Abstract
Background
Although bone morphogenetic protein 6 (BMP6) signaling pathway has been implicated in many types of cancer, its role of tumorigenesis seems to be controversial and its ubiquitin-modifying mechanisms have not been fully addressed. Our study was designed to investigate how BMP6 signaling pathway is regulated by ubiquitin-modifying systems and to address molecular and clinical significance in colorectal cancers.
Methods
Human deubiquitnase (DUB) siRNA library was used to screen the specific DUB, named PSMD14, involved in BMP6 signaling pathway. Immunoblot, immunoprecipitation and ubiquitination assays were used to analyze targets of the PSMD14. A role of PSMD14-mediated BMP6 signaling pathway for malignant cancer progression was investigated using in vitro and in vivo model of colorectal cancers as well as clinical samples of colorectal cancer patients.
Findings
The deubiquitinase PSMD14 acts as a positive regulator for the initiation of the BMP6 signaling pathway through deubiquitinating K48-linked ALK2 type I receptor ubiquitination mediated by Smurf1 E3 ligase, resulting in increased stability of the ALK2. This role of PSMD14 is independent of its intrinsic role in the 26S proteasome system. Furthermore, either PSMD14 or ALK2 depletion significantly decreases tumorigenesis of HCT116 colorectal cancer cells in a xenograft model as well as cancer stemness/chemoresistance, and expression of the PSMD14 and ALK2 gene are correlated with malignant progression and the survival of colorectal cancer patients.
Interpretation
These findings suggest that the PSMD14-ALK2 axis plays an essential role in initiation of the BMP6 signaling pathway and contributes to tumorigenesis and chemoresistance of colorectal cancers.
Key words: PSMD14, Deubiqutinase, ALK2 receptor, Smurf1, BMP6, Colorectal cancers
1. Introduction
Bone morphogenetic proteins (BMPs), members of the transforming growth factor (TGF)-β family, play fundamental roles in a variety of cellular processes through regulating cell lineage commitment, morphogenesis, differentiation, proliferation and apoptosis of a wide range of cells and thus contributes to the maintenance of homeostasis during organ and tissue development [1,2]. Aside from these intrinsic functions of BMPs, accumulating evidence demonstrated that BMPs participate in cancer formation and progression through acting as a tumor suppressor or tumor promoter in a context-dependent manner [1,3].
The BMP family is composed of about 20 ligands, which are classified into at least four groups based on sequence similarity and affinities for specific receptors, and these BMPs mainly transmit the signal through the Smad-dependent canonical signaling pathway [2,3]. The BMPs bind to heteromeric combinations of type 1 [ALK1 (ACVRL1), ALK2 (ACVR1), ALK3 (BMPR1A), and ALK6 (BMPR1B)] and type 2 (ACTRII, ACTRIIB, and BMPRII) transmembrane receptors with serine-threonine kinase activities. The activated receptors subsequently phosphorylate the receptor-regulated Smad1, Smad5, and Smad8, respectively. The phosphorylated Smad protein forms a heterotrimeric complex with the common Smad, Smad4, and the complex subsequently translocates into the nucleus, modulating the expression of BMP-induced target genes.
Since the regulation of this intrinsic canonical signaling pathway has been reported by various groups, emerging evidence emphasizes the importance of ubiquitin-mediated post-translational modification of TGF-β/BMP signaling components [4], [5], [6]. Ubiquitination to target proteins has been found to be involved in a vast range of cellular process through regulating protein stability, signal transduction, endocytosis, trafficking, and enzyme activities [7], [8], [9], [10] and this versatile modification is mainly controlled by E3 ubiquitin ligases and deubiquitinases (DUBs) [11], [12], [13], [14], [15], [16]. In particular, deubiquitinases (DUBs) reverse the ubiquitination process by removing ubiquitin from target proteins [17] and have been recognized as critical regulators of several signaling pathways that are involved in a variety of human cancers [6,[18], [19], [20], [21], [22]].
However, compared to DUBs in the TGF-β signaling pathway, much less is known regarding which DUBs regulate the BMP signaling pathways and what are the underlying mechanisms of the DUBs. Among the DUBs, USP15 has been reported to deubiquitinate ALK3 (BMPRIA) through direct interaction, thereby promoting the phosphorylation of receptor-activated Smad1 [23]. Another group reported that USP15 opposes receptor-activated Smad (R-Smad) monoubiquitylation, which targets the DNA-binding domain of R-Smads and prevents promoter recognition, leading to enhanced activity of the TGF-β and BMP pathways [24]. Furthermore, AMSH, another DUB, has been reported to antagonize the function of Smad6, the inhibitory Smad of BMP signaling [25] and USP9X has been reported to negatively regulate the TGF-β/BMP pathway by deubiquitinating and stabilizing the Smurf1 E3 ligase [26]. Aside from these DUBs previously reported in the regulation of the BMP signaling pathway, much is still unknown about novel DUBs targeting other BMP receptors, except for ALK3 [23]. Therefore, investigation of other DUBs regulating BMP signaling components can contribute to detailed understanding of the molecular mechanism of the BMP signal transduction pathway.
In this study, we focused on the BMP6 signaling pathway in human colon cancers. The reason is that the role of BMP6 and related DUBs are not clearly addressed in colon cancers, although upregulation of BMP6 has been associated with the shorter survival of colorectal cancer patients through the analysis of the Gene Expression Omnibus public dataset [27]. Herein we identified a new DUB, named PSMD14 (POH1), that regulates the ALK2 receptor involved in BMP6 signaling pathway and revealed its molecular mechanism of regulating the ALK2 protein. Furthermore, we provide experimental evidence to demonstrate a role of PSMD14 in tumor growth and cancer stem cells/chemoresistance.
2. Materials and methods
2.1. Plasmids
All cloning procedures were performed after PCR gene amplification. Full-length human HA-ALK2 and HA-ALK3 complementary DNAs (cDNAs) were kind gifts from Dr. Susumu Itoh (Showa Pharmaceutical University, Tokyo, Japan). Using the HA-ALK2, HA-ALK3 plasmids as templates, ALK2 and ALK3 cDNA were subcloned into the EcoRI and XhoI sites of the pcDNA3-Flag vector (Invitrogen, Camarillo, USA) respectively. Full length human ALK6 cDNA was cloned into the EcoRI and XhoI site of the pcDNA3-HA and pcDNA3-Flag vector. Full length human PSMD14 cDNA was cloned into the EcoRI and XhoI site of pcDNA3-HA, pcDNA3-Flag vector. Plasmids expressing human wild-type HA-ubiquitin (HA-Ubi), HA-UbiK48, and HA-UbiK63 were subcloned into the pcDNA3-HA vector from His-Ubi, His-UbiK48, and His-UbiK63, respectively. These templates (His-Ubi, His-UbiK48 and His-UbiK63) were previously described [28]. The plasmids expressing other ubiquitin mutants (HA-UbiK6, HA-UbiK11, HA-UbiK27, HA-UbiK29, and HA-UbiK33) were provided by Dr. Jaewhan Song (Yonsei University, Korea). Human full length Smurf1 cDNA was cloned into the NotI and XhoI site of the pcDNA3-Flag vector and HindIII and XhoI site of the pcDNA3.1 vector (Invitrogen) respectively, resulting in Flag-Smurf1 and pcDNA-Smurf1. Human full length Smurf2 cDNA was cloned into the EcoRI and NotI site of the pcDNA3-Flag vector. Human full-length CHIP (STUB1) cDNA was cloned into the EcoRI and XhoI site of the pcDNA3-Flag vector. Human full length TRIM33 cDNA was cloned into the BamHI and XhoI site of the pcDNA3-Flag vector. BRE-Luc luciferase reporter plasmids were previously described [29]. cDNA of PSMD14 point mutant forms (H113Q, C120S, C120A) were generated by PCR using the QuikChange Mutagenesis kit (Stratagene, La Jolla, USA). To generate the reporter plasmids, ABCA7-Luc and ABCC4-Luc, promoter regions of abca7 and abcc4 genes were amplified by PCR using genomic DNA as a template and cloned into the BamHI and XhoI site of the pGL3-basic vector, respectively. All mutant forms generated by PCR were verified by sequencing. PCR primer sequences are described in the Table S1.
2.2. Cell culture, reagents and antibodies
HCT116 cells, a human colorectal cancer cell line, and DLD-1 cells, a human colorectal adenocarcinoma cell line, were maintained in RPMI (HyClone, Logan, UT) with 10% FBS (HyClone). RKO cells, a human colon carcinoma cell line, were maintained with DMEM (HyClone) with 10% FBS. HT29 cells, a human colorectal adenocarcinoma cell line, were maintained with McCoy's 5a medium (Thermo Fisher Scientific, Waltham, USA) with 10% FBS. Caco-2 cells and LS174T, a human colorectal adenocarcinoma cell lines, were maintained with EMEM (Thermo Fisher Scientific, Waltham, USA) with 10% FBS. All cell lines used in this study were purchased form the American Type Culture Collection (Manassas, USA) and routinely tested for mycoplasma contamination by PCR. Recombinant human BMP2, BMP4, BMP6 and BMP7 were purchased from R&D System (Mineapolis, USA). MG132 (M-1157) was purchased from A.G. Scientific, Inc. (San Diego, CA). Bortezomib (#2204) was obtained from Cell Signaling (Danvers, USA). To analyze drug resistance, oxaliplatin (O951; Sigma-Aldrich, St Louis, USA) and DAPT (D5942; Sigma-Aldrich) were purchased. Company names, catalog numbers, clone numbers, species and dilution ratio of the antibodies used for immunoblot, immunofluorescence, immunohistochemistry and FACS are described in Table S2.
2.3. Transfection of siRNA
We used Human ON-TARGETplus siRNA Library–Deubiquitinating Enzymes–SMARTpool (#G-104705) to screen for regulators of the BMP signaling pathway in colorectal cancer, obtained from Dharmacon (Lafyette, USA). Each DUB siRNA was transfected into HCT116 cells respectively. Every human DUB knockdown cell line was treated with 100 ng/ml BMP6 (R&D system) and incubated for 2 h before comparing levels of the target protein with the control. siRNAs were reverse-transfected using Lipofectamine RNAiMAX (Invitrogen).
2.4. Transfection and reporter assay
Plasmids were transiently transfected into HEK293 cells using PEI (Polyethyleneimine) and HCT116 cells using Lipofectamine 2000 (Invitrogen). PSMD14 knockdown cells were treated for the indicated time with BMP6 (100 ng/ml). Luciferase activity was performed with the dual-luciferase reporter assay system (Promega, Madison, USA). All experiments were independently repeated in at least triplicates for reproducibility.
2.5. RNA extraction and quantitative real-time RT-PCR
Total RNA of cell lines was isolated using the TRIzol reagent (Invitrogen). Reverse transcription was carried out by PrimScript Reverse Transcriptase (TaKaRa, Shiga, Japan). The primer sequences for each BMP target gene, Gapdh, and 48 ABC transporter genes are described in Table S3. For quantitative real-time RT-PCR, we used an iCycler real-time PCR machine and iQ SYBR Green Supermix (Bio-Rad, Hercules, USA) to measure gene expression levels. Expression of the genes was measured under the following cycling conditions: 45 cycles of 95 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s. Each quantitative real-time RT-PCR experiment was performed as described [30]. All reactions were independently repeated at least three times for reproducibility of experiment.
2.6. Construction of small hairpin RNAs and lentiviral/retroviral infection
The small hairpin RNA (shRNA) sequences specifically targeting endogenous human PSMD14 or human ALK2 are described in Table S4. Specific shRNAs were obtained from Mission-shRNA (Sigma-Aldrich). A lentiviral packaging system purchased from Invitrogen was used to express each shRNA lentivirus. Green fluorescence protein (GFP) targeting shRNA was used as a negative control for lentiviral infection. Generation of recombinant lentiviurses expressing each shRNA or retroviruses expressing Flag-PSMD14 was performed according to the protocols previously described [30].
2.7. Subcellular fractionation, immunoblotting and immunoprecipitation
The Subcellular Proteome Extraction Kit (Thermo scientific, Waltham, MA) was used to obtain the membrane, cytosolic, and nuclear fractions, respectively. For the immunoblot assay, lysis buffer (1% Triton X-100, 20 mM Hepes at pH 7.4, 150 mM NaCl, 12.5 mM b-glycerol phosphate, protein inhibitor cocktail, 10 mM NaF, 1 mM NaOV, 1.5 mM MgCl2, 2 mM DTT, 2 mM EGTA, 1 mM PMSF) was used to lyse cells. SDS-PAGE gel was used to separate protein extracts. Subsequently, protein extracts were transferred to a PVDF membrane 0.45 μm filter (Merk Millipore, Burlington, USA) and immunoblot analysis was performed. For immunoprecipitation, protein lysates were incubated with protein G agarose beads (GenDEPOT, Katy, USA) at 4 °C for 15 h. Protein lysates were washed three times with lysis buffer, and separated from the beads by boiling for 5 min with 2X sample buffer. Immunoblot and immunoprecipitation analysis was performed using the indicated antibodies.
2.8. Immunofluorescence
For immunofluorescence assays, PKH26 (PKH26GL, 1:1000; Sigma-Aldrich) was administered to cells for 10 min and cold methanol at −20 °C was used to fix cells for 10 min, and subsequently, 5% BSA in PBS solution was used for blocking at room temperature for 30 min. Next, samples were incubated with primary antibodies at 4 °C for 15 h. Mouse polyclonal anti-PSMD14 antibody (SAB1408741, 1:100; Sigma-Aldrich), rabbit polyclonal ALK2 antibody (orb314588, 1:100; Biorbyt, San Francisco, USA) were used to detect endogenous PSMD14 and ALK2, respectively. After washing with PBS 4 times for 10 min each, samples were stained with the following secondary antibodies at 4 °C for 15 h. Alexa Fluor-594-conjugated goat anti-rabbit IgG (1:200 for anti-ALK2; Invitrogen,), Alexa Fluor-488-conjugated goat anti-mouse IgG (1:200 for anti-PSMD14; Invitrogen) were used. Samples were stained with DAPI (D9542, 1:1000; Sigma-Aldrich) for 10 min and mounted on slides. A laser-scanning confocal microscope (Carl-Zeiss, Oberkochen, Germany) was used to observe samples.
2.9. Animal studies
All procedures for animal experiments were approved by the Animal Research Center of Sungkyunkwan University (Suwon, Korea) and performed in a manner compliant with all relevant ethical regulations regarding animal research. A pathogen-free barrier in the facility center was used to house all animals with 12 h light/dark cycles. Human HCT116 colorectal cancer cells (5 × 105) were infected with lentiviruses, injected into the flank fat pad of 5-week-old NOD/SCID female mice to measure tumor growth. Tumor volume was measured by the formula: (length) x (width)2 x 0.5.
2.10. Extraction of tissue proteins and immunohistochemistry of tumor xenografts
After injections of PSMD14-depleted, ALK2-depleted or control HCT116 cells into NOD/SCID mice, isolated tumor xenografts were hemisected for immunohistochemistry and immunoblot assay. For IHC, paraffin-blocked xenograft tumors were cut into 4 μm slices horizontally and staining was performed by primary antibodies described in Table S2. For protein extraction, chopped tumor samples were homogenized in RIPA buffer (89900; Thermos Fisher Scientific). Extracted samples were used for immunoblot analysis.
2.11. Human colorectal tumor microarray and immunohistochemistry
Paraffin-embedded human colorectal tumor microarrays (TMAs) containing normal and tumor colon tissues were purchased from US Biomax (Rockville, USA; https://www.biomax.us/Tissue_Arrays/tissue-arrays/Colon). For immunohistochemistry, each TMA slide was stained with rabbit anti-PSMD14 antibody (HPA002114, 1:100; Sigma-Aldrich), rabbit anti-ALK2 antibody (PA5-13881, 1:100; Thermos Fisher Scientific). Methylene blue was used to counterstain the TMA slides. After staining, slides were observed by a microscope and scored.
2.12. In vivo ubiquitination assay
In vivo ubiquitination assays were performed according to the protocols previously described [30]. Lysates were incubated at 4 °C for 15 h with the indicated antibodies and protein G agarose beads. The beads were washed four times with lysis buffer and samples were boiled for 5 min with 2X sample buffer. Immunoprecipitation samples were transferred onto PVDF membranes and the membranes were denaturated by 6 M guanidine hydrochloride buffer (20 mM Tris-HCl pH7.5 buffer containing 6 M guanidium chloride, 5 mM β-mercaptoethanol) at 4 °C for 30 min. Subsequently, membranes were washed with washing buffer three times. After the denaturation and washing steps, membranes were blocked in 5% BSA for 2 h and incubated with anti-FK2-HRP antibody (BML-PW9910; Enzo Life Sciences, Farmingdale, USA) at 4 °C overnight. Each ubiquitination was examined by an immunoblotting assay.
2.13. Colony forming assay
For soft agar colony formation, a 6-well plate was prepared in advance with 0.5% base agar that prevents cells from attaching to the plate. 1 × 104 HCT116 cells were seeded into the prepared 6-well plate with 0.35% top agar. Cells were incubated for 14 days at 37 °C and colonies with a diameter of >100 μm were counted. Each experiment was performed in triplicates.
2.14. Cell proliferation analysis
Cells were seeded in 12-well plates with 2 × 104 cells /well and cultured for 1–4 or 5 days. After the indicated time, cells were harvested and counted with a hemocytometer. All experiments were performed in triplicate for reproducibility. BrdU and MTT assays were performed in HCT116 cells, 1 × 104 cells were seeded onto a 96-well plate and incubated at 37 °C for 2 days. The BrdU assay was performed using a BrdU kit purchased from BD Biosciences (San Jose, CA). In the MTT assay, after the incubation of cells, the MTT solution (11465007001; Sigma-Aldrich) was added to each well and incubated for 1 h at 37 °C. Then, the media was discarded and 200 μl of DMSO was added into each well. Absorbance values at 490 nm were determined by a VersaMax ELISA microplate reader.
2.15. Chemoresistance analysis
HCT116 cells with lentiviruses were seeded in 96-well plates and incubated at 37 °C for 2 days. Cells were treated with 20 μM oxaliplatin and 30 μM DAPT for 12 h. After treatment of the anti-cancer drug, the MTT assay was performed to measure cell viability.
2.16. Flow cytometry
For FACS analysis, dissociated single cells were subjected to fluorescence-activated cell sorting (FACS) analysis using cell surface markers for CD44 (11-0441-91; Thermo Fisher Scientific) and CD133 (130-090-826; Miltenyi Biotec, Auburn, USA). The proportion of CD44-positive (+) and CD133-positive (+) populations were measured by FACS analysis using FACSCanto II (BD Biosciences) and data were analyzed by FlowJo 7.6.5. software.
2.17. Wound healing assay
HCT116 cells were plated in 12 well plates with 5 × 104 cells /well. Wounds were made by scratching with a pipette tip and BMP6 (100 ng/ml) was treated with indicated time points.
The quantification of wound areas was performed by Image J software.
2.18. Statistical analysis
The quantitative data in this study are presented as the means ± s.d. and were analyzed by a two-tailed unpaired Student's t-test or one-way and two-way ANOVA to compare the difference between groups. P < 0.05 was considered statistically significant. The public GSE21510, GSE68468 and GSE17538 datasets were downloaded from NCBI's Gene Expression Omnibus. The Kaplan-Meier (KM) Plotter Tool (http://kmplot.com/analysis) was used to show correlation between PSMD14 or ALK2 expression and the overall survival rates of colorectal cancer patients. Statistical significance was calculated by a log-rank test. GraphPad Prism 5 software (GraphPad, La Jolla, USA) was used in this study to calculate statistical significance.
3. Results
3.1. Identification of PSMD14 involvement in regulation of BMP signaling
To identify deubiquitinases (DUBs) involved in regulation of the BMP signaling pathway, we performed a loss-of-function screen for human DUBs in human HCT116 colorectal cancer cells using a human DUB siRNA library (Fig. S1a). After HCT116 cells were transfected with the respective siRNA and subsequently treated with BMP6 for 2 h, we examined the expression levels of the ID3 protein that is a major target protein of the BMP signaling pathway [31] by immunoblot analysis (Fig. S1b). The reason we chose the HCT116 cells and BMP6 in this study was because BMP6 mRNA was significantly increased in HCT116 cells compared to other cancer cell lines (Fig. S1c), and increased BMP6 has been reported to be associated with the shorter survival of colorectal cancer patients [27]. Through three consecutive screenings in HCT116 and RKO colon cancer cells, we found that depletion of the DUB enzyme PSMD14 downregulates ID3 expression in both cell lines (Fig. S1d), suggesting that PSMD14 may act as a putative regulator of BMP6-mediated signaling pathway in human colorectal cancer.
The deubiquitinase PSMD14, named POH1 or rpn1, has been known as a component of the 19S proteasomal lid of the 26S proteasome complex [32,33]. Aside from this intrinsic role, PSMD14 has been reported to be involved in diverse processes, including double-strand DNA breaks, embryonic stem cell differentiation, cell viability and cell cycle [34], [35], [36], [37]. Furthermore, PSMD14 specifically deubiquitinates several target proteins such as c-Jun, Mitf, ErbB2 receptor, and the E2F1 transcription factor [38], [39], [40], [41]. To date, there has been no report about a role for PSMD14 and its substrate in the regulation of the BMP signaling pathway.
To reveal a novel function of PSMD14 in the BMP signaling pathway, we first examined whether PSMD14 regulates ID3 expression in different colorectal cancer cell lines such as HCT116 and RKO cells (Fig. 1a). Depletion of PSMD14 significantly reduced expression of the ID3 protein in both cells upon BMP6 treatment (Fig. 1a). In addition, quantitative real-time RT-PCR analysis (qRT-PCR) showed that PSMD14 depletion decreases mRNA levels of Id3 as well as other target genes such as Id1 and SMAD6, in both cell lines upon BMP6 treatment (Fig. 1b). Together with these results, decreased activity of the BRE-Luc reporter upon BMP6 treatment in PSMD14-depleted HCT116 and RKO cells (Fig. 1c) suggests that PSMD14 is involved in BMP6 signaling pathway through targeting the upstream components required for transcriptional expression of BMP6-mediated target genes. Notably, decreased phosphorylation of Smad1/5 in PSMD14-depleted cells showed that PSMD14 targets upstream components required for Smad phosphorylation in BMP6 signaling (Fig. 1a). These findings were also supported by the reduced phosphorylation of Smad1/5 upon PSMD14 depletion in Smad4-null HT29 colon cancer cells which are defective in BMP6-mediated ID3 expression (Fig. 1d).
Fig. 1.
Characterization of DUB enzymes involved in regulation of BMP signaling in human colorectal cancer cells. (a) After HCT116 and RKO cells were reverse-transfected with specific siRNA against PSMD14 and control siRNA (siCON) and treated with 100 ng/ml BMP6 for the indicated times, immunoblotting assays were performed with the indicated antibodies. (b) Expressions of ID1, ID3, and SMAD6 mRNAs were analyzed by quantitative real-time RT-PCR (qRT-PCR) in PSMD14-knockdown and control HCT116 or RKO cells. Expression of the mRNAs detected by qRT-RCR were normalized to Gapdh mRNA. (c) BRE-Luc luciferase reporter assays were performed in PSMD14-knockdown and control HCT116 and RKO cells. In (b) and (c), bar graphs show the mean ± s.d. from three independent experiments. **P <0.01 (one-way ANOVA followed by Dunnett's test, n = 3, compared to the indicated controls). (d) After Smad4-null HT29 cells were reverse-transfected with PSDM14-specific siRNA or siCON and treated with BMP6, immunoblots were performed with the indicated antibodies. (e) HCT116 or RKO cells overexpressing Flag-PSMD14 were immunoblotted with the indicated antibodies upon treatment of BMP6. (f) Expressions of ID1, ID3, and SMAD6 mRNAs were analyzed by qRT-PCR in PSMD14-overexpressing HCT116 and RKO cells. In all immunoblot analyses, β-actin expression was used as a loading control and the images are representative of three independent experiments.
To further confirm whether Smad1/5 phosphorylation and expression of the downstream target genes are regulated by PSMD14, we generated stable HCT116 and RKO cells overexpressing the PSMD14 gene. PSMD14 overexpression increased basal Smad1/5 phosphorylation in the absence of BMP6 and also sustained the phosphorylation status up to 2 h upon treatment of BMP6 (Fig. 1e), eventually resulting in increased mRNA expression of downstream target genes (Fig. 1f) compared to the control cells. On the other hand, the total amount of Smad1/5/8 proteins were not affected by PSMD14 depletion or overexpression (Fig. 1a and e). Therefore, these results suggest that the deubiquitinase PSMD14 is involved in the BMP6 signaling pathway through targeting upstream components of Smad phosphorylation.
3.2. The role of PSMD14 in BMP6 signaling is independent of the 26S proteasome system
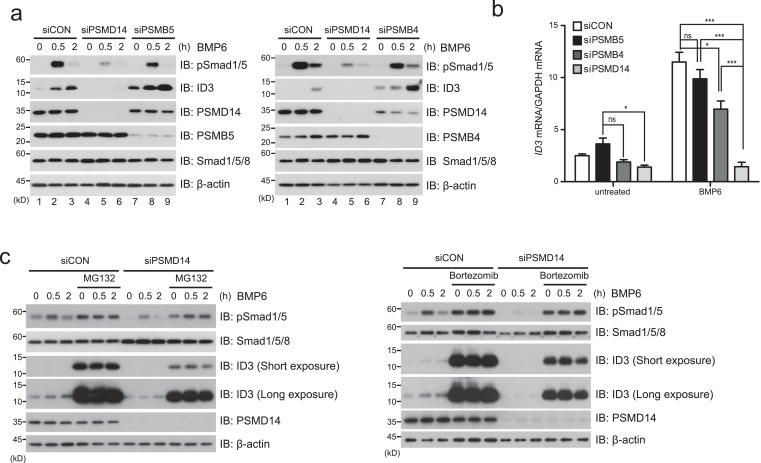
Because PSMD14 is known as a component of the 26S proteasome complex [32,33], it is possible that PSMD14 may regulate the upstream components required for Smad phosphorylation through the 26S proteasome system. To address whether the role of PSMD14 regulating the BMP6 signal is dependent or independent of the 26S proteasome system, we generated PSMB4- or PSMB5-depleted HCT116 cells because both genes encode essential components of the 26S proteasome system [42,43]. If the role of PSMD14 in BMP6 signaling is dependent on the 26S proteasome system, PSMB4- or PSMB5-depleted cells should show the same effects as PSMD14-depleted cells. In comparison to PSMD14-depleted HCT116 cells, PSMB5- and PSMB4-depleted cells did not reduce Smad1/5 phosphorylation (Fig. 2a; lane 7-9), suggesting that PSMD14 in the BMP6 signaling pathway acts independently of the 26S proteasome system. However, basal expression of the ID3 protein in PSMB5- and PSMB4-knockdown cells was slightly increased in the absence of BMP6 whereas Id3 mRNA expression was not (Fig. 2a and b). Although we do not know the exact reason, it is possible that basal expression of the ID3 protein might be regulated by the 26S proteasome at the post-transcriptional level. In fact, basal expression of ID3 increased upon pre-treatment with proteasome inhibitors such as MG132 and bortezomib (Fig. 2c). In addition, although Id3 mRNA expression in PSMB5- or PSMB4-depleted cells upon BMP6 treatment was not remarkably reduced compared to PSMD14-knockdown cells (Fig. 2b), the significant reduction of Id3 mRNA in PSMB4-depleted cells, compared to control or PSMB5-depleted cells, could be speculated to be due to the decreased expression of PSMD14 protein in PSMB4-depleted cells (Fig. 2a; right panel, lane 7-9). Notably, MG132 or bortezomib treatment did not affect the total levels of Smad1/5 in siCON- or siPSMD14-expressing HCT116 cells upon BMP6 treatment, and profoundly increased Smad1/5 phosphorylation in both cells even in the absence of BMP6 (Fig. 2c), implying that upstream proteins could be PSMD14 target proteins. These results provide robust evidence that PSMD14 is an important protein regulating the BMP6 signaling pathway beyond its intrinsic role as a component of the 26S proteasome complex.
Fig. 2.
The role of PSMD14 in the BMP6 signaling pathway is independent of the 26S proteasome system. (a) After PSMD14, PSMB5 or PSMB4 expression was depleted by specific siRNA against each mRNA in HCT116 cells and cells were subsequently treated with 100 ng/ml BMP6, cells were immunoblotted with the indicated antibodies. (b) Expression of ID3 mRNA was analyzed by qRT-PCR in PSMD14, PSMB4 or PSMB5-knockdown HCT116 cells, which were treated with BMP6. Expressions of ID3 mRNA were normalized to Gapdh mRNA. Bar graphs show the mean ± s.d. from three independent experiments. *P <0.05, **P <0.01, ***P< 0.001, ns; not significant (one-way ANOVA followed by Dunnett's test, n = 3, compared to the indicated controls). (c) After PSMD14-knockdown HCT116 cells were pre-treated with protease inhibitor MG132 or bortezomib, cells were treated with BMP6 for the indicated times and subsequently immunoblotted with the indicated antibodies. Expression of β-actin was used as a loading control. The images in all immunoblot analyses are representative of three independent experiments.
3.3. PSMD14 stabilizes the ALK2 protein via direct interaction
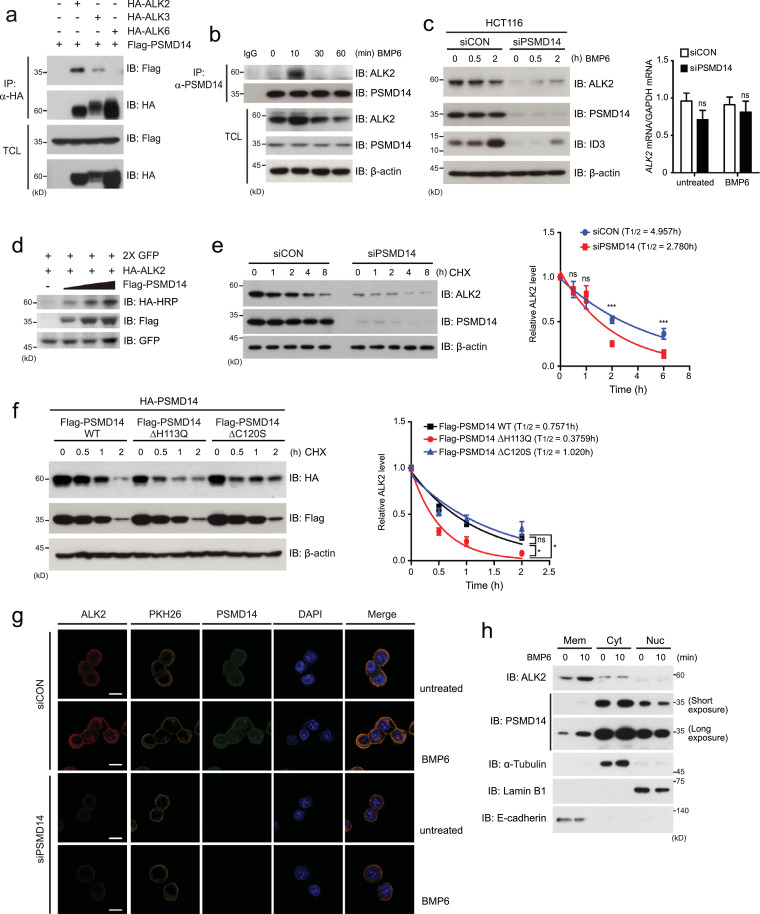
Since our present findings suggest that PSMD14 targets upstream components for Smad phosphorylation, we hypothesized that PSMD14 may regulate BMP receptors with serine-threonine kinase activity. To test our hypothesis, we first examined the interaction of PSMD14 with BMP type II receptors and type I receptors because BMP6 transmits its signal through the combination of three type I receptors (ALK2, ALK3 and ALK6) and three type II receptors (ActRII, ActRIIB and BMPRII) [2,44]. Co-immunopreciptitation (Co-IP) assays revealed stronger binding of PSMD14 to ALK2 than the other type I receptors, ALK3 and ALK6 (Fig. 3a). In addition, PSMD14 significantly interacted with the activin type II receptor ACTRIIB, compared to BMPRII or TGF-β type II receptor (TβRII) (Fig. S2a). Because ALK2 is an important type I receptor in BMP6 signaling and directly phosphorylates downstream Smad1/5/8, further experiments in this study focused on ALK2 regulation by PSMD14. In fact, immunoprecipitation assays strongly supported the endogenous interaction between PSMD14 and ALK2 upon BMP6 treatment (Fig. 3b). PSMD14 bound to ALK2 and increased the level of ALK2 protein at 10 min post BMP6 treatment and this complex was subsequently dissociated (Fig. 3b).
Fig. 3.
PSMD14 stabilizes ALK2 protein through direct binding. (a) After HEK293FT cells were co-transfected with the indicated plasmids, cells were immunoprecipitated (IP) with anti-HA antibody and subsequently immunoblotted (IB) with the anti-Flag or anti-HA antibody. (b) After HCT116 cells were treated with 100 ng/ml BMP6, endogenous PSMD14 was immunoprecipitated with anti-PSMD14 antibody and subsequently immunoblotted with the indicated antibodies against endogenous ALK2 and PSMD14 proteins. (c) Expression of endogenous ALK2 and ID3 proteins in PSMD14-depleted or control (siCON) HCT116 cells were examined by immunoblot analysis (left). Expression of ALK2 mRNA in PSMD14-depleted or control HCT116 cells was analyzed by qRT-PCR (right). (d) After a plasmid encoding HA-ALK2 was co-transfected into 293FT cells with dose-dependent expression of Flag-PSMD14, cells were immunoblotted with the indicated antibodies. (e) PSMD14-depleted or control (siCON) HCT116 cells were treated with 20 μM cycloheximide (CHX; protein synthesis inhibitor) for the indicated times. Cells were immunoblotted with anti-ALK2 and anti-PSMD14 antibodies. (f) After plasmids encoding wild-type (WT) Flag-PSMD14 and catalytic inactive DUB mutants of PSMD14 were respectively transfected into HCT116 cells, cells were treated with 20 μM CHX for the indicated times and immunoblotted. In (e) and (f), ALK2 levels were quantified by using ImageJ software and normalized to β-actin expression. The data were statistically analyzed and the bars represent the mean ± s.d. from three independent experiments. *P <0.05, ***P< 0.001 (one-way ANOVA followed by Dunnett's test, n = 3, compared to the indicated controls). (g)PSMD14-depleted and control (siCON) HCT116 cells were treated with 100 ng/ml BMP6 for 10 min and immunostained with the indicated antibodies to detect endogenous PSMD14 (green) and ALK2 (red). PKH26 molecule (yellow) and DAPI (blue) were used to stain the cell membrane and nuclei. Scale bar; 10 μm. (h) After HCT116 cells were treated with BMP6 for 10 min, cells were fractionated into membrane (Mem), cytoplasmic (Cyt) and nuclear (Nuc) extracts, which were subsequently immunoblotted with the indicated antibodies. Expressions of α-Tubulin, Lamin B1 and E-cadherin were used as markers and loading controls of cytosolic, nuclear and membrane fractions, respectively. Except for (h), expression of β-actin was used as a loading control of immunoblot analysis. The images in this figure are representative of three independent experiments.
These results prompted us to investigate whether PSMD14 with its DUB activity can regulate the stability of the ALK2 protein. To verify this possibility, we examined the expression of ALK2 mRNA and protein in PSMD14-depleted (siPSMD14) HCT116 or RKO cells in the absence or presence of BMP6, compared to control cells (siCON). The ALK2 mRNA expression was unchanged in both PSMD14-depleted cells, compared to control cells, but ALK2 protein levels significantly decreased upon PSMD14 depletion (Fig. 3c and S2b). Furthermore, dose-dependent expression of Flag-PSMD14 in HCT116 cells increased expression of the ALK2 protein (Fig. 3d). To further confirm these results, we examined the half-life of the ALK2 protein in the absence or presence of PSMD14 upon treatment of protein synthesis inhibitor cycloheximide (CHX) after a plasmid encoding HA-ALK2 protein was transfected into HCT116 cells together with or without Flag-PSMD14. The half-life of ALK2 was significantly increased by the expression of PSMD14 (Fig. S2c). In addition, the half-life of endogenous ALK2 was remarkably decreased in the PSMD14-knockdown HCT116 cell line treated with CHX (Fig. 3e).
Next, to address whether the DUB activity of PSMD14 is required for the stability of the ALK2 protein, we generated PSMD14 mutants in which the histidine at residue 113 and the cysteine at residue 120 were substituted into glutamic acid and serine or alanine, respectively, resulting in PSMD14-ΔH113Q, PSMD14-ΔC120S and PSMD14-ΔC120A [34,39]. After these PSMD14 mutants and wild-type PSMD14 were respectively co-transfected into HCT116 cells with HA-ALK2 and treated with CHX for the indicated times, the half-life of HA-ALK2 was measured. The mutants, PSMD14-ΔH113Q and PSMD14-ΔC120A, significantly decreased HA-ALK2 stability whereas PSMD14-ΔC120S did not (Fig. 3f and S2d). In addition, immunofluorescence analysis showed that ALK2 levels were significantly increased in siCON-expressing HCT116 cells upon BMP6 treatment together with co-localization of PSMD14 and ALK2 into the plasma membrane (Fig. 3g). Like the results displayed by the above immunoblots, basal ALK2 proteins levels were profoundly decreased in PSMD14-depleted HCT116 cells without BMP6 (Fig. 3g). To further verify the immunofluorescence results showing that a part of PSMD14 protein is localized into the plasma membrane, we performed the subcellular fractionation and immunoblot assays. After HCT116 cells were treated with BMP6 for 10 min, cells were separated into membrane, cytoplasmic and nuclear fractions, respectively. Because PSMD14 has been known as a component of 26S proteasome, PSMD14 was mainly localized into cytosol (Fig. 3h). However, small amounts of PSMD14 were observed in the membrane fraction at the basal state (Fig. 3h). Interestingly, the amounts of PSMD14, localized into the membrane, were increased by BMP6 treatment (Fig. 3h), although we do not know the exact reason. In contrast, ALK2 proteins were mainly localized into the membrane and increased by BMP6 treatment (Fig. 3h). Based on our current findings, the increased ALK2 levels upon BMP6 treatment seem to be due to the ALK2 stabilization by PSMD14 in the membrane. Furthermore, PSMD14 was also observed in the nuclear fraction, suggesting an unknown function of PSMD14 in the nucleus. These results collectively suggest that PSMD14 binds to ALK2 and is involved in the stability of ALK2 through its DUB activity.
3.4. PSMD14 deubiquitinates K48-linked polyubiquitination of the ALK2 protein
Although it has been reported that the ALK3 protein is deubiquitinated by USP15 [23], the deubiquitination of ALK2 and ALK6 as well as their polyubiquitination are poorly understood. To address whether PSMD14 deubiquitinates the ALK2 protein, we first examined the ubiquitination patterns of three BMP type I receptors, ALK2, ALK3, and ALK6, after plasmids encoding Flag-ALK2, Flag-ALK3 and Flag-ALK6 were respectively co-transfected into HCT116 cells in the absence or presence of HA-tagged ubiquitin (HA-Ubi). Immunoprecipitation with anti-Flag antibody and immunoblot analysis with HA-HRP antibody showed polyubiquitination of ALK2 and ALK3 proteins, but not ALK6 (Fig. 4a). These results led us to examine whether ALK2 or ALK3 is deubiquitinated by PSMD14. Deubiquitination assays showed that PSMD14 specifically deubiquitinates polyubiquitinated ALK2 and causes increased expression levels of ALK2 protein in total cell lysates, but not ALK3 (Fig. 4b).
Fig. 4.
PSMD14 stabilizes ALK2 protein through deubiquitinating Smurf1-mediated polyubiquitination of ALK2 (a, b) Plasmids encoding Flag-ALKs, Flag-PSMD14 or HA-Ubi were co-transfected into HEK293FT cells according to the indicated combinations. Ubiquitination of Flag-ALK2, Flag-ALK3, and Flag-ALK6 were examined by immunoprecipitation (IP) and immunoblots (IB) with the indicated antibodies. (c) Flag-ALK2 and Flag-PSMD14 were co-transfected into HEK293FT cells with a plasmid encoding wild-type or lysine mutant (K48 or K63) HA-Ubi in the indicated combinations. When the K48 mutant of His-Ubi was transfected, MG132 was treated to prevent ALK2 degradation. (d) The catalytically inactive DUB mutant of Flag-PSMD14 (H113Q, C120A or C120S) and wild-type Flag-PSMD14 were co-transfected into HEK293FT cells with Flag-ALK2 and HA-Ubi in the indicated combinations. (e) After PSMD14-depleted and control HCT116 cells were treated with 100 ng/ml BMP6 for the indicated times, ubiquitination of endogenous ALK2 protein was examined by IP and IB with the indicated antibodies. IgG was used as a negative control for IP. (f) After a plasmid encoding HA-ALK2 was co-transfected into HEK293FT cells with plasmids encoding Flag-Smurf1, Flag-Smurf2, Flag-CHIP, or Flag-TRIM33, respectively, co-immunoprecipitation assays were performed with the indicated antibodies. (g) Flag-Smurf1 or Flag-Smurf2 was co-transfected into HEK293FT cells with HA-Ubi and Flag-ALK2 in the indicated combinations. (h) Flag-ALK2 was co-transfected into HEK293FT cells with wild-type His-Ubi or a lysine mutant (K48 or K63) of HA-Ubi in the absence or presence of Flag-Smurf1. (i) Flag-Smurf1 and the K48 lysine mutant of HA-Ubi were co-transfected into HEK293FT cells with wild-type Flag-PSMD14 or the catalytic inactive DUB mutant of PSMD14 (H113Q). In (a)-(d), ubiquitination of Flag-ALK proteins were examined by immunoprecipitation (IP) and immunoblots (IB) with the indicated antibodies. In (g)-(i), ubiquitination of the ALK2 protein was analyzed by IP and IB with the indicated antibodies. In (h) and (i), cells were pre-treated with MG132 to prevent protein degradation. Expression of β-actin was used as a loading control. The images in immunoblot analyses are representative of three independent experiments.
To identify which polyubiquitination pattern of ALK2 is deubiquitinated by PSMD14, wild-type ubiquitin (HA-Ubi), the K48 ubiquitin mutant (HA-Ubi-K48) in which six lysine residues except for lysine 48 are substituted into arginines, and the K63 ubiquitin mutant (HA-Ubi-K63) in which only lysine 63 is left intact, were transfected into HCT116 cells with or without Flag-PSMD14. When the plasmid encoding the HA-Ubi-K48 mutant was transfected with the indicated combinations, cells were pre-treated with MG132 for 8 h to prevent proteasomal degradation of Flag-ALK2. The K48-linked polyubiquitination of the ALK2 protein was significantly deubiquitinated by PSMD14 (Fig. 4c). Other polyubiquitination patterns of ALK2 mediated by other ubiquitin mutants, including HA-Ubi-K11, were not affected by PSMD14 (Fig. S3). The PSMD14 mutants with impaired deubiquitinase activity, i.e. Flag-PSMD14-ΔH113Q and Flag-PSMD14-ΔC120A, did not deubiquitinate the Flag-ALK2 as much as wild-type PSMD14 (Fig. 4d). However, Flag-PSMD14-ΔC120S still deubiquitinated the ALK2 protein (Fig. 4d), which was similar to our previous finding that this mutant did not reduce the stability of ALK2 upon cycloheximide treatment (Fig. 3f), indicating that substitution of cysteine at residue 120 into serine does not affect the deubiquitinase activity of PSMD14.
To further verify our results, we examined whether the deubiquitination of endogenous ALK2 protein is regulated by PSMD14. Polyubiquitination of endogenous ALK2 protein was reduced at 10 min after BMP6 treatment in siCON-expressing HTC116 cells, with increased expression of ALK2 in total cell lysates, and this reduced polyubiquitination was subsequently recovered after 10 min (Fig. 4e). This time point was similar to the time point of endogenous ALK2 interaction with PSMD14 (Fig. 3b). In comparison to control cells, reduction of polyubiquitinated ALK2 was not observed at the same time point in PSMD14-knockdown HCT116 cells, but the polyubiquitination pattern of endogenous ALK2 was significantly increased and sustained upon BMP6 treatment (Fig. 4e). The levels of endogenous ALK2 protein in PSMD14-depleted cells were profoundly reduced even compared to the basal state without BMP6 (Fig. 4e), which is due to the increased polyubiquitination of endogenous ALK2 causing proteasomal degradation. Considering these results, it is evident that PSMD14 regulates the stability of ALK2 through deubiquitinating the K48-linked polyubiquitination of ALK2.
3.5. K48-linked polyubiquitination of ALK2 by Smurf1 is counteracted by PSMD14
Our current findings prompted us to investigate which E3 ubiquitin ligase induces K48-linked polyubiquitination of ALK2 regulated by PSMD14. Although E3 ubiquitin ligases have been extensively studied in the TGF-β/BMP signaling pathway [45], [46], [47], [48], [49], [50], E3 ligases targeting the ALK2 protein have not been clearly addressed. Based on published literature, we generated expression plasmids encoding Smurf1, Smurf2, CHIP, and TRIM33, respectively, and examined the interaction of each E3 ligase with ALK2 in HEK293 cells. Co-immunoprecipitation assays revealed that Smurf1 and CHIP proteins directly bind to ALK2 protein (Fig. 4f). Next, we examined which protein is required for ALK2 stability because ALK2 protein is degraded through K48-linked polyubiquitination. Ectopic expression of Smurf1 in HCT116 cells significantly reduced the expression of ALK2, but CHIP or Smurf2 expression did not (Fig. S4a). In addition, Smad1/5 phosphorylation and ALK2 and ID3 protein expression was significantly increased in Smurf1-depleted HCT116 cells, compared to siCON-expressing control cells (Fig. S4b). Although expression of Smurf2, which did not directly bind to ALK2, was used as a negative control, Smurf2 depletion in HCT116 cells also increased Smad1/5 phosphorylation and ID3 expression to the same levels as Smurf1-depleted cells and slightly augmented the expression of ALK2 (Fig. S4b). Although we do not know the exact reason, based on the finding that Smurf1 interacts with Smurf2 [51], it is possible that Smurf2 acts as a single complex through binding to Smurf1 and indirectly affects ALK2 stability.
To further verify the role of Smurf1 as an E3 ligase inducing the K48-linked polyubiquitination of ALK2, we next examined the ubiquitination pattern of ALK2 by Smurf1 expression. The ubiquitination assays indicated that Smurf1 induces K48-linked polyubiquitination of the ALK2 protein (Fig. 4g, lane 3 and 4 h, lane 2, 4). The increased K48-linked polyubiquitination of ALK2 by Smurf1 was clearly deubiquitinated by wild-type PSMD14, but not the PMSD14 mutant (PSMD14-H113Q) with impaired DUB activity (Fig. 4i, lane 3-5). Therefore, these results indicate that K48-linked polyubiquitination of ALK2 mediated by the Smurf1 protein is deubiquitinated by PSMD14, resulting in the augmentation of ALK2 stability.
3.6. The PSMD14-ALK2 axis is required for tumorigenesis of colorectal cancer cells
Although accumulating evidence emphasizes the importance of the BMP signaling pathway in cancer, evidence on its role in colorectal cancers has been ambivalent. For example, BMP2 and BMP4 have been reported as tumor suppressors in colorectal cancers whereas BMP7 has been reported to be responsible for an oncogenic function [52], [53], [54], [55]. The role of the BMP6 signaling pathway remains unknown in the field of colorectal cancers. Therefore, our findings that PSMD14 targets the ALK2 receptor in colorectal cancers regarding the initiation of BMP6-mediated signaling pathway led us to verify the function of PSMD14 in the tumorigenesis of colorectal cancers. To examine whether BMP6 is involved in the proliferation of HCT116 colorectal cancer cells, HCT116 cells expressing siRNA against GFP (shGFP) were treated with BMP6 for the indicated times. BMP6 treatment significantly increased the numbers of shGFP-expressing HCT116 cells, compared to untreated cells (Fig. 5a). These results prompted us to hypothesize that the PSMD14 and ALK2 proteins are required for BMP6-induced proliferation of HCT116 cells.
Fig. 5.
PSMD14 and ALK2 are required for BMP-mediated colon cancer tumorigenesis. (a) For the analysis of cell proliferation, 1 × 103 cells for each cell line were cultured in 12-well plates in the absence or presence of 100 ng/ ml BMP6 for the indicated times. Cell numbers were counted at the indicated time points. (b) BrdU incorporation assays were used to detect cell proliferation. (c) MTT assays were performed to analyze cell viability. (d) For colony forming assays, cells were seeded in 6-well plates with soft-agar media and incubated for 14 days. Colonies were counted and described as graphs demonstrating the percentage of colonies in the field. In (a)-(d), the data were statistically analyzed and the error bras represent the mean ± s.d. *P <0.05, **P <0.01, ***P< 0.001, ns; not significant (one-way ANOVA followed by Dunnett's test. n = 3 per group, compared to the indicated controls). (e)1 × 106PSMD14-depleted, ALK2-depleted or control HCT116 cells were injected in the NOD-SCID mouse group (n = 5 per group). The tumorigenesis experiment was processed for 30 days. (f) Cell lysates isolated from each tumor xenograft were immunoblotted with the indicated antibodies. Expression of β-actin was used as a loading control. The images in immunoblot analyses are representative of three independent experiments. (g) Tumor sizes (n = 5 per group) were calculated every 5 days. (h) After isolating each tumor from mice (n = 5 per group), tumor weights were measured and calculated as the average per group. In (g) and (h), the data were statistically analyzed and the error bras represent the mean ± s.d. ***P< 0.001, ns; not significant (one-way ANOVA followed by Dunnett's test. n = 5 per group, compared to the indicated controls). In (a)-(h), PSMD14- and ALK2-depleted HCT116 cells were generated by the infection of recombinant lentiviruses expressing shRNAs targeting ALK2 or PSMD14.
To verify our hypothesis, PSMD14 and ALK2 genes were respectively silenced by the infection of two independent lentiviruses expressing different PSMD14 or ALK2-specific shRNAs in the HCT116 cells, resulting in HCT116-shPSMD14 #1, HCT116-shPSMD14 #3, HCT116-shALK2 #3, and HCT116-shALK2 #5 cells, and thus cell proliferation was analyzed in the absence or presence of BMP6. Cell numbers, BrdU and MTT assays consistently indicated that both PSMD14 and ALK2 depletion significantly decrease the proliferation and viability of HCT116 cells irrespective of BMP6 treatment, compared to shGFP-expressing control cells (Fig. 5a–c). In addition, colony forming assays showed that PSMD14 and ALK2 depletion in HCT116 cells inhibit the formation of tumor colonies. Although these results support that the PSMD14-ALK2 axis in the BMP6 signaling pathway is involved in tumor growth, it is possible that this axis may affect the migration of colorectal cancers. To answer this question, we performed wound healing assays and also examined the markers of epithelial-mesenchymal transition (EMT) in PSMD14-depleted or control HCT116 cells. The wound healing assays were done with the same numbers of PSMD14-depleted cells as the control cells, because PSMD14-depleted HCT116 cells showed the reduced growth. PSMD14 depletion did not show the significant reduction of cell migration (Fig. S5a and b). In addition, expressions of EMT markers such as E-cadherin and Vimentin were unchanged between PSMD14-depleted and the control HCT116 cells irrespective of BMP6 treatment (Fig. S5c). These results suggest that the PSMD14-ALK2 axis is required for cell growth, but not cell migration.
Next, we examined the possibility that the PSMD14-ALK2 axis regulates the proliferation of colorectal cancer cells through modulating other target proteins of PSMD14 such as c-Jun, Mitf, and ErbB2 which have been previously reported [38], [39], [40]. To this end, we investigated expressions of ErbB2, c-Jun, and Mitf in both PSMD14- and ALK2-depleted HCT116 cells upon BMP6 treatment, together with siCON-expressing control cells. The expression of ErbB2 gene was slightly decreased by PSMD14 depletion whereas it was not affected by ALK2 depletion (Fig. S6a), suggesting that the regulation of ErbB2 protein is independent of the PSMD14-ALK2 axis. In contrast, c-Jun expression was significantly increased in PSMD14-depleted HCT116 cells irrespective of BMP6, but it was not affected by ALK2 depletion, compared to control cells. These results are different from the previous results that ectopic expression of PSMD14 (POH1) in HEK293 cells has been known to stabilize c-Jun [39]. This discrepancy seems to be due to cell-context dependency or some artifacts of overexpression system, because previous report did not show the loss-of-function experiments regarding the PSMD14. In our opinion, the increased level of c-Jun in PSMD14-depleted HCT116 cells might be due to the defect of 26S proteasome system. In fact, the expression of c-Jun in PSMB4- or PSMB5-depleted HCT116 cells, like the experiments of Fig. 2a, was similarly increased as much as in PSMD14-depleted ones (Fig. S6b), suggesting that the change of c-Jun protein in PSMD14-depleted HCT116 cells may be due to the decreased activity of 26S proteasome. We also examined the expression of Mitf, which is an interacting protein of PSMD14 (POH1) as a transcription factor for osteoclast differentiation. Previous results suggest that PSMD14 interacts with Mitf, and prevents Mitf ubiquitination, thus increasing the Mitf activity in osteoclast and RAW264.7 cells [40]. However, Mitf expression was not affected in PSMD14- or ALK2-depleted HCT116 cells, compared to control cells (Fig. S6a). Therefore, the regulation of Mitf by PSMD14 in osteoclasts may not be relevant for colorectal cancer cells. Collectively, our results indicate that the PSMD14-ALK2 axis is not involved in the regulation of c-Jun, ErbB2 and Mitf proteins upon BMP6 treatment.
Next, to address the in vivo role of PSMD14 and ALK2, either PSMD14- or ALK2-depleted HCT116 cells were subcutaneously injected into the flanks of SCID mice, and we analyzed tumor volume and tumor weight of these cancer cells at 30 days post-injection. Both PSMD14 and ALK2 depletions significantly decreased tumor growths compared to control cells (Fig. 5e). The depletion of PSMD14 or ALK2 in these tumor xenografts was confirmed by immunoblot analysis and immunohistochemistry (IHC) (Fig. 5f and S7a). Immunoblot and IHC of tumor xenografts regarding the expression of Ki67 supported the reduced growth by PSMD14 and ALK2 depletions (Fig. 5f and S7b). In addition, PSMD14 depletion in tumor xenografts decreased the expression of endogenous ALK2 protein, together with the decreased expression of target proteins, including phospho-Smad5 and ID3, modulated by the PSMD14-ALK2 axis (Fig. 5f, S7a and b). In addition, tumor volume and weight were clearly reduced in PSMD14- and ALK2-depleted xenograft models (Fig. 5g and h). These results demonstrate that the PSMD14-ALK2 axis is responsible for the tumorigenesis of colon cancers.
3.7. PSMD14 is required for chemoresistance through ABC transporters and cancer stemness
Since cancer stemness and chemoresistance have been recognized as key factors inducing tumorigenesis and tumor recurrence [56], [57], [58], [59], [60], we next examined whether the PSMD14-ALK2 axis in the BMP6 signaling pathway is involved in cancer stemness and chemoresistance of colorectal cancers. FACS analysis indicated that both PSMD14- and ALK2-depleted HCT116 cells, which are widely used in cancer stemness, significantly decrease CD133+/CD44+ cancer stem cell (CSC) populations of HCT116 cells at the basal state without BMP6 treatment (Fig. 6a and b). In addition, the increased CSC populations of shGFP-expressing control HCT116 cells upon BMP6 treatment was not observed in PSMD14- and ALK2-depleted cells (Fig. 6a and b). Furthermore, sphere forming assays showed that both PSMD14- and ALK2-depletion inhibit the formation of tumor spheres of HCT116 cells (Fig. 6c), supporting our FACS data, and immunoblot analysis also revealed that expression of stem cell transcription factors induced by BMP6, including Oct4, Sox2, and Nanog, are significantly reduced in PSMD14- and ALK2-depleted HCT116 cells (Fig. 6d). Next, we examined whether PSMD14 and ALK2 in the BMP6 signaling pathway is crucial for chemoresistance of colorectal cancer cells, because cancer stemness is highly associated with chemoresistance [61], [62], [63]. Both PSMD14 and ALK2 depletion profoundly reduced cell viability of HCT116 colon cancer cells following treatment with anti-cancer drugs, oxaliplatin and DAPT (Fig. 6e). To further verify an importance of PSMD14-ALK2 axis in chemoresistance, we examined whether expressions of 48 ATP binding cassette (ABC) transporter genes regulated by BMP6. The reason is that ABC transporters are key mediators of the efflux of anticancer agents form cancer cells and play a crucial role in acquired resistance of cancer cells to anti-cancer drugs [64], [65], [66], [67]. In addition, it still remains unknown about the relationship of BMP6 signaling pathway with ABC transporters. Quantitative RT-PCR analysis indicated that expressions of 9 ABC transporter genes are significantly upregulated in HCT116 cells more than five folds upon BMP6 treatment, compared to expression of Id3 mRNA (Fig. S8a). Among 9 ABC transporters, expressions of both ABCA7 and ABCC4 genes were commonly decreased in both PSMD14- and ALK2-depleted HCT116 cells (Fig. 6f, g and S8b), suggesting that ABCA7 and ABCC4 are key targets regulated by BMP6-ALK2-PSMD14 axis. In addition, reporter assays using ABCA7 or ABCC4 promoter-luciferase plasmid supported that these two genes are induced by BMP6 treatment (Fig. S8c) and both ABCA7 and ABCC4 depletion reduced cell viability of HCT116 colon cancer cells following treatment with anti-cancer drugs, oxaliplatin and DAPT (Fig. 6h). These results indicate that the PSMD14-ALK2 axis plays a crucial role in cancer stemness and chemoresistance of colon cancers through regulating the BMP6 signaling pathway.
Fig. 6.
PSMD14 depletion reduces BMP6-mediated colorectal cancer stemness. (a, b) FACS analysis of CD133+/CD44+ cells in PSMD14- or ALK2-depleted HCT116 cells, which were treated with 100 ng/ml BMP6 for 48 h. The proportion of the CD133+/CD44+ fraction was described with the density plots (a) and a bar graph (b). shGFP-expressing HCT116 cells were used as a control. (c) Sphere forming assay of PSMD14- or ALK2- depleted HCT116 cells. Spheres with a diameter above 50 μm were counted and described in a bar graph. Scale bars, 50 μm. (d)PSMD14- or ALK2-depleted HCT116 cells were treated with BMP6. Expression of pluripotent transcription factors were analyzed by immunoblotting with the indicated antibodies. siCON-expressing HCT116 cells were used as a control. (e, h) 2 × 104 cells of PSMD14-, ALK2-, ABCA7- or ABCC4-depleted HCT116 were respectively treated with 20 μM oxaliplatin and 30 μM DAPT and their viabilities were measured at 6 h. shGFP-expressing HCT116 cells were used as a control. (f, g)PSMD14- or ALK2-depleted HCT116 cells were treated with BMP6 for 6 h. Expressions of ABCA7 and ABCC4 were measured by quantitative RT-PCR. The data were statistically analyzed by two-way ANOVA followed by Bonferroni's multiple comparison test (n = 3, ***P< 0.001 compared to the indicated controls. ns; not significant). The bars represent the mean ± s.d. The images in this figure are representative of three independent experiments. In (b), (c), (e) and (h) were statistically analyzed by one-way ANOVA followed by Dunnett's test (n = 3, **P <0.01, ***P< 0.001 compared to the indicated controls. ns; not significant).
To further support the importance of the PSMD14-ALK2 axis in chemoresistance, however, we have to demonstrate why ABCA7 and ABCC4 genes are induced at 6 h upon BMP6 treatment, because endogenous ALK2 expression was increased at early time and subsequently decreased after 1 h upon BMP6 treatment (Fig. 3b). To address this discrepancy, we examined time-dependent expressions of ALK2 and ABC transporter proteins, ABCA7 and ABCC4, in HCT116 cells upon BMP6 treatment. Immunoblot analysis indicated that ALK2 expression is subjected to be dually regulated (Fig. S8d). That is, after ALK2 expression was increased at 10 min post BMP6 treatment and gradually decreased until 2 h, it was re-upregulated at 4 h (Fig. S8d). In contrast, ABCA7 and ABCC4 genes were maximally induced after 4 h (Fig. S8d). Based on these results, we questioned how ALK2 expression is re-increased at 4 h post BMP6 treatment and thus hypothesized that BMP6, secreted by autocrine pathway after initial BMP6 treatment, is responsible for the increased expression of ALK2 at late stage. To verify this hypothesis, we examined time-dependent expression of bmp6 mRNA in HCT116 cells by qRT-PCR. After initial BMP6 treatment on HCT116 cells, endogenous bmp6 mRNA was significantly increased after 4 h and decreased at 24 h (Fig. S8e). Therefore, ALK2 expression in HCT116 cells seems to be dually regulated at early and late stages, due to exogenous initial treatment of BMP6 and the increased BMP6 by autocrine pathway.
3.8. PSMD14 expression is associated with poor prognosis of colorectal cancer patients
Since our present data indicates that PSMD14 plays an essential role in the BMP6 signaling pathway through deubiquitinating the ALK2 receptor, and this PSMD14-ALK2 axis is required for tumorigenesis and cancer stemness of colon cancers, we next investigated the correlation between PSMD14 and ALK2 expression in colorectal cancer cell lines and public microarray datasets of cancer patients. Immunoblot analysis showed that both PSMD14 and ALK2 expressions are significantly correlated in colorectal cancer cell lines we tested (Fig. 7a). Analysis of the expression of PSMD14 and ALK2 in a public microarray dataset (GSE21510) of 148 colorectal cancer patients indicated that both PSMD14 and ALK2 are significantly upregulated in the cancer tissues of patients compared to normal tissues (Fig. 7b and c). Moreover, cancer tissues highly expressing the PSMD14 gene also showed significantly increased ALK2 expression (Fig. 7d). Another public dataset (GSE68468) of 288 colorectal cancer patients revealed that expression of both PSMD14 and ALK2 are clearly increased in cancer tissues of patients, compared to polyps of patients and normal tissues (Fig. 7e). Immunohistochemistry of tissue microarray (TMA) of normal and tumor tissues derived from 70 colon cancer patients corroborated our findings, demonstrating higher expression and correlation of both genes (Fig. 7f-h and S9). Furthermore, analysis of the overall survival of 224 colorectal cancer patients profoundly demonstrated an association of higher expression of PSMD14 and ALK2 with poor survival of colorectal cancer patients (Fig. 7i and j). Therefore, our findings suggest that PSMD14 and ALK2 expression are significantly correlated with the progression of colorectal cancers and survival of cancer patients, emphasizing the crucial role of BMP6 signaling pathway in colon cancers.
Fig. 7.
Higher expression of PSMD14 and ALK2 confer poor prognosis in human colorectal cancer. (a) Cell lysates isolated from the indicated colorectal cancer cell lines were immunoblotted with the ALK2 and PSMD14 antibodies. Expression of β-actin was used as a loading control. The images in immunoblot analyses are representative of three independent experiments. (b, c) Using the Kaplan-Meier (KM) plotter tool, expression of PSMD14 and ALK2 mRNA between normal and tumor tissues of colorectal cancer patients in a public GEO dataset (GSE21510; n = 148) were analyzed. (d) To verify the correlation of ALK2 mRNA expression in normal colon tissues and tumor tissues with the expression levels of PSMD14, a public GSE dataset (GSE21510) was analyzed by the KM plotter tool. In (b)-(d), the boxes represent the interquartile range, centre is the median, and the minimum and maximum values are represented in the whiskers. ***P<0.001, ns; not significant (Student t-test, compared to normal tissues or PSMD14-low samples). (e) Correlation of ALK2 and PSMD14 mRNAs in normal colon tissues, polyps in colon, and colorectal tumor tissues of human colorectal cancer patient samples (GSE68468, n = 288). Samples were classified into three groups (double low expression of ALK2 and PSMD14, single high expression of ALK2 or PSMD14, and double high expression of ALK2 and PSMD14) in normal tissues, polyps and tumors. (f, g) Scatter dot plot analysis represents the scores of the expression of PSMD14 and ALK2 protein in the matched normal and tumor tissues of human colon cancer patients (normal tissue n = 70, cancer tissue n = 70) analyzed by immunohistochemistry. ***P<0.001 (Student t-test, compared to normal tissues). (h) Scatter dot plot shows Spearman correlations between ALK2 and PSMD14 protein expression according to scoring within all samples. *** P < 0.001 (Student t-test). The Spearman r indicates the Spearman correlation coefficients. (i, j) Correlation of PSMD14 or ALK2 mRNA expression with overall survival rates of human colorectal cancer patients were analyzed by a KM plot analysis in a public GSE dataset (GSE17538; n = 224 patients). P = 0.0372, P = 0.0021 (Log-rank test). HR = hazard ratio.
4. Discussion
Accumulating evidence has revealed essential functions of the BMP signaling pathway in diverse cellular contexts, including embryonic development, cellular lineage commitment, morphogenesis, differentiation, proliferation, and apoptosis. However, the ubiquitinating/deubiquitinating modifications of BMP signaling components has not been as fully understood as the TGF-β signaling pathway. We here identified the deubiquitinating enzyme PSMD14 regulating the stability of the ALK2 type I receptor in the BMP6 signaling pathway and thus demonstrated the importance of the PSMD14-ALK2 axis in the BMP6 signaling pathway regarding the proliferation, cancer stemness, and chemoresistance of colon cancers (Fig. 8).
Fig. 8.
Schematic representation of the proposed mechanism of BMP6 signaling pathway regulated by PSMD14-ALK2 axis in colorectal cancers. ALK2 type I receptor is polyubiquitinated by E3 ligase Smurf1 and subsequently degraded in the absence of BMP6. Upon treatment of BMP6, PSMD14 binds to ALK2 protein and deubiquitinates the K48-linked polyubiquitin chains of ALK2 protein, resulting in the increase of ALK2 stability and thus leads to the initiation of BMP6 signaling pathway. This initiation of BMP6 signaling pathway facilitates tumor growth, cancer stemness and chemoresistance in colorectal cancers.
Although several studies have suggested that PSMD14 is responsible for important biological processes such as DNA repair, cell differentiation, transcriptional control and cell cycle, which are crucial pathways in cancer progression, beyond being a component of the 26S proteasome complex [34], [35], [36], [37], [38], [39], [40], [41], it remains unknown whether PSMD14 is involved in the BMP signaling pathway and which components are the substrates regulated by PSMD14. Thus, this is the first report to demonstrate that the deubiquitinating enzyme PSMD14, independent of the 26S proteasomal system, is involved in the initiation of BMP6 signaling through deubiquitinating the ALK2 type I receptor.
Our studies provide evidence that the ALK2 type I receptor may be liable to be degraded at the basal state without BMP6 treatment. That is, the instability of the ALK2 protein at the basal state is due to K48-linked polyubiquitination of ALK2 by the Smurf1 E3 ligase, and BMP6 treatment initiates BMP6 signaling by stabilization of the ALK2 receptor through removing the K48-linked polyubiquitin chains by the PSMD14 deubiquitinating enzyme. In the BMP signaling pathway, the Smurf1 E3 ubiquitin ligase is known to degrade the Smad1/5 proteins through K48-linked polyubiquitination [50]. However, it has not been clearly addressed whether the ALK2 receptor is a substrate of the Smurf1 protein in the BMP pathway. Therefore, our experiments, including co-immunoprecipitation, Smurf1 depletion, and ubiquitination assays, also suggest that the ALK2 receptor is a substrate of Smurf1. These results are consistent with a recent report that AMP-activated kinase downregulates ALK2 through enhanced interaction between Smad6 and Smurf1 in osteogenic differentiation [68].
More interestingly, it appears that the antagonistic activity of PSMD14 on ALK2 ubiquitination is specific to the ALK2 type I receptor. Although ALK2 and ALK3 respectively bound to PSMD14, ubiquitination assays revealed that PSMD14 specifically deubiquitinates only ALK2. In contrast, another type I receptor ALK6 did not interact with nor is deubiquitinated by PSMD14. However, PSMD14 depletion in HCT116 and RKO cells showed decreased Smad1/5 phosphorylation or reduced ID3 expression upon BMP2 or BMP4 treatment requiring ALK3 or ALK6 as a type I receptor as well as BMP7 (Fig. S10a). These findings suggest the possibility that ALK3 and ALK6-mediated BMP signaling pathways are differently regulated by PSMD14 in comparison to ALK2. The specificity between ALK2 and PSMD14 may be another clue to demonstrate a mechanism underlying cellular context-dependent BMP signaling pathway in diverse conditions. That is, it is possible that cell-type specific expression of a certain DUB is one of the contextual determinants of BMP signaling requiring different combinations of type I and type II receptors in the initiation step.
In this study, we focused on the BMP6-mediated signaling pathway among the BMP ligands, because analysis of public datasets indicated that the upregulation of BMP6 is highly related to shorter survival and human colon cancer cell lines such as HCT116 and RKO cells also showed better responsiveness to BMP6 than other BMP ligands, including BMP2, BMP4 and BMP7 (Fig. S10b). Furthermore, the specific function of the BMP6 signaling pathway and its related DUBs have not been fully understood in colon cancers. Our results emphasize the importance of the BMP6 signaling pathway in terms of growth and therapeutics of colon cancers. ALK2- and PSMD14-depleted HCT116 cells decreased tumor growth in a xenograft model and also reduced the population of cancer stem cells and chemoresistance. In particular, our results showed that chemoresistance of colorectal cancer cells is highly associated with BMP6 signaling pathway inducing ABC transporter genes. The requirement of the BMP6-PSMD14-ALK2 signaling axis regarding cancer stemness and chemoresistance of colorectal cancer cells had not been reported prior to our current findings. In terms of recent progress regarding anti-cancer therapy targeting cancer stem cells, PSMD14 and ALK2 proteins may be valuable targets to develop therapeutic molecules treating colon cancers. Moreover, our studies suggest that PSMD14 might be a significant prognostic biomarker designating the stages from polyps to malignant colon cancers or predicting the survival of colon cancer patients.
In conclusion, we here demonstrate that initiation of the BMP6 signaling pathway is driven by the deubiquitinating enzyme PSMD14 targeting the ALK2 receptor, and show the importance of the ALK2-PSMD14 axis in tumorigenesis, cancer stemness and chemoresistance of colon cancers. Thus, modulation of the ALK2 and PSMD14 axis in the BMP6 signaling pathway may be an important therapeutic strategy for the treatment of colon cancers.
Research in context
Evidence before this study
Although bone morphogenetic proteins (BMPs), members of the transforming growth factor (TGF)-β family, play fundamental roles in a variety of cellular processes, their roles in cancer progression are likely to be context-dependent and the ubiquitn-mediated post-translational modification mechanisms of BMP signaling components are not clearly addressed as much as those of TGF-β signaling pathway. In particular, it remains unknown about how the initiation of BMP signaling pathways is regulated by the ubiquitin-mediated modification system regarding colorectal cancers and what the related deubiquitinating enzymes (DUB) are.
Added value of this study
In this study, we identified an essential role of deubiquitinating enzyme PSMD14 in the initiation of BMP6 signaling pathway through deubiquitinating the Smurf1-mediated BMP type I receptor ALK2 polyubiquitination and provide a basis for underlying mechanism of BMP6 pathway via ALK2-PSMD14 axis in tumorigenesis and stemness/chemoresistance of human colorectal cancer. In addition, we provide experimental evidence that BMP6 pathway regulated by ALK2-PSMD14 axis induces chemoresistance of human colorectal cancer cells through upregulating ABC transporter genes, and higher expressions of PSMD14 and ALK2 genes are clinically associated with poor prognosis of human colorectal cancer patients.
Implications of all the available evidence
Our findings are the first report about the regulatory mechanism of the initiation of BMP6 signaling pathway by the deubiquitinating enzyme PSMD14, independent of the 26S proteasomal system, and the related E3 ubiquitin ligase Smurf1 in colorectal cancers. We also provide evidence showing the importance of BMP6-ALK2-PSMD14 signaling axis regarding tumorigenesis, cancer stemness and chemoresistance of colorectal cancer cells. These finding implicate that PSMD14 and ALK2 proteins may be valuable targets to develop therapeutic molecules treating colon cancers and their expressions may be used as significant prognostic biomarkers predicting the survival of colon cancer patients.
Funding sources
This work was supported by the National Research Foundation grant of Korea (2018R1A2A2A14023152, SRC 2017R1A5A1014560) funded by the Ministry of Science and ICT, and in part by a grant from the National R&D program for Cancer Control (HA15C0003 to S.H.P) funded by Ministry for Health and Welfare, Republic of Korea. The funders had no role in study design, all experiments, data analysis, decision to publish or preparation of the manuscript. The corresponding author (Seok Hee Park) had full access to all of the data in the study and had final responsibility for the decision to submit for publication.
CRediT authorship contribution statement
Dongyeob Seo: Conceptualization, Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Su Myung Jung: Conceptualization, Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Jin Seok Park: Formal analysis. Jaewon Lee: Formal analysis. Jihoon Ha: Formal analysis. Minbeom Kim: Formal analysis. Seok Hee Park: Conceptualization, Data curation, Funding acquisition, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
We thank Dr. Min Sung Choi for critical reading of the manuscript and Dr. Susumu Itoh for plasmids expressing HA-ALK2 and HA-ALK3.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.039.
Appendix. Supplementary materials
References
- 1.Ehata S., Yokoyama Y., Takahashi K., Miyazono K. Bi-directional roles of bone morphogenetic proteins in cancer: another molecular Jekyll and Hyde? Pathol Int. 2013;63:287–296. doi: 10.1111/pin.12067. [DOI] [PubMed] [Google Scholar]
- 2.Miyazono K., Kamiya Y., Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147:35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 3.Katagiri T., Watabe T. Bone morphogenetic proteins. Cold Spring Harb Perspect Biol. 2016:8. doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herhaus L., Sapkota G.P. The emerging roles of deubiquitylating enzymes (DUBs) in the TGFbeta and BMP pathways. Cell Signal. 2014;26:2186–2192. doi: 10.1016/j.cellsig.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imamura T., Oshima Y., Hikita A. Regulation of TGF-beta family signalling by ubiquitination and deubiquitination. J Biochem. 2013;154:481–489. doi: 10.1093/jb/mvt097. [DOI] [PubMed] [Google Scholar]
- 6.Liu S., de Boeck M., van Dam H., Ten Dijke P. Regulation of the TGF-beta pathway by deubiquitinases in cancer. Int J Biochem Cell Biol. 2016;76:135–145. doi: 10.1016/j.biocel.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z.J., Sun L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Husnjak K., Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 9.Kulathu Y., Komander D. Atypical ubiquitylation – the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 10.Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 11.Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 13.Buetow L., Huang D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshaies R.J., Joazeiro C.A. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 15.Gallo L.H., Ko J., Donoghue D.J. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijman S.M., Luna-Vargas M.P., Velds A. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Diefenbacher M.E., Popov N., Blake S.M. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J Clin Invest. 2014;124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Song N., Liu L. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat Commun. 2017;8:14866. doi: 10.1038/ncomms14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolaou K., Tsagaratou A., Eftychi C., Kollias G., Mosialos G., Talianidis I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21:738–750. doi: 10.1016/j.ccr.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Mancera P.A., Rust A.G., van der Weyden L. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi T., Kimura J., Miki Y., Yoshida K. The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha) J Biol Chem. 2007;282:33943–33948. doi: 10.1074/jbc.M706282200. [DOI] [PubMed] [Google Scholar]
- 23.Herhaus L., Al-Salihi M.A., Dingwell K.S. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open Biol. 2014;4 doi: 10.1098/rsob.140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inui M., Manfrin A., Mamidi A. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 25.Itoh F., Asao H., Sugamura K., Heldin C.H., ten Dijke P., Itoh S. Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J. 2001;20:4132–4142. doi: 10.1093/emboj/20.15.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Y., Avello M., Schirle M. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288:2976–2985. doi: 10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen H.F., McCrudden C.M., Grills C. Combinatorial use of bone morphogenetic protein 6, noggin and SOST significantly predicts cancer progression. Cancer Sci. 2012;103:1145–1154. doi: 10.1111/j.1349-7006.2012.02252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y.S., Park J.S., Kim J.H. Smad6-specific recruitment of Smurf E3 ligases mediates TGF-beta1-induced degradation of MyD88 in TLR4 signaling. Nat. Commun. 2011;2:460. doi: 10.1038/ncomms1469. [DOI] [PubMed] [Google Scholar]
- 29.Ishida W., Hamamoto T., Kusanagi K. Smad6 is a smad1/5-induced smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J Biol Chem. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.H., Jung S.M., Yang K.M. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat Cell Biol. 2017;19:1260–1273. doi: 10.1038/ncb3609. [DOI] [PubMed] [Google Scholar]
- 31.Miyazono K., Miyazawa K. Id: a target of BMP signaling. Sci STKE. 2002;2002:pe40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- 32.Verma R., Aravind L., Oania R. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 33.Yao T., Cohen R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 34.Buckley S.M., Aranda-Orgilles B., Strikoudis A. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler L.R., Densham R.M., Jia J. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012;31:3918–3934. doi: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byrne A., McLaren R.P., Mason P. Knockdown of human deubiquitinase PSMD14 induces cell cycle arrest and senescence. Exp Cell Res. 2010;316:258–271. doi: 10.1016/j.yexcr.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 37.Gallery M., Blank J.L., Lin Y. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol Cancer Ther. 2007;6:262–268. doi: 10.1158/1535-7163.MCT-06-0542. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Buus R., Clague M.J., Urbe S. Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS ONE. 2009;4:e5544. doi: 10.1371/journal.pone.0005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nabhan J.F., Ribeiro P. The 19 s proteasomal subunit POH1 contributes to the regulation of c-Jun ubiquitination, stability, and subcellular localization. J Biol Chem. 2006;281:16099–16107. doi: 10.1074/jbc.M512086200. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz T., Sohn C., Kaiser B., Jensen E.D., Mansky K.C. The 19S proteasomal lid subunit POH1 enhances the transcriptional activation by Mitf in osteoclasts. J Cell Biochem. 2010;109:967–974. doi: 10.1002/jcb.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B., Ma A., Zhang L. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:8704. doi: 10.1038/ncomms9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C.Y., Li C.Y., Hsu H.P. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res. 2017;7:2103–2120. [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Lin D., Lin Y. Proteasome beta-4 subunit contributes to the development of melanoma and is regulated by miR-148b. Tumour Biol. 2017;39 doi: 10.1177/1010428317705767. 1010428317705767. [DOI] [PubMed] [Google Scholar]
- 44.Rosenzweig B.L., Imamura T., Okadome T. Cloning and characterization of a human type II receptor for bone morphogenetic proteins. Proc Natl Acad Sci U S A. 1995;92:7632–7636. doi: 10.1073/pnas.92.17.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonni S., Wang H.R., Causing C.G. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 46.Kavsak P., Rasmussen R.K., Causing C.G. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 47.Kuratomi G., Komuro A., Goto K. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem J. 2005;386:461–470. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Xin H., Xu X. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol Cell Biol. 2004;24:856–864. doi: 10.1128/MCB.24.2.856-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami G., Watabe T., Takaoka K., Miyazono K., Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H., Kavsak P., Abdollah S., Wrana J.L., Thomsen G.H. A Smad ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 51.Fukunaga E., Inoue Y., Komiya S. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem. 2008;283:35660–35667. doi: 10.1074/jbc.M710496200. [DOI] [PubMed] [Google Scholar]
- 52.Alarmo E.L., Parssinen J., Ketolainen J.M., Savinainen K., Karhu R., Kallioniemi A. BMP7 influences proliferation, migration, and invasion of breast cancer cells. Cancer Lett. 2009;275:35–43. doi: 10.1016/j.canlet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Camara-Clayette V., Koscielny S., Roux S. BMP7 expression correlates with secondary drug resistance in mantle cell lymphoma. PLoS ONE. 2013;8:e73993. doi: 10.1371/journal.pone.0073993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowanetz M., Valcourt U., Bergstrom R., Heldin C.H., Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol. 2004;24:4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Q., Zhong Y.S., Ren Z. Analysis of the role of the BMP7-Smad4-Id2 signaling pathway in SW480 colorectal carcinoma cells. Mol Med Rep. 2011;4:627–631. doi: 10.3892/mmr.2011.481. [DOI] [PubMed] [Google Scholar]
- 56.Al-Hajj M., Becker M.W., Wicha M., Weissman I., Clarke M.F. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Al-Hajj M., Clarke M.F. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 58.Baccelli I., Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hermann P.C., Huber S.L., Heeschen C. Metastatic cancer stem cells: a new target for anti-cancer therapy? Cell Cycle. 2008;7:188–193. doi: 10.4161/cc.7.2.5326. [DOI] [PubMed] [Google Scholar]
- 60.Ho M.M., Ng A.V., Lam S., Hung J.Y. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 61.Dean M., Fojo T., Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 62.Merlos-Suarez A., Barriga F.M., Jung P. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 63.Singh A., Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole S.P., Bhardwaj G., Gerlach J.H. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 65.Doyle L.A., Yang W., Abruzzo L.V. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharom F.J. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9:105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 67.Szakacs G., Paterson J.K., Ludwig J.A., Booth-Genthe C., Gottesman M.M. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 68.Lin H., Ying Y., Wang Y.Y. AMPK downregulates ALK2 via increasing the interaction between Smurf1 and Smad6, leading to inhibition of osteogenic differentiation. Biochim Biophys Acta Mol Cell Res. 2017;1864:2369–2377. doi: 10.1016/j.bbamcr.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.