Abstract
A total of 17,439 mature miRNAs (~ 21 nt) earlier generated through RNA seq in the pomegranate were used for in silico analysis. After complexity reduction, a total of 1922 representative mature miRNAs were selected and used as query sequences against pomegranate genome to retrieve 2540 homologous contigs with flanking regions (~ 800). By using pre-miRNA prediction web server, a total of 1028 true contigs harbouring pri-miRNAs encoding 1162 pre-miRNAs were identified. Survey of these sequences for SSRs yielded a total of 1358 and 238 SSRs specific to pri-miRNA and pre-miRNAs, respectively. Of these, primer pairs were designed for 897 pri-miRNA and 168 pre-miRNA SSRs. In pri-miRNA sequences, hexa-nucleotides repeats were found to be most abundant (44.18%) followed by mono- (18.41%) and di-nucleotide (17.01%), which is also observed in pre-miRNA sequences. Further, a set of 51 randomly selected pre-miRNA-SSRs was examined for marker polymorphism. The experimental validation of these markers on eight pomegranate genotypes demonstrated 92.15% polymorphism. Utility of these functional markers was confirmed via examination of genetic diversity of 18 pomegranate genotypes using 15 miRNA-SSRs. Further, potential application of miRNA-SSRs for discovery of trait specific candidate genes was showed by validating 51 mature miRNA against publically available 2047 EST sequences of pomegranate by target and network analysis. In summary, the current study offers novel functional molecular markers for pomegranate genetic improvement.
Electronic supplementary material
The online version of this article (10.1007/s12298-020-00790-6) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA-SSRs, Functional markers, Genetic diversity, Pomegranate
Introduction
MicroRNAs (miRNAs) are the endogenous non-coding RNAs of ~ 19 to 24 nt in length (Bartel 2009; Voinnet 2009). Since the discovery of the first miRNA (lin-4) in Caenorhabditis elegans (Lee et al. 1993), hundreds of miRNAs have been identified in plants, animals and viruses. In plants, miRNAs are reported to play important role in various biological processes such as reproductive development (Mishra and Bohra 2018), biotic and abiotic stress response, signal transduction, and protein degradation (Bartel 2004). The recent advances in next-generation sequencing (NGS) technologies and bioinformatics tools have facilitated large-scale analysis of genome-wide miRNAs across different organisms.
In plants, miRNAs are transcribed by RNA polymerase II to long RNA transcripts (ranging from hundreds of nucleotides to tens of kilobases in length), called primary miRNAs (pri-miRNAs, Zhang et al. 2008). These pri-miRNAs may contain sequences encoding multiple miRNAs, which are then cleaved to short stem-loop structured precursor miRNAs (pre-miRNAs) of ~ 70 nucleotides in length by the enzyme Dicer-like 1 (DCL1) and are subsequently processed into an miRNA:miRNA* duplex (miRNA* is a small RNA on the opposite arm of the miRNA in the hairpin with partial complementarity to the miRNA). The miRNA duplex finally processed into single stranded mature miRNAs (Russo and Giordano 2009), that are assembled into the RNA-induced silencing complex (RISC), which regulates gene expression negatively at the post-transcriptional level by targeting mRNA cleavage or inhibiting mRNA translation (Zhang et al. 2008; Kusenda et al. 2009).
In recent years, computational analysis has facilitated SSR discovery in pre-miRNA sequences in plants (Joy and Soniya 2012, Joy et al. 2013; Mondal and Ganie 2013). The microsatellites present in pre-miRNAs in different species did not show noticeable locational preferences and are found anywhere in pre-miRNAs, suggesting that SSRs are the important component of pre-miRNAs. These studies also suggested potential role for SSR and SNP markers located in miRNAs vis a vis expression of quantitative traits (Ganie and Mondal 2015). Polymorphisms including SSRs or SNPs on mature miRNAs, may impair interaction between putative target gene and miRNA (Ferrao et al. 2015). Most recently, Joy et al. (2018) have reported the significance of SSRs in pre-miRNAs. The authors also provided clues for involvement of SSRs in alternative splicing events to produce mature RNA isoforms in response to stressed environments.
A comprehensive analysis of the SSR prediction in 8619 pre-miRNA sequences from 87 species was performed covering Arthropoda, Nematoda, Platyhelminthes, Urochordata, Vertebrata, Mycetozoa, Protistate, Viridiplantae, and Viruses (Chen et al. 2010). Various miRNA families are known to influence different aspects of plant development and stress response (Lima et al. 2012). For instance, researchers have found salt responsive (trait specific) miRNA-SSRs in rice genome (Mondal and Ganie 2013; Ganie and Mondal 2015) linking them to phenotype and expression of genes. There is a need for development of DNA markers associated with miRNAs, so that the developed DNA markers may facilitate improvements of traits influenced by miRNAs (Kumar et al. 2017).
Several reports describe the benefits of pomegranate (Punica garnatum L.) natural products for humans; however lack of genomic information is a major bottleneck in pomegranate research (Saminathan et al. 2016). Hence, development of genome wide miRNA-SSR markers leverages genomic resources to support functional genomics of pomegranate. To this end, evolving computational tools are extremely useful for organisms for which extensive DNA/genome sequence information is available. Due to its agricultural and medicinal significance, the whole genome sequence of pomegranate has been assembled recently (Qin et al. 2017; Yuan et al. 2018). In pomegranate, five miRNAs were reported through analysis of partial genome sequence data (Kanupriya et al. 2013). More recently, Saminathan et al. (2016) reported genome-wide discovery of microRNAs in pomegranate covering seedling to fruit development stages through high-throughput sequencing. However, the information on large-scale development of novel miRNA-SSR markers in pomegranate is still lacking. The miRNAs are known to play key roles in different crops for development and stress response, regulation of anthocyanin accumulation in tomato (Jia et al. 2015).
The miRNA expression has been used successfully as a biomarker in medical diagnostics such as for breast cancer (Shah et al. 2010; Fu et al. 2011). Genomic sequences corresponding to miRNAs have significant potential for developing novel functional markers. Pre-miRNA stem-loop regions are preferred for SSR designing, since this regions are relatively highly conserved between closely related species (Mendes et al. 2009), resulting in good marker transferability. In view of this, here we first time report large scale development of novel miRNA-SSRs that could greatly strengthen future genomics research in pomegranate.
Materials and methods
Plant materials and DNA extraction
Eighteen pomegranate genotypes were used in this study, which are maintained at field gene bank ICAR-National Research Centre on Pomegranate, Solapur, Maharashtra, India (Table 1). All of them are promising cultivars. Initially, a sub-set of eight diverse pomegranate genotypes namely Ganesh, Kalpitiya, Co-White, Dholka, Yercaud, P-23, Daru 17 and Nana were used for the validation of newly designed miRNA-SSR markers. The fresh leaf samples were collected from the all genotypes and washed with sterile distilled water. Genomic DNA was extracted from the leaf samples by following modified CTAB method (Ravishankar et al. 2000). The quality and concentration of genomic DNA was determined using QiaExpert (Qiagen India Pvt. Ltd). Final dilution of 20 ng/μl was made for all the DNA sample to perform polymerase chain reaction (PCR).
Table 1.
Details of eighteen pomegranate genotypes used in the study
| Sl. no. | Genotype name | Type | Parentage | Location/source |
|---|---|---|---|---|
| 1 | Bhagawa | Commercial variety | Selection (Ganesh × Gul-e-Shah Red) | India (Rahuri) |
| 2 | Solapur Lal | New variety | Hybridization (Bhagawa × [(Ganesh × Nana) × Daru] | India (Solapur) |
| 3 | G-137 | Commercial variety | Clonal selection (Ganesh) | India (Rahuri) |
| 4 | Ganesh | Commercial variety | Selection (Alandi) | India (Rahuri) |
| 5 | Arakta | Commercial variety | Selection F2 (Ganesh × Gul-EShah Red) | India (Rahuri) |
| 6 | Mridula | Commercial variety | Selection F2 (Ganesh × Gul-EShah Red) | India (Rahuri) |
| 7 | Dholka | Commercial variety | Selection | India (Gujarat) |
| 8 | Jyoti | Commercial variety | Selection F2 (Bassein Seedless × Dholka) | India (Karnataka) |
| 9 | Jodhpur Red | Cultivar | Selection from local type | India (Rajasthan) |
| 10 | Co-white | Commercial variety | Selection from local type | India (Maharashtra) |
| 11 | Bedana sri | Cultivar | Selection | India (Rajasthan) |
| 12 | Nimali | Commercial variety | Introduction | Sri Lanka |
| 13 | P-13 | Commercial variety | Selection (Muscat) | India (Rahuri) |
| 14 | P-23 | Commercial variety | Selection (Muscat) | India (Rahuri) |
| 15 | P-26 | Commercial variety | Selection (Muscat) | India (Rahuri) |
| 16 | Yercaud HRS | Cultivar | Selection (Yercaud local) | India (Tamil Nadu) |
| 17 | Ruby | Commercial variety | Hybridization {[(Ganesh × Kabul) × Yercaud]-F2} × {(Ganesh × Gule Shah Red)-F2} | India (Karnataka) |
| 18 | Super Bhagawa | Commercial variety | Selection (Bhagawa) | India (Rahuri) |
In silico dataset
A total of 17,439 mature miRNA sequences of pomegranate representing 30 conserved miRNA families were retrieved from NCBI database (www.ncbi.nlm.nih.gov) (Saminathan et al. 2016). First all the mature miRNA sequences (~ 21 nt) were subjected to cluster analysis through CDHIT-EST tool with default parameters (Li and Godzik 2006), to obtain a subset of representative mature miRNA sequences for further analysis.
Identification of pri-miRNAs and pre-miRNAs from pomegranate genome
After complex reduction a set of mature miRNA sequences were used as query for BLASTn homology search (mismatch < 1, with no gap and e-value ≤ 0.01) against pomegranate genome cv. Dabenzi (Qin et al. 2017). The highly similar contigs with flanking sequences of ~ 700 to 800 bp around query were retrieved. All these contigs were tested for pri-miRNA regions using sequence-structure motif base: pre-miRNA prediction webserver (http://www.regulatoryrna.org/webserver/SSMB/pre-miRNA/home.html). The contigs with no miRNA regions were excluded from the analysis. The non-redundant set of pri-miRNA contigs were then obtained using CAP3 program (Huang and Madan 1999). Further, all the pre-miRNAs of these contigs were analyzed through BLASTx against non-redundant (NR) protein database to remove protein-coding sequences (Altschul et al. 1997). Finally, non-protein coding pre-miRNA sequences and their respective pri-miRNA contigs were deployed for microsatellite search.
Mining of SSRs from pri-miRNAs and pre-miRNAs
Mining of microsatellites with 2–6 bp repeats, which are specific to pri-miRNA and pre-miRNA sequences were performed using MISA tool (MIcroSAtellite identification tool; http://pgrc.ipk-gatersleben.de/misa/). SSR motifs were surveyed at genome-wide scale by considering minimum repeat length of 12 bp through defining 12, 6, 4, 3, 3 and 2 for mono, di, tri, tetra, penta and hexa nucleotides, respectively. Compound SSRs were defined as two SSRs interrupted within 100 bases. The MISA statistics was analysed further to draw frequency distribution graphs using Microsoft Excel. For designing flanking primers to the SSRs present in primary and precursor miRNA sequences, online web based high throughput primer designing tool Batch Primer 3v1.0 (https://wheat.pw.usda.gov/demos/BatchPrimer3) was used. Primer pairs were designed to amplify products of 100–400 bp with the following parameters: primer length (bp) 18–20 bp with 19 bp as optimum; GC content (%) 40–60, with the optimum value being 50%; Tm (°C) 52–60, with 55 as the optimum. Other parameters used were as that of default program values.
Amplification of pre-miRNA-SSRs
Newly designed miRNA-SSRs were first tested for PCR amplification in eight pomegranate genotypes using Prime-96™ Thermal Cycler (Himedia, India). PCR analysis was performed in 10 μl reaction volume containing 1.0 μl of 10X PCR buffer, 1 μl (2 mM dNTP mix), 0.5 μl each of forward and reverse primers (10 pmol), 0.2 μl of Taq DNA polymerase 5U/μl (Himedia, India) and 1 μl (20 ng) of template DNA. PCR condition was set as initial denaturation at 94 °C for 5 min, followed by 36 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min and a final extension at 72 °C for 7 min. PCR products were separated by electrophoresis in 3% metaphor agarose gels containing 0.5 μg/ml ethidium bromide in 1X TBE buffer at 130 V for 4 h, visualized and photographed in gel documentation system (Vilbert dourmet, France).
Gene target prediction for miRNAs
To understand the biological functions of selected pre-miRNAs, first their miRNA families were determined through homology search against miRBase. Then, putative target genes for these families were identified based on target analysis for mature miRNAs against publically available 2047 EST sequences of pomegranate by using web server for prediction of plant miRNA targets FASTA search option of (TAPIR) (http://bioinformatics.psb.ugent.be/webtools/tapir) (Bonnet et al. 2010). Functional annotations for Gene Ontology (GO) of target genes and enrichment analysis and KEGG pathways were performed using Blast2GO Pro (https://www.blast2go.com/blast2go-pro). Based on alignment duplex target score (with cut off value ≤ 0.4) and duplex free energy ratio of hybridization (with cut off value ≥ 0.7) for target genes obtained from TAPIR, miRNA regulatory networks were built among miRNA, and their target genes using Gephi 0.9.2 software (Bastian et al. 2009).The miRNA target score is taking into account the number of mismatches, gaps (introduced by bulges and loop structures) and number of G-U pairs located in position 2 and 12th of 5′ seed regions of miRNAs to the target mRNA. The duplex miRNA-mRNA free energy ratio is the ratio of free energy of duplex to the free energy of the same duplex having only perfect matches (Allen et al. 2005; Schwab et al. 2005).
Statistical analysis for miRNA-SSR marker data
In-order to check the level of polymorphism for newly designed miRNA-SSR primers, a set of eight pomegranate genotypes were screened initially and separated on 3% metaphor gels. A subset of 15 highly informative miRNA-SSRs was then selected to analyze diversity among 18 pomegranate genotypes. Amplification products were separated on fragment analyser QIAxcel Advanced (Qiagen India Pvt. Ltd), fragments were scored and analyzed using QIAxcel Screen Gel Software. The genetic diversity parameters such as major allelic frequency, polymorphic information content (PIC), heterozygosity, gene diversity were computed using PowerMarker v.3.25 (Liu and Muse 2005). Concerning distance based clustering, DARwin v. 6.0.13 (Perrier and Jacquemoud-Collet 2006) was employed to generate genetic distance (GD) matrix, which was then used to create unweighted neighbor joining (NJ) tree with 1000 bootstraps. Factorial analysis was also performed with GD matrix created using DARwin software.
Results
Characterization and frequency distribution of miRNA-SSRs in the pomegranate genome
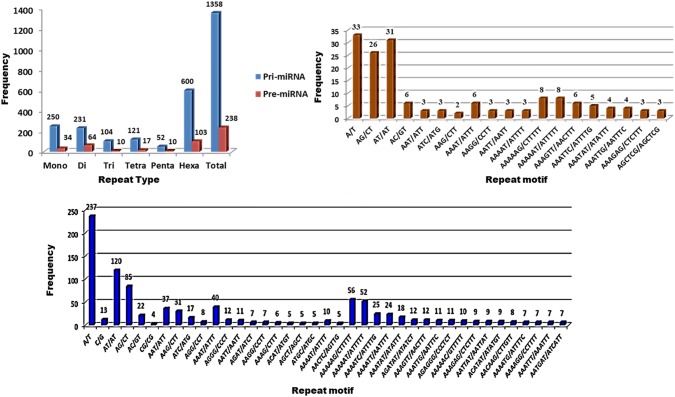
Analysis of 17,439 mature miRNA sequences through CDHIT-EST tool yielded a total of 1922 non-redundant mature miRNAs. These sequences were then used as query sequences for homology BLASTn search against pomegranate genome cv. Dabenzi at an e-value threshold ≤ 0.01. Total 2540 contigs were selected to extract ~ 700 to 800 bp flanking sequences around miRNA complementary regions. MiRNA prediction tool identified 1676 contigs (~ 721 to 821) that code for true pri-miRNAs. These sequences were further analysed to identify 1162 pre-miRNA sequences through BLASTX search against nr protein database. We found 1028 pri-miRNAs sequences coding for 1162 pre-miRNAs. Finally, all the pri- and pre-miRNA sequences were surveyed for presence of SSRs. These sequences represented approximately 701.8 kilo base (kb) for pri-miRNA and 119.8 kb for pre-miRNA sequences of pomegranate genome. MISA search tool discovered a total of 1358 SSR motifs from 656 (63.8%) pri-miRNAs and 238 SSRs from 204 (17.55%) pre-miRNA sequences. The distribution frequency of one SSR locus per 0.52 kb and 0.50 kb was observed for pri-miRNAs and pre-miRNAs, respectively. We found 385 (58.6%) pri-miRNA and 31 (15.1%) pre-miRNA sequences with more than one SSRs (Table 2). Of the total 1358 pri-miRNA-SSRs and 238 pre-miRNA-SSRs, 366 (26.95%) and 34 (14.28%) were compound motifs, respectively. As illustrated by overall frequency distribution graphs, in pri-miRNAs hexa-nucleotides repeats were more dominant (44.18%) followed by mono- (18.41%) and di-nucleotide (17.01%). Similarly, hexa-nucleotides were most abundant (43.27%) followed by di (26.9%) and mono (14.3%) nucleotides (Fig. 1). Further analysis of the frequency distribution graphs for SSR repeat types suggested abundance of A/T (94.80%) in pri-miRNAs followed by AT/AT (51.95%) and AG/CT (36.80%) and AAAAAG/CTTTTT (9.33%) and AAAAAT/ATTTTT (8.66%) among di- and hexa-nucleotides, respectively (Fig. 1c). Similarly, higher frequency for these motif types was also observed in pre-miRNA sequences (Fig. 1b).
Table 2.
Characterization of microsatellites in pri- and pre-miRNAs sequences of pomegrante genome
| Parameters | Pri-miRNA | Pre-miRNA |
|---|---|---|
| Number of sequences examined | 1028 | 1162 |
| Examined sequences size (bp) | 701,888 | 119,835 |
| Total number of identified SSRs | 1358 | 238 |
| Number of sequences with SSRs | 656 | 204 |
| Number of sequences with more than 1 SSRs | 385 | 31 |
| Number of compound SSRs | 366 | 34 |
Fig. 1.
Frequency distribution of different SSR repeats in pre- and pri-miRNA sequences of pomegranate
Development of miRNA-SSR markers and polymorphism survey
Using web based high throughput primer designing Batch primer 3 tool, we successfully designed 897 primer pairs for miRNA-SSRs from 1028 pri-miRNA sequences. The detailed information of these 897 primer pairs is presented as supplementary data (Supl. Table S1). Of the 897 primer pairs, 486 (54.18%) were hexanucleotide repeats; while 192 (21.40%), 102 (11.37%), 81 (9.03%) and 36 (4.01%) were di, tetra, tri and penta nucleotide repeats, respectively. Out of 897 pri-miRNA-SSRs, 168 primers were exclusively from pre-miRNA sequences. Further, we synthesized a set of 51 SSRs for experimental validation. Initially, the primers were assayed on eight pomegranate genotypes viz, Ganesh, Kalpitiya, Co-White, Dholka, Yercaud, P-23, Daru 17 and Nana on 3% metaphor agarose gels. As a result, 47 (92.15%) primers showed scorable amplicons with significant level of DNA polymorphism.
The number for alleles ranged from 1 to 2, with an average of 1.91. Similarly, PIC values ranged from 0 to 0.53, with an average of 0.29 (Supl. Table S2). Based on these results, a subset of 15 SSR primers were genotyped on a panel of 18 pomegranate genotypes for genetic diversity estimation. These markers revealed 3–9 number of alleles/locus with an average of 5.8. The frequency of major alleles (MAF) per locus ranged from 0.22 to 0.67, with average of 0.37. Similarly, PIC values varied from 0.48 to 0.81, with an average of 0.68. The heterozygosity values ranged from 0.0 to 0.94 with average of 0.57. Average gene diversity for these markers were 0.72 among 18 pomegranate genotypes (Table 3). A representative gel image obtained using metaphor and fragment analyzer illustrating the SSR profiles of pomegranate genotypes is shown in Fig. 2a and b.
Table 3.
Diversity analysis of 18 pomegranate genotypes using 15 pre-miRNA-SSR markers
| Sl. no. | Marker | Family | MAF | NA | PIC | Heterozygosity | GD |
|---|---|---|---|---|---|---|---|
| 1 | Pre miR_SSR32 | ath-MIR5664 | 0.50 | 4 | 0.58 | 0.00 | 0.64 |
| 2 | Pre miR_SSR33 | ath-MIR169c | 0.67 | 5 | 0.48 | 0.06 | 0.52 |
| 3 | Pre miR_SSR2 | ath-MIR157c | 0.25 | 9 | 0.81 | 0.89 | 0.83 |
| 4 | Pre miR_SSR3 | ath-MIR156b | 0.42 | 6 | 0.58 | 0.78 | 0.65 |
| 5 | Pre miR_SSR8 | crm-mir-244 | 0.44 | 3 | 0.57 | 0.00 | 0.64 |
| 6 | Pre miR_SSR9 | ath-MIR156b | 0.28 | 9 | 0.81 | 0.94 | 0.83 |
| 7 | Pre miR_SSR13 | ath-MIR156b | 0.47 | 5 | 0.61 | 0.94 | 0.67 |
| 8 | Pre miR_SSR17 | ath-MIR5651 | 0.28 | 9 | 0.80 | 0.94 | 0.82 |
| 9 | Pre miR_SSR16 | ath-MIR156b | 0.28 | 5 | 0.75 | 0.72 | 0.78 |
| 10 | Pre miR_SSR30 | gga-mir-7439 | 0.39 | 4 | 0.67 | 0.00 | 0.72 |
| 11 | Pre miR_SSR45 | ath-MIR167d | 0.22 | 7 | 0.80 | 0.89 | 0.82 |
| 12 | Pre miR_SSR41 | ath-MIR157c | 0.44 | 4 | 0.62 | 0.61 | 0.68 |
| 13 | Pre miR_SSR42 | ath-MIR156b | 0.22 | 7 | 0.81 | 0.83 | 0.83 |
| 14 | Pre miR_SSR44 | ath-MIR5645d | 0.31 | 6 | 0.74 | 1.00 | 0.78 |
| 15 | Pre miR_SSR26 | sly-MIR172c | 0.44 | 4 | 0.61 | 0.00 | 0.67 |
| Mean | 0.37 | 5.8 | 0.68 | 0.57 | 0.72 |
MAF Major allele frequency, NA no. of obtained alleles, PIC polymorphic information content, GD gene diversity, miR miRNA
Fig. 2.
Gel images showing allelic variations as revealed by novel miRNA-SSR markers. a Assaying eight pomegranate genotypes with the markers miRNA-SSR 40, -41, -42, -43 and -44 on 3% metaphor gel, L, DNA mass ladder. Lane 1–8, eight set of pomegranate genotypes as mentioned in materials and methods, b Assaying miRNA-SSRs 42 and miRNA-SSR 44 on 18 pomegranate genotypes using fragment analyzer. Lanes 1–18, pomegranate genotypes as listed in Table 1
Diversity analysis
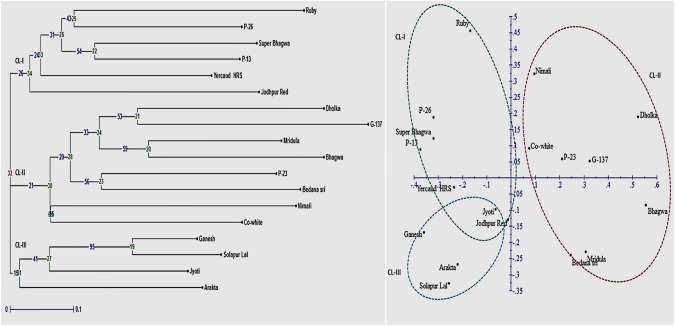
A subset of 15 miRNA-SSR markers was selected from the 51 pre-miRNA primers for analyzing diversity in 18 genotypes. The pooled marker data for 18 genotypes were used to construct UPGMA based NJ tree, which suggested presence of three clusters (Fig. 3a). The factorial analysis was also undertaken to elucidate overall diversity in the selected genotypes. A close agreement was observed between the results arising from NJ tree and factorial analysis. All genotypes of CL-I as depicted in NJ tree were represented in quadrant II, similarly for CL-II were in quadrant I and IV. Whereas, genotypes of CL-III were grouped under quadrant III. The genetic relationship between genotypes belonging to each clusters were well resolved in factorial plot (Fig. 3b). The NJ analysis revealed cluster 1 comprising six genotypes, whereas clusters 2 and 3 had eight and four genotypes, respectively. As evident in both NJ tree and factorial plots, higher diversity was observed for genotypes belonging to CL-II, followed by CL-I and CL-III.
Fig. 3.
Neighbor joining tree and factorial analysis depicting genetic relationships among 18 pomegranate genotypes based on 15 miRNA-SSR marker data
Functional classification and pathway enrichment analysis for target genes of miRNA-SSR genes
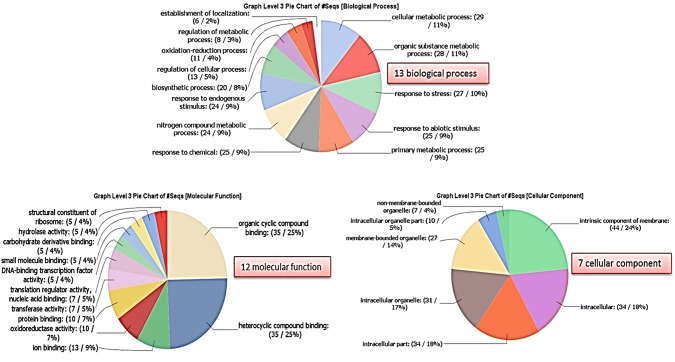
To demonstrate the functional utility of miRNA-SSR markers for genetic analysis, mature RNAs of 51 miRNA-SSRs used for diversity analysis were deployed for target analysis using TAPIR program with 2417 pomegranate EST sequences fetched as the reference transcript library. As a result, a total of 128 putative gene targets were identified (Supl. Table S3). Further, the GO annotations of all the predicted targets were analyzed to understand the functional features of these gene targets. The most significant BLAST hits for each target across different species were obtained by BLASTx search and used for GO annotation. The GO terms were classified for the gene targets and 13, 12 and 7 genes involved in biological process (BP), molecular function (MF), and cellular component (CC), respectively (Fig. 4). In the BP category, cellular and organic substance metabolic processes were the major functional groups, followed by response to stress and abiotic stimulus, primary metabolic process, and response to chemical. With respect to MF category, predicated targets showed participation in organic and hetero cyclic compound binding, followed by ion binding, protein binding and oxidoreductase activity. In the CC category, greater proportion of genes (44) was associated with intrinsic component of membrane, followed by intracellular and intracellular part, and membrane bounded organelle. Many of the predicated targets were annotated to be involved in cellular metabolic process, response to stress and various compound binding.
Fig. 4.
Distribution of GO terms in the biological, molecular function and cellular component category, level 3
Further, enrichment analysis through KEGG pathways revealed, out of 128 target genes 18 genes coding for 14 enzymes that are part of 11 important pathways viz, purine metabolism, thiamine metabolism, phenylpropanoid biosynthesis, amino sugar and nucleotide sugar metabolism, mTOR signalling pathway, glutathione metabolism, drug metabolism-other enzymes, ascorbate and aldarate metabolism, phosphonate and phosphinate metabolism, glycerol phospholipid metabolism and pyrimidine metabolism (Supl. Table S4).
miRNA-mediated regulatory networks
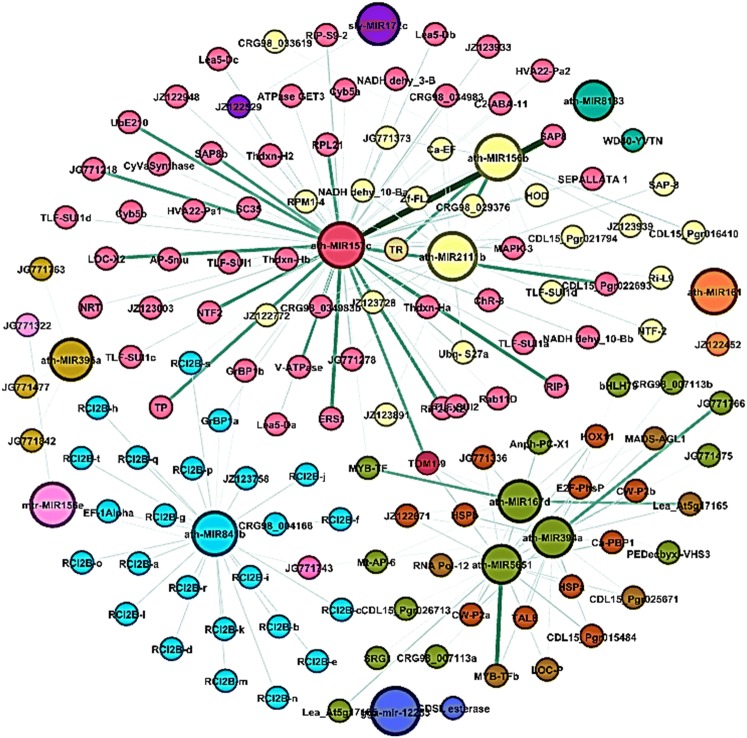
To further explore the relationship among the miRNAs and their targets, miRNA-mediated regulatory networks were constructed (Fig. 5). Thirteen independent networks for miRNA families showed multiplicity behavior for few families i.e., one miRNA can target more than one genes. Exploring these results ath-MIR157c has revealed maximum targets (79 genes), followed by ath-MIR394a (26 genes), ath-MIR841b (25 genes), ath-MIR2111b (23 genes), ath-MIR5651 (20 genes), ath-MIR167d (19) and ath-MIR156b (12). However, the lowest targets were observed for ath-MIR161 (1), sly-MIR172c (1), gga-MIR12253 (1), ath-MIR8183 (1), mtr-MIR156e (2) and ath-MIR396a (3). The ath-MIR157c was the largest regulatory network and shared maximum genes with MIR2111b (18 genes) and ath-MIR156b (6) for tropinone reductase, Zf-FLZ domain protein, homeodomain-like protein, mitogen-activated protein kinase 3, NADH dehydrogenase (ubiquinone) 1 beta sub-complex, ubiquitin-40S ribosomal protein S27, RPM1-interacting protein 4, thioredoxin H-type, calcium-binding EF-hand family protein and protein chromatin remodeling 8. Similarly, ath-MIR394a regulatory network shared many common targets with ath-MIR5651 and ath-MIR167d. Further, network analysis for the individual miRNAs families based on alignment score and minimum free energy ratio of hybridization revealed, ath-MIRNA 157c showed strongest interaction with stress associated protein 8, ath-MIR394a with unknown gene (JG771766.1), ath-MIR2111b (Zf-FLZ, UBq-S27a and NADH dehy_10Ba), ath-MIR5651 (MYB-TF), ath-MIR167d (MYB-TF and Lea_At5g17165), ath-156b (TR and CRG98_029376) and for ath-MIR841b many genes (Supl. Fig. 1).
Fig. 5.
A comprehensive miRNA-target genes regulatory network identified in pomegranate for 13 miRNA families (darker green lines for each family indicates stronger interactions for miRNAs with their target genes based on minimum free energy ratio of hybridization)
SSR markers in target genes
Mining of SSR motifs in 128 target genes showed 143 SSR motifs, with maximum frequency of mono-nucleotide repeats (65) followed by di-nucleotides (63) and tri-nucleotides (15). It was observed that 53 of target gene sequences have more than one SSR motifs. A total of 29 compound SSR motifs were observed. Finally, 58 functional EST-SSR markers were designed that can be deployed for mapping genes/QTLs for quality traits in pomegranate (Supl. Table S5).
Discussion
Recent genomic advances in pomegranate leading to the establishment of draft genome sequence (Qin et al. 2017; Yuan et al. 2018), and coding and non-coding transcriptome sequences (Ono et al. 2011; Ophir et al. 2014; Saminathan et al. 2016) have opened unprecedented opportunities for genome wide identification of functional DNA markers for downstream applications. A variety of functional DNA markers have been widely used for trait mapping in many plant species. These include DNA markers derived from genes, such as genic EST-SSRs (Ophir et al. 2014), EST-SNPs (Ophir et al. 2014; Harel-Beja et al. 2015), TRAPs (Hu and Vick 2003), resistance gene analogues (RGAs) (Hinchliffe et al. 2005) and SRAPs (Soleimani et al. 2012). However, gene conservation greatly limits the genetic polymorphism of these functional markers. In contrast, miRNA-based molecular markers are more polymorphic with enhanced possibilities to predict phenotypes that are controlled by miRNAs. To allow identification of miRNAs at genome scale, computational tools are gaining widespread popularity owing to their fast, inexpensive, and effective nature. However, for identifying miRNA genes, several criteria have been laid down by various workers, which are primarily done by the alignment of small RNA reads on the reference genome (Axtell 2014). For instance conserved miRNAs have been investigated by using genomic survey sequence (GSS) and expressed sequence tag (EST) sequences in public data sets of many plant species viz, Glycine max L. Merr (Zhang et al. 2008), Nicotiana tabacum L. (Frazier et al. 2010), Solanum tuberosum L. (Xie et al. 2011), Brassica oleracea L. (Wang et al. 2012) and cucurbit species (Hu et al. 2014). Recent advances in RNA sequencing have facilitated the identification of huge number of putative miRNAs during seedling to fruit development stages in pomegranate (Saminathan et al. 2016). The large-scale development of SSR markers specific to miRNA genes has not been reported in pomegranate. Since, miRNA-SSRs are of potential utility for identifying master miRNAs that regulate various genes for fruit quality traits. These miRNAs can be later targeted for genome editing to gain desired level of phenotypes in pomegranate. Given this, here we report genome-wide development of first set of miRNA-SSRs specific to fruit developmental stages. We assayed a set of miRNA-SSR markers on pomegranate genotypes in order to show their immediate utility for genetic analysis. A similar approach showing development and validation of miRNA-SSRs has been reported recently in other crops like rice (Ganie and Mondal 2015), Arabidopsis thaliana (Kumar et al. 2017), Brassica (Singh et al. 2017) and Medicago truncatula (Min et al. 2017).
Distribution of miRNA-SSRs in the pomegranate genome
In the present study, a total of 17,439 mature miRNA sequences of pomegranate genome (Saminathan et al. 2016) were retrieved for identification of SSRs. The removal of redundant sequences resulted in identification of 1922 mature miRNAs with corresponding 1028 pri-miRNAs (Qin et al. 2017), which enabled identification of 1358 pri-SSR motifs. Similarly, Min et al. (2017) performed homology search from 356 non redundant miRNAs to extract corresponding pri-miRNA sequences from Medicago truncatula genome and identified 189 SSRs. Here, we have discovered 1162 pre-miRNAs coded by 1028 pri-miRNAs and identified 238 SSRs. Similarly, Kumar et al. (2017) discovered 147 miRNA-SSRs from 169 pre-miRNA transcripts from the genome of Arabidopsis thaliana. In this study, approximately 701.8 kilobase (kb) for pri-miRNA and 119.8 kb for pre-miRNA sequences were surveyed for presence of SSR markers. We found one SSR locus per 0.52 kb and 0.50 kb for pri-miRNA and pre-miRNA sequences. Earlier, Ravishankar et al. (2015) have estimated the density of one genomic SSRs for every 5.56 kb based on partial genome sequence of pomegranate. Also, we computed frequency distribution for different SSR motifs in miRNA sequences of pomegranate. We found hexa-nucleotide repeats to be most abundant (44.18%) followed by mono- (18.41%) and di-nucleotides (17.01%) in pri-miRNAs, which is also evident in pre-miRNA sequences. Similarly, Joy and Soniya (2012) carried out preliminary analysis to discover SSRs in primary miRNAs of black pepper and isolated SSR containing transcripts. Chen et al. (2010) analyzed SSRs in 8619 pre-miRNAs from 87 species. However, authors found mono-nucleotide repeats to be the most abundant class followed by di-nucleotide repeats in comparison to tri-, tetra-, penta- and hexa-nucleotide repeats. Similarly, mono-nucleotide (70.41%) and di-nucleotide repeats (14.20%) were found more pronounced in pri-miRNAs of Medicago trucatula, and di-nucleotide in the pre-miRNAs of Arabidopis thaliana genome (Min et al. 2017; Kumar et al. 2017).
To the best of our knowledge, this is the first report on large scale development of 897 SSR primers specific to pri-miRNAs and 168 for pre-miRNAs in pomegranate genome and other crops as well. For instance, Min et al. (2017) developed 169 primers from 130 pri-miRNAs in M. trucatula. With respect to motif types in miRNA genes of pomegranate, A/T (94.80%) were found more abundant among all the microsatellite repeats, followed by AT/AT (51.95%) and AG/CT (36.80%) in di- and AAAAAG/CTTTTT (9.33%) and AAAAAT/ATTTTT (8.66%) in hexa-nucleotides. Similarly, Ganie and Mondal (2015) reported for di-nucleotides (AT)n was found to be present in the maximum frequency followed by (CT)n and (AG)n in rice miRNA genes.
Development of miRNA-SSR markers and polymorphism survey
We successfully validated a set of randomly selected 51 pre-miRNA SSRs, out of which 47 (92.15%) showed high level of polymorphism among eight pomegranate genotypes. All 47 pre-miRNA-SSRs had clear and reproducible specific amplifications on metaphor gels, with an average PIC value of 0.29. Fu et al. (2013) observed the average PIC value of 0.43 for 34 miRNA-SSRs when screened on six Brassica species. Our results suggest high level of genetic polymorphism and diversity among selected pomegranate genotypes. Greater informativeness of miRNA-SRRs over genomic SSRs is also evident. For example, Ravishankar et al. (2015) reported 97.6% polymorphism in pomegranate for SSRs with up to 14 alleles with an average PIC value of 0.54. Similarly, Fu et al. (2013) demonstrated high polymorphism with miRNA-based marker system in Brassica.
Concerning allelic variations, a total of 87 alleles were obtained for 15 miRNA-SSRs across 18 pomegranate genotypes with alleles per locus ranging from 3 to 9. Our results remain in congruence with the findings of Mondal and Ganie (2013) in rice, where the authors scored a total of 88 alleles while assaying 16 miRNA-SSRs on 24 genotypes. The same group later reported a total of 114 alleles with 20 miRNA-SSRs in 24 rice genotypes, with an average of 5.7 alleles per marker (Mondal and Ganie 2013). Out of 15 polymorphic pre-miRNA-SSRs in our study, the highest alleles (7–9), PIC values (0.80–0.81), heterozygosity (0.83–0.94) and gene diversity (0.82–0.83) were observed for Pre miR_SSR 2, 9, 17, 45 and 42 of ath-MIR157c, ath-MIR156b, ath-MIR5651 and ath-MIR167d, respectively. Similarly. Mondal and Ganie (2013) reported highest number of alleles (8) for miR156g-SSR, miR 166d-SSR and miR-171a SSRs while screening rice genotypes. However, all the 15 markers showed higher PIC (≥ 0.48) and gene diversity values (≥ 0.52), which implies towards their great potential for genetic diversity and genetic mapping applications. According to Bandelj et al. (2004), DNA markers are considered informative when their PIC values greater than 0.5 and markers with PIC values greater than 0.7 were suitable for genetic mapping. Informative SSR markers have widespread applications in plant improvement (Bohra et al. 2011, 2015, 2017).
Genetic relationship
The genetic relationships of 18 pomegranate genotypes were determined using 15 polymorphic pre-miRNA-SSRs. Entire pomegranate genotypes could be grouped into three distinct clusters (CLI, CLII and CLIII). It is interesting to note that the genotypes Ruby, Super Bhagawa and Jodhpur Red were found to be the most diverse in CLI. Similarly, G-137 followed by Dholka and Bhagawa were found most diverse in cluster II. In cluster III, newly bred bio-fortified variety Solapur Lal was found most diverse followed by Arakta and Ganesh. These results highlighted the variations in pre-miRNAs can regulate a variety of genes underlying varietal characters. Notably the highest variation observed in the cluster CLII could be due to the inclusion of one exotic line Nimali (Sri Lanka) in this group. According to Ganie and Mondal (2015), genetic diversity remains higher among the exotic rice material than the Indian landraces, based on the corresponding PIC values of 0.31 and 0.29 for miRNA-SSRs. Further, the pomegranate genotypes viz, Dholka, Bedana Sri and Nimali of CLII have much higher adoption to arid and semiarid regions with tropical climate. However, cluster III represented by Ganesh, Solapur Lal and Arakta, which share pedigree relationships. Although in this study, we did not observe clear-cut grouping for most of the genotypes according to the geographical locations/distribution and pedigree relationships as depicted in NJ and factorial analysis plot. However, few genotypes showed pedigree linages in different clusters i.e. Ganesh, Solapur Lal and Arakta in CLIII, Mridula and Bhagawa (CLII), and selections P-26 and P-23 from Muscat (CLI). Lack of a clear-cut pattern could be due to limited number of miRNA-SSR markers used in this study. The mixed grouping of most of genotypes irrespective of their varietal characteristics may be due to complex regulation brought about by these miRNAs at post-transcriptional level. Fu et al. (2013) reported marker polymorphisms elucidated by miRNA-based markers indicate sequence changes in the miRNA loci, which may cause differential regulation of targeted genes. Overall these makers showed higher average PIC (0.68) and gene diversity (0.72) values within genotypes suggesting utility of these markers for genetic mapping applications.
Functional classification and pathway enrichment analysis for target genes of miRNAs
The complementarities between the miRNA and their target `3`genes provide the hierarchy of biological process regulation (Singh et al. 2017; Mishra and Bohra 2018). In view of this, the candidate mature miRNAs of 51 pre-miRNAs were selected for target analysis with 2417 pomegranate EST sequences serving as the reference transcript library. A total of 128 targets were identified that are classified into three GO categories with 13 biological process, 12 in molecular function, and 7 in cellular component. In the biological process category, the majority of the targets participated in cellular and organic substance metabolic processes. In molecular function category, predicated targets showed association with organic and hetero-cyclic compound binding. In the cellular component category, majority of target genes were assigned to intrinsic component of membrane, followed by intracellular and intracellular part, and membrane bounded organelle. Our results concur with an earlier study on genome wide identification of miRNAs in pomegranate and their comprehensive target analysis against known poplar (Populus trichocarpa) transcript database (Saminathan et al. 2016). Further, enrichment analysis through KEGG pathways revealed that out of 128 target genes 18 genes coding for 14 enzymes that are part of 11 pathways viz, purine metabolism, thiamine metabolism, phenylpropanoid biosynthesis, amino sugar and nucleotide sugar metabolism, mTOR signalling pathway, glutathione metabolism, drug metabolism-other enzymes, ascorbate and aldarate metabolism, phosphonate and phosphinate metabolism, glycerol phospholipid metabolism and pyrimidine metabolism. Taken together, these leads support the participation of the KEGG pathways in pomegranate fruit development. Recently, Saminathan et al. (2016) explained elaborately for enrichment of 41 miRNA families together with their target candidate genes involved in 107 major pathways related to pomegranate fruit development.
miRNA-families and their regulatory networks
To explore the relationship among the miRNAs and their targets, regulatory networks were constructed. Results suggest that ath-MIR157c has maximum targets (79 genes). The possible reasons may be miR157 family was the most abundant, followed by miR156, miR166, and miR168, with variants within each family in pomegranate genome during fruit development stages as indicated by Saminathan et al. (2016). This ath-MIR157c regulatory network shared maximum target genes with MIR2111b (18 genes) and ath-MIR156b (6). Singh et al. (2017) also found miR156 shared targets genes with miR157 in their study in brassica for abiotic stress tolerance. Chen et al. (2015) reported SlymiR157 and SlymiR156 differentially modulate ripening and softening in tomato (Solanum lycopersicum). Xu et al. (2013) reported, miR2111 was induced under P-deficiency stress. Similarly, mir157 showed differential expression upon P deficiency in common bean (Valdes-Lopez et al. 2010). Further, network analysis for individual miRNAs families with miRNA-targets revealed, ath-MIR157c with strongest interaction with stress associated protein 8, ath-MIR394a with unknown gene (pomegrante_JG771766.1), ath-MIR2111b (Zf-FLZ domain containing protein, ubiquitin-40S ribosomal protein and NADH dehydrogenase), ath-MIR5651 (MYB-like transcription factor), ath-MIR167d (MYB-like transcription factor and late embryogenesis abundant protein_At5g17165), ath-MIRNA156b (Tropinone reductase and hypothetical protein CRG98_029376) and for ath-MIR841b many genes suggesting possible positive targets. The miR156 and miR157 were reported to target transcription factors to bring about development stress response in cassava (Patanun et al. 2013). The miR394 was reported to target F-box genes to bring stress response in grapevine (Han et al. 2014). The miR2111 was reported to be downregulated in peanut in responses to bacterial wilt (Ralstonia solanacearum) (Zhao et al. 2015). The miR167 targets ARF transcription factors into bring stress responses to cold and drought in rice, cowpea and maize (Jeong et al. 2011; Barrera-Figueroa et al. 2011; Sheng et al. 2015). Similarly, regulatory network graphs were explored for understanding, miRNA and their target genes in various crops viz., bread wheat (Nigam et al. 2015), radish (Wei et al. 2016), maize (Wu et al. 2016) and brassica (Singh et al. 2017), With respect to overall regulatory role of some of these important miRNA families in various crops, miR157 known to target SPL genes to regulate floral organ size and ovule production in cotton (Liu et al. 2017), over expression of gma- miR394a in Arabidopsis enhanced drought tolerance (Ni et al. 2012), miR2111 was induced under P-deficiency stress (Xu et al. 2013) and over expression of OsmiR156k in rice reduced cold tolerance (Cui et al. 2015).
Although a number of SSR markers are currently available in various plant species, discovery of trait specific SSR markers that can be deployed in marker assisted breeding remains a formidable task. Mondal and Ganie (2013) first time reported identification of miR172b-SSR, which could enable differentiating salt tolerant and susceptible genotypes of rice. Therefore, miRNA-SSR holds great potential for identifying master miRNAs that can regulate hundreds of genes for traits of intrest. Later these miRNAs can be targeted for genome editing to gain yield advantages. To complement this, further we designed 58 functional EST-SSRs markers from 128 target genes of pomegranate for gene/QTL mapping studies. Similarly, Singh et al. (2017) reported characterization of 700 SSRs from 621 miRNA target genes of Brassica for marker development.
Conclusion
We identified a total of 1028 pri-miRNAs sequences that encode 1162 pre-miRNAs. Homology search was made using 1922 non-redundant mature miRNAs as query against pomegranate whole genome sequence cv. Dabenzi. SSR survey of these sequences facilitated identification of 1358 SSRs in pri-miRNAs, of which 238 SSRs were found in pre-miRNA sequences. Hexa-nucleotide repeats were the most abundant in both pri-miRNAs and pre-miRNA sequences. In summary, we identified genome-wide miRNA-SSRs i.e. 897 specific to pri-miRNAs and 168 for pre-miRNAs. Besides miRNA-SSRs, a set of 58 EST-SSRs was mined from 128 target genes of miRNAs. The study provides resource of highly informative functional molecular markers, which can assist for trait mapping and selections in pomegranate. Most notably, this is the first report on genome-wide identification and characterization of miRNA-SSRs in pomegranate, which could serve as a reference for identifying more sequences from non-coding repertoire of the genome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are highly grateful to the Indian Council of Agricultural Research (ICAR), New Delhi for extending financial support through ICAR-National Research Centre on Pomegranate, Solapur to carry out this work.
Abbreviations
- pri-miRNA
Primary micro RNA
- pre-miRNA
Precursor micro RNA
- SSRs
Simple sequence repeats
- CTAB
Cetyl trimethyl ammonium bromide
- NCBI
National Centre for Biotechnology Information
- UPGMA
Unweighted pair group method of arithmetic averages
- NJ
Neighbor joining
- PIC
Polymorphic information content
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Prakash G. Patil, Email: patilbt@gmail.com
N. V. Singh, Email: nripendras72@gmail.com
Shilpa Parashuram, Email: shilpa9193@gmail.com.
Abhishek Bohra, Email: abhi.omics@gmail.com.
Dhanajay M. Mundewadikar, Email: nikhilbio0695@gmail.com
Vipul R. Sangnure, Email: sangnurevipul@gmail.com
K. Dhinesh Babu, Email: ckdhinesh36@gmail.com.
Jyotsana Sharma, Email: jyotisharma128@yahoo.com.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3444. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. Short stack: comprehensive annotation and quantification of small RNA genes. RNA. 2014;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelj D, Jakse J, Javornik B. Assessment of genetic variability of olive varieties by microsatellite and AFLP markers. Euphytica. 2004;136:93–102. [Google Scholar]
- Barrera-Figueroa BE, Gao L, Diop NN, Wu Z, Ehlers JD, Roberts PA, Close TJ, Zhu J, Liu R. Identification and comparative analysis of drought associated microRNAs in two cowpea genotypes. BMC Plant Biol. 2011;11:127. doi: 10.1186/1471-2229-11-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. In: International AAAI conference on weblogs and social media. Palo Alto, California
- Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN, Kumar N, Farmer AD, Srivani G, Upadhyaya HD, Gothalwal R, Ramesh R, Singh D, Saxena KB, Kavi Kishor PB, Singh NK, Town CD, May GD, Cook DR, Varshney RK. Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol. 2011;11:56. doi: 10.1186/1471-2229-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, Singh IP, Yadav AK, Pathak A, Soren KR, Chaturvedi SK, Nadarajan N. Utility of informative SSR markers in the molecular characterization of cytoplasmic genetic male sterility-based hybrid and its parents in pigeonpea. Natl Acad Sci Lett. 2015;38:13–19. [Google Scholar]
- Bohra A, Jha R, Pandey G, Patil PG, Saxena RK, Singh IP, Singh D, Mishra RK, Mishra A, Singh F, Varshney RK, Singh NP. New hypervariable SSR markers for diversity analysis, hybrid purity testing and trait mapping in pigeonpea [Cajanus cajan (L.) Millspaugh] Front Plant Sci. 2017;8:377. doi: 10.3389/fpls.2017.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, He Y, Billiau K, Van de PY. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26:1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
- Chen M, Tan Z, Zeng G, Peng J. Comprehensive analysis of simple sequence repeats in pre-miRNAs. Mol Biol Evol. 2010;27:2227–2232. doi: 10.1093/molbev/msq100. [DOI] [PubMed] [Google Scholar]
- Chen W, Kong J, Lai T, Manning K, Wu C, Wang Y, Qin C, Li B, Yu Z, Zhang X, He M, Zhang P, Gu M, Yang X, Mahammed A, Li C, Osman T, Shi N, Wang H, Jackson S, Liu Y, Gallusci P, Hong Y. Tuning LeSPL-CNR expression by SlymiR157 affects tomato fruit ripening. Sci Rep. 2015;5:7852. doi: 10.1038/srep07852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui N, Sun X, Sun M, Jia B, Duanmu H, Lv D, Duan X, Zhu Y. Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa) Mol Breed. 2015;35:214. [Google Scholar]
- Ferrao LFV, Caixeta ET, Pena G, Zambolim EM, Cruz CD, Zambolim L, Cruz CD, Zambolim L, Ferrao MAG, Sakiyama NS. New EST–SSR markers of Coffea arabica: transferability and application to studies of molecular characterization and genetic mapping. Mol Breed. 2015;35:1–5. [Google Scholar]
- Frazier TP, Xie FL, Freistaedter A, Burklew CE, Zhang BH. Identification and characterization of microRNAs and their target genes in tobacco (Nicotiana tabacum) Planta. 2010;232:1289–1308. doi: 10.1007/s00425-010-1255-1. [DOI] [PubMed] [Google Scholar]
- Fu SW, Chen L, Man YG. miRNA biomarkers in breast cancer detection and management. J Cancer. 2011;2:116–122. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Ma BI, Naliese AN, Mason S, Xiao M, Wei L, An ZE. MicroRNA-based molecular markers: a novel PCR-based genotyping technique in Brassica species. Plant Breed. 2013;132:375–381. [Google Scholar]
- Ganie SA, Mondal TK. Genome-wide development of novel miRNA-based microsatellite markers of rice (Oryza sativa) for genotyping applications. Mol Breed. 2015;35:51. [Google Scholar]
- Han J, Fang J, Wang C, Yin Y, Sun X, Leng X, Song C. Grapevine microRNAs responsive to exogenous Gibberellin. BMC Genom. 2014;15:111. doi: 10.1186/1471-2164-15-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel-Beja R, Sherman A, Rubinstein M, Eshed R, Bar-Ya’akov I, Trainin T, Ophir R, Holland D. A novel genetic map of pomegranate based on transcript markers enriched with QTLs for fruit quality traits. Tree Genet Genomes. 2015;11:109. [Google Scholar]
- Hinchliffe DJ, Lu Y, Potenza C, Segupta-Gopalan C, Cantrell RG, Zhang J. Resistance gene analogue markers are mapped to homeologous chromosomes in cultivated tetraploid cotton. Theor Appl Genet. 2005;110:1074–1085. doi: 10.1007/s00122-005-1928-5. [DOI] [PubMed] [Google Scholar]
- Hu J, Vick BA. Target region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol Rep. 2003;21:289–294. [Google Scholar]
- Hu JH, Sun LL, Zhu ZX, Zheng Y, Xiong W, Ding Y. Characterization of conserved microRNAs from five different cucurbit species using computational and experimental analysis. Biochimie. 2014;102:137–144. doi: 10.1016/j.biochi.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell. 2011;23:4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Shen J, Liu H, Li F, Ding N, Gao C, Pattanaik S, Patra B, Li R, Yuan L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta. 2015;242:283–293. doi: 10.1007/s00425-015-2305-5. [DOI] [PubMed] [Google Scholar]
- Joy N, Soniya EV. Identification of an miRNA candidate reflects the possible significance of transcribed microsatellites in the hairpin precursors of black pepper. Funct Integr Genomics. 2012;12:387–395. doi: 10.1007/s10142-012-0267-2. [DOI] [PubMed] [Google Scholar]
- Joy N, Asha S, Mallika V, Soniya EV. De novo transcriptome sequencing reveals a considerable bias in the incidence of simple sequence repeats towards the downstream of ‘pre-miRNAs’ of black pepper. PLoS ONE. 2013;8(3):e56694. doi: 10.1371/journal.pone.0056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy N, Maimoonath BYP, Soniya EV. A deeper view into the significance of simple sequence repeats in pre-miRNAs provides clues for its possible roles in determining the function of microRNAs. BMC Genet. 2018;19:29. doi: 10.1186/s12863-018-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanupriya C, Radhika V, Ravishankar KV. Mining of miRNAs in pomegranate (Punica granatum L.) by pyrosequencing of part of the genome. J Hortic Sci Biotechnol. 2013;88:735–742. [Google Scholar]
- Kumar A, Chauhan A, Kompelli SK, Gahlaut V, Ijaq J, Singh KP, Prasad Gajula MNV, Suravajhala P, Mishra AK, Balyan HS, Gupta PK. Genome-wide mining, characterization and development of miRNA-SSRs in Arabidopsis thaliana. BioRxiv. 2017 [Google Scholar]
- Kusenda B, Mraz M, Mayer J, Pospisilova S. MicroRNA biogenesis, functionality and cancer relevance. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2009;150:205–215. doi: 10.5507/bp.2006.029. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lima JC, Loss-Morais G, Margis R. MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet Mol Biol. 2012;35:1069–1077. doi: 10.1590/s1415-47572012000600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu N, Tu L, Wang L, Hu H, Xu J, Zhang X. MicroRNA157 targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol. 2017;17:7. doi: 10.1186/s12870-016-0969-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37:2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X, Zhang Z, Liu Y, Wei X, Liu Z, Wang Y, Liu W. Genome-wide development of microRNA-based SSR markers in Medicago truncatula with their transferability analysis and utilization in related legume species. Int J Mol Sci. 2017;18:2440. doi: 10.3390/ijms18112440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Bohra A. Non-coding RNAs and plant male sterility: current knowledge and future prospects. Plant Cell Rep. 2018;37:177–191. doi: 10.1007/s00299-018-2248-y. [DOI] [PubMed] [Google Scholar]
- Mondal TK, Ganie SA. Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa) Gene. 2013;535:204–209. doi: 10.1016/j.gene.2013.11.033. [DOI] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. Overexpression of gma-MIR394a confers tolerance to drought in transgenic Arabidopsis thaliana. Biochem Biophys Res Commun. 2012;427:330–335. doi: 10.1016/j.bbrc.2012.09.055. [DOI] [PubMed] [Google Scholar]
- Nigam D, Puneet K, Sanjeev K, Dwijesh K, Mishra C, Rai A. Computational analysis of miRNA-target community network reveals cross talk among different metabolisms. Genomics Data. 2015;5:292–296. doi: 10.1016/j.gdata.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono NN, Britton MT, Fass JN, Nicolet CM, Lin D, Tian L. Exploring the transcriptome landscape of pomegranate fruit peel for natural product biosynthetic gene and SSR marker discovery. J Integr Plant Biol. 2011;53:800–813. doi: 10.1111/j.1744-7909.2011.01073.x. [DOI] [PubMed] [Google Scholar]
- Ophir R, Sherman A, Rubinstein M, Eshed R, Schwager MS, Harel-Beja R, Bar-Ya’akov I, Holland D. Single-nucleotide polymorphism markers from de-novo assembly of the pomegranate transcriptome reveal germplasm genetic diversity. PLoS ONE. 2014;9:e88998. doi: 10.1371/journal.pone.0088998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patanun O, Lertpanyasampatha M, Sojikul P, Viboonjun U, Narangajavana J. Computational identification of microRNAs and their targets in Cassava (Manihot esculenta Crantz.) Mol Biotechnol. 2013;53:257–269. doi: 10.1007/s12033-012-9521-z. [DOI] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. DARwin software. Paris: Centre de Cooperation Internationale en Recherche Agronomique Pour le Developpement (CIRAD); 2006. [Google Scholar]
- Qin G, Xu C, Ming R, Tang H, Guyot R, Kramer EM, Hu Y, Yi X, Qi Y, Xu X, Gao Z, Pan H, Jian J, Tian Y, Yue Z, Xu Y. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017;91:1108–1128. doi: 10.1111/tpj.13625. [DOI] [PubMed] [Google Scholar]
- Ravishankar KV, Anand L, Dinesh MR. Assessment of genetic relatedness among mango cultivars of India using RAPD markers. J Hort Sci Biotechnol. 2000;75:198–201. [Google Scholar]
- Ravishankar KV, Chaturvedi K, Puttaraju N, Gupta S, Pamu S, Flachowsky H. Mining and characterization of SSRs from pomegranate ( L.) by pyrosequencing. Plant Breeding. 2015;134:247–254. [Google Scholar]
- Russo G, Giordano A. miRNAs: from biogenesis to networks. Methods Mol Biol. 2009;563:303–352. doi: 10.1007/978-1-60761-175-2_17. [DOI] [PubMed] [Google Scholar]
- Saminathan T, Bodunrin A, Singh NV, Devarajan R, Nimmakayala P, Jeff M, Aradhya M, Reddy UK. Genome-wide identification of microRNAs in pomegranate (Punica granatum L) by high-throughput sequencing. BMC Plant Biol. 2016;16:122. doi: 10.1186/s12870-016-0807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Shah AA, Leidinger P, Blin N, Meese E. miRNA: small molecules as potential novel biomarkers in cancer. Curr Med Chem. 2010;17:4427–4432. doi: 10.2174/092986710794182980. [DOI] [PubMed] [Google Scholar]
- Sheng L, Chai W, Gong X, Zhou L, Cai R, Li X, Zhao Y, Jiang H, Cheng B. Identification and characterization of novel maize miRNAs involved in different genetic background. Int J Biol Sci. 2015;11:781–793. doi: 10.7150/ijbs.11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I, Smita S, Mishra DC, Kumar S, Singh BK, Rai A. Abiotic stress responsive miRNA-target network and related markers (SNP, SSR) in Brassica juncea. Front Plant Sci. 2017;8:1943. doi: 10.3389/fpls.2017.01943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani MH, Talebi M, Sayed-Tabatabaei BE. Use of SRAP markers to assess genetic diversity and population structure of wild, cultivated, and ornamental pomegranates (Punica granatum L.) in different regions of Iran. Plant Syst Evol. 2012;298:1141–1149. [Google Scholar]
- Valdes-Lopez O, Yang S, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G. MicroRNA expression profiles in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010;187:805–818. doi: 10.1111/j.1469-8137.2010.03320.x. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang JY, Yang XD, Xu HB, Chi XY, Zhang M, Hou XL. Identification and characterization of microRNAs and their target genes in Brassica oleracea. Gene. 2012;505:300–308. doi: 10.1016/j.gene.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Wei Z, Xie Y, Xu L, Wang Y, Zhu X, Wang R, Zhang Y, Muleke EM, Liu L. Identification of microRNAs and their target genes explores miRNA-mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus L) Front Plant Sci. 2016;7:1054. doi: 10.3389/fpls.2016.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ding D, Shi C, Xue Y, Zhang Z, Tang G, Tang J. MicroRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol. 2016;16:73. doi: 10.1186/s12870-016-0755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie FL, Frazier TP, Zhang BH. Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum) Gene. 2011;473:8–22. doi: 10.1016/j.gene.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Xu F, Liu Q, Chen L, Kuang J, Walk T, Wang J, Liao H. Genome-wide identification of soybean microRNAs and their targets reveals their organ specificity and response to phosphate starvation. BMC Genom. 2013;14:66. doi: 10.1186/1471-2164-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Fang Y, Zhang T, Fei Z, Han F, Liu C, Liu M, Xiao W, Zhang W, Wu S, Zhang M, Ju Y, Xu H, Dai H, Liu Y, Chen Y, Wang L, Zhou J, Guan D, Yan M, Xia Y, Huang X, Liu D, Wei H, Zheng H. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol J. 2018;16:1363–1374. doi: 10.1111/pbi.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- Zhao C, Xia H, Cao T, Yang Y, Zhao S, Hou L, Zhang Y, Li C, Zhang X, Wang X. Small RNA and degradome deep sequencing reveals peanut microRNA roles in response to pathogen infection. Plant Mol Biol Rep. 2015;33:1013–1029. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.