG. [Posterior crystalline corneal dystrophy]. Klin Monbl Augenheilkd 1984;185(2): 128–131. [DOI] [PubMed] [Google Scholar]
- 7.Henríquez-Recine MA, Marquina-Lima KS, Vallespín-García E, García-Miñaur S, Benitez Del Castillo JM, Boto de Los Bueis A. Heredity and in vivo confocal microscopy of punctiform and polychromatic pre-Descemet dystrophy. Graefes Arch Clin Exp Ophthalmol 2018;256(9): 1661–1667. [DOI] [PubMed] [Google Scholar]
- 8.Weiss JS, Moller HU, Aldave AJ, et al. IC3D classification of corneal dystrophies-edition 2. Cornea 2015;34(2):117–159. [DOI] [PubMed] [Google Scholar]
- 9.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31(13):3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 2019;47(D1):D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012;7(10):e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baralle M, Baralle D, De Conti L, et al. Identification of a mutation that perturbs NF1 agene splicing using genomic DNA samples and a minigene assay. J Med Genet 2003;40(3):220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagani F, Stuani C, Tzetis M, et al. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet 2003;12(10):1111–1120. [DOI] [PubMed] [Google Scholar]
- 17.Muro AF, Caputi M, Pariyarath R, Pagani F, Buratti E, Baralle FE. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol Cell Biol 1999;19(4):2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frausto RF, Le DJ, Aldave AJ. Transcriptomic Analysis of Cultured Corneal Endothelial Cells as a Validation for Their Use in Cell-Replacement Therapy. Cell transplantation 2016;25(6):1159–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Shoji W, Takano H, et al. Increased susceptibility of MER5 (peroxiredoxin III) knockout mice to LPS-induced oxidative stress. Biochem Biophys Res Commun 2007;355(3):715–721. [DOI] [PubMed] [Google Scholar]
- 20.Shibata E, Nanri H, Ejima K, et al. Enhancement of mitochondrial oxidative stress and up-regulation of antioxidant protein peroxiredoxin III/SP-22 in the mitochondria of human pre-eclamptic placentae. Placenta 2003;24(6):698–705. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Wi SM, Min Y, Lee KY. Peroxiredoxin-3 Is Involved in Bactericidal Activity through the Regulation of Mitochondrial Reactive Oxygen Species. Immune Netw 2016; 16(6):373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Hu Y, Liang H, Sun Z, Feng S, Deng H. Silencing PRDX3 Inhibits Growth and Promotes Invasion and Extracellular Matrix Degradation in Hepatocellular Carcinoma Cells. J Proteome Res 2016;15(5):1506–1514. [DOI] [PubMed] [Google Scholar]
- 23.Hu JX, Gao Q, Li L. Peroxiredoxin 3 is a novel marker for cell proliferation in cervical cancer. Biomed Rep 2013;1(2):228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinnula VL, Lehtonen S, Sormunen R, et al. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol 2002;196(3):316–323. [DOI] [PubMed] [Google Scholar]
- 25.Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, Chae HZ. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res 2001;21(3B):2085–2090. [PubMed] [Google Scholar]
- 26.Whitaker HC, Patel D, Howat WJ, et al. Peroxiredoxin-3 is overexpressed in prostate cancer and promotes cancer cell survival by protecting cells from oxidative stress. Br J Cancer 2013;109(4):983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurkunas UV, Rawe I, Bitar MS, et al. Decreased expression of peroxiredoxins in Fuchs’ endothelial dystrophy. Invest Ophthalmol Vis Sci 2008;49(7):2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croxatto J F-SD, Malbran ES. Clinicopathologic findings in punctiform and polychromatic pre-Descemet’s corneal dystrophy. Invest Ophthalmol Vis Sci 2002;43(13):E-Abstract 1724. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.