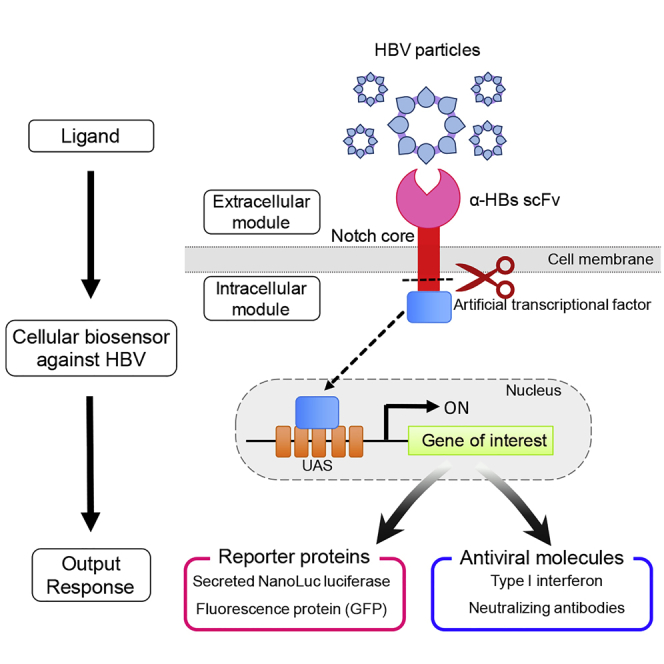
Summary
SynNotch receptor technology is a versatile tool that uses the regulatory notch core portion with an extracellular scFv and an intracellular transcription factor that enables to program customized input and output functions in mammalian cells. In this study, we designed a novel synNotch receptor comprising scFv against HBs antigen linked with an intracellular artificial transcription factor and exploited it for viral sensing and cellular immunotherapy. The synNotch receptor expressing cells sensed HBV particles and membrane-bound HBs antigens and responded by expressing reporter molecules, secNL or GFP. We also programmed these cells to dispense antiviral responses such as type I interferon and anti-HBV neutralizing mouse-human chimeric antibodies. Our data reveal that synNotch receptor signaling works for membrane-bound ligands such as enveloped viral particles and proteins borne on liposomal vesicles. This study establishes the concepts of “engineered immunity” where the synNotch platform is utilized for cellular immunotherapy against viral infections.
Subject Areas: Immunity, Viral Microbiology, Bioengineering
Graphical Abstract
Highlights
-
•
We designed synNotch receptor with scFv to sense HBs antigen
-
•
SynNotch-receptor-transduced cells can express reporter and antiviral molecules
-
•
Membrane-bound viral surface proteins can activate synNotch signaling
-
•
SynNotch technology can foster “engineered immunity” against viral infections
Immunity; Viral Microbiology; Bioengineering
Introduction
Biosensors are devices that detect biological analytes in the test environment and notify with a specific output response. Cellular biosensors possess unique advantages over conventional biosensors, including the possibility of “in situ” use within the test environment (Gui et al., 2017). Although programming of prokaryotic cells with synthetic genetic circuits to use as “whole cell biosensors” has long been in vogue, the recent advancements in synthetic biology have opened the doors to engineer mammalian cells to yield a user-defined output in response to a specific input signal (Baeumler et al., 2017, Xia et al., 2019). With the availability of modular receptor platforms, human cells can be re-wired to possess novel input-output relationships, allowing them to be exploited as biosensors with a wide range of functionality.
The synthetic biology tool kit for engineering cellular biosensors currently comprises of modular receptor technologies such as receptors activated solely by synthetic ligands (RASSL), transcriptional activation following arrestin translocation (Tango), chimeric antigen receptors (CAR), synthetic Notch receptors (synNotch), and modular extracellular sensor architecture (MESA) (Brenner et al., 2017). Among these, the synNotch platform offers high pliability in programming input-output relationship, centered over surface receptor-ligand interaction. This technology provides the scope of customizing the extracellular receptor domains for sensing and the intracellular artificial transcription factor to transcribe specific response genes. In cells engineered with this technology, engagement of the extracellular domain with its corresponding ligand induces the cleavage of the transcriptional regulatory domain, which activates the transcriptional circuit specific for the expression of pre-programmed genetic material (Morsut et al., 2016). As this technology offers immense scope in customization, we exploited it to develop bio-sensor cells with antiviral activity against hepatitis B virus (HBV).
Among all blood-borne viral infections, HBV poses the major problem with respect to the magnitude of prevalence, as it infects more people than human immunodeficiency virus (HIV) and hepatitis C virus (HCV) put together (Gomes et al., 2019). Chronicity is the hallmark feature of HBV infection caused by a persistent viral reservoir, which leads to severe complications such as end-stage liver diseases such as liver cirrhosis and hepatocellular carcinoma. Chronic HBV infection poses formidable diagnostic and therapeutic challenges especially with regard to monitoring the course of infection (Liu et al., 2019). Current antiviral chemotherapy for HBV infection comprises non-nucleoside reverse transcriptase inhibitors (NRTIs), entecavir (ETV) and tenofovir disoproxil fumarate (TDF) (Clark and Hu, 2015). Despite the putative anti-viral therapy, NRTIs do not offer a complete cure for the disease because long-term treatment with NRTIs is associated with adverse reactions and the emergence of drug-resistant sub-strains. It is therefore necessary to develop alternative approaches for treating HBV infection by sensing the fluctuation of viral antigens in vivo and executing de novo anti-viral actions.
Exploiting in vivo viral sensing, we engineered whole-cell biosensors for HBV by re-wiring the cellular genetic circuits to offer pre-programmed outputs, upon sensing the whole virus or sub-viral particles. The synNotch receptor utilizes mechanotransduction to convert ligand binding (input) into proteolytic release of the sequestered transcription factor that enters the nucleus to express the desired genetic element (output). The input system comprises of the extracellular receptor, a single-chain variable fragment (scFv) against HBV surface antigen (HBs) (αHBs-scFv), which detects and engages with subviral particles and also the Dane particles in the extracellular milieu with high specificity and consequently cleaves the notch core to release the Gal4-VP64 transcription factor (Morsut et al., 2016). Subsequent nuclear translocation of the released Gal4-VP64 proceeds to activate the expression of either of the pre-programmed genetic elements, secNL or GFP reporter molecules. Moreover, we exploited our system for innate or acquired anti-viral immune responses, i.e. interferon-beta (IFNβ) or an anti-HBV neutralizing mouse-human chimeric antibody. We demonstrate the robust performance of these cells in antigen sensing and dispensing different reporters or macromolecules of immune response and hence propose this system as a unique platform that can be utilized for developing novel HBV diagnostics and therapeutics.
Results
Engineering HBV Biosensor Cells Utilizing the synNotch Platform
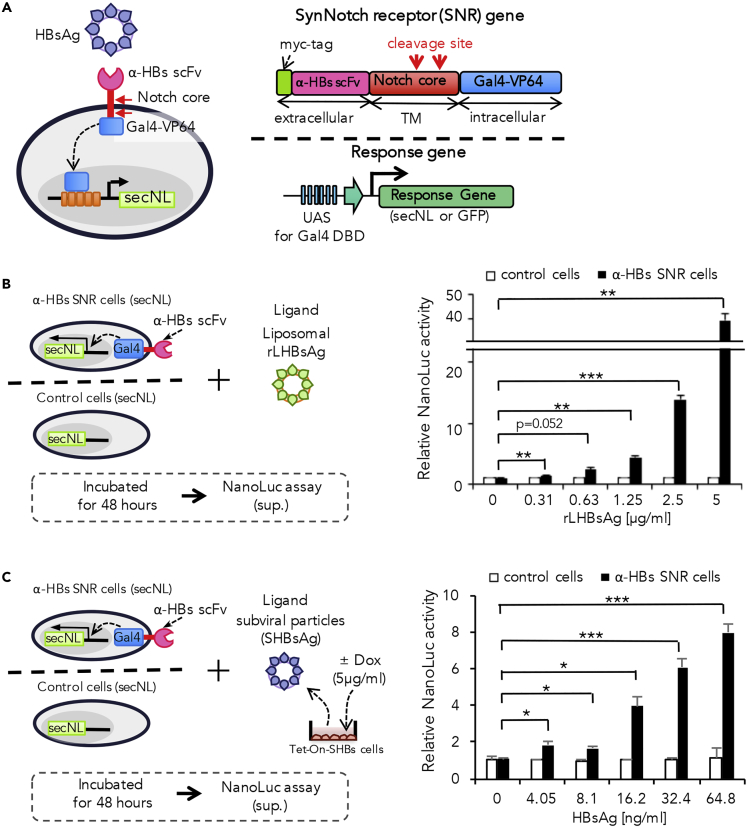
α-HBs-scFv was combined with notch core to generate the desired synNotch receptor, designated here as anti-HBs synNotch receptor (α-HBs SNR). The α-HBs SNR is composed of myc-tagged α-HBs scFv as the extracellular domain and a trans-membranous notch core region fused to Gal4-VP64 fusion protein as the artificial transcription factor. We transduced α-HBs SNR gene to Jurkat cells, which were designed to express reporter genes through the upstream activating sequences (UASs) with a tandem array of five optimized Gal4-binding sites. We introduced either secNL or GFP as the response biomarkers upon antigen stimulation (Figure 1A). We tested two α-HBs scFvs (S1and S2; GenBank accession numbers: AF410257/AF410258 and AB027447/AB027448, respectively) against a common antigenic “a” determinant region within SHBs (Figure S1A). These antibodies could recognize both subviral and infectious viral particles. Cell-surface expressions of these two α-HBs SNRs were verified by flow cytometry analysis with anti-myc antibody. We observed that α-HBs SNRs with both S1 and S2 were adequately expressed on the cell surface at higher rates as 92%–97% of transduced cells (Figure S1B). To test the functionality of these two α-HBs SNR cells generated, the cells were exposed to rLHBs, and secNL activity was monitored in the cell supernatant. We found that the S2 α-HBs SNR cells showed remarkable induction of secNL upon rLHBs stimulation, whereas the other exhibited only marginal induction (Figure S1C). Parallel experiments demonstrated that stimulation through the S2 α-HBs SNR caused a dose-dependent increase of the secNL reporter expression (Figure 1B). Therefore, we used the S2 α-HBs SNR (hereafter α-HBs SNR) in our subsequent analyses.
Figure 1.
Establishment of HBV-Sensing Cells Using Anti-HBs scFv synNotch Receptor (α-HBs SNR) and Sensing HBs Protein by α-HBs SNR Cells
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.isci.2020.100867.
(A) Schematic diagram of α-HBs SNR cells against HBsAg. The α-HBs SNR cell has α-HBs SNR on cell surface and Gal4-response gene in the nucleus. SynNotch receptor has myc-tagged scFv for HBsAg binding, the Notch core activation domain, and an artificial transcription factor (TF), Gal4-VP64. Reporter genes (secreted NanoLuc luciferase (secNL) or GFP) are designed to be regulated by Gal4 TF via an upstream activating sequence (UAS).
(B) The α-HBs SNR cells expressed secreted NanoLuc luciferase (secNL) in response to liposomal recombinant LHBsAg (rLHBsAg) in dose-dependent manner. The control or α-HBs SNR cells and rLHBsAg were incubated at 37°C, and the NanoLuc activity in the cell supernatant was measured after 48 h. Control cells indicate Jurkat T cells harboring only secNL reporter gene without α-HBs SNR. Each value is normalized to those obtained from without rLHBsAg and represents the mean ± standard deviation (SD) of three independent experiments. Horizontal lines represent axis break of the y-axis. **p: <0.01, ***p: <0.001.
(C) The α-HBs SNR cells sensed HBsAg secreted by Tet-ON-SHBs cells in dose-dependent manner. α-HBs SNR cells and HBsAg derived from untreated or 5μg/mL doxycycline (Dox)-treated Tet-ON-SHBs cells were incubated at 48 h and then the NanoLuc signal in the supernatant was measured. Control cells indicate Jurkat T cells harboring only secNL reporter gene without α-HBs SNR. Amount of HBsAg was calculated using HBs ELISA kit. Each value is normalized to those obtained from without HBsAg and represents the mean ± SD of three independent experiments. *p: <0.05, ***p: <0.001.
We then tested if the α-HBs SNR cell can respond to HBs antigen on subviral particles. For this, we generated HepG2 cells with tetracycline-inducible HBs gene expression, designated here as Tet-ON-SHBs cells. Treatment of the Tet-ON-SHBs cells with doxycycline (Dox) caused the expression of HBsAg and/or subviral particles in cell supernatant in a dose-dependent manner (Figure S2A). When α-HBs SNR cells were exposed to the secreted HBsAg and/or subviral particles derived from Tet-ON-SHBs cells, we observed a dose-dependent increase of the secNL reporter (Figure 1C). The α-HBs SNR cells responded exclusively to stimulation by HBV but not to virus-like particles (VLP) derived from other viral species (Figure S2B). As α-HBs SNR cells are basically Jurkat cells, they do not express the entry receptors such as sodium taurocholate co-transporting polypeptide (NTCP) for HBV infection. Therefore, all interactions between HBV and α-HBs SNR cells principally happen only at the extracellular scFv domain.
As synNotch receptor is conventionally considered to be activated by the mechanical force induced by surface ligands, this observation of activation by membrane-bound soluble ligands was surprising. We hypothesized that the viral particle could attach to its extracellular receptor on a cell and this “receptor-ligand complex” might act as a surface ligand to activate the synNotch receptor of an adjacent cell and not that of the same cell. To check this, we challenged decreasing cell densities of α-HBs SNR cells with the equivalent amount of viral antigen, expecting that the cells would be spatially dispersed far to interact and activate another cell. Interestingly, we observed significant luciferase activity in all the lower cell densities tested (Figure S2C), suggesting the possibility of synNotch activation by membrane-bound soluble ligands such as surface antigens of enveloped viruses.
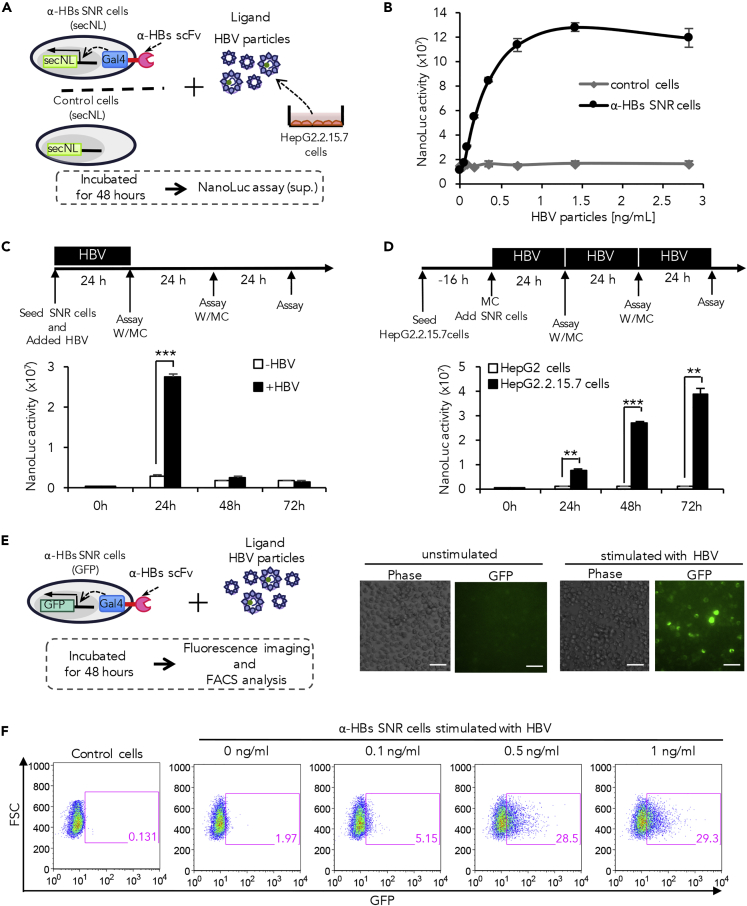
α-HBs SNR Cells Sense Subviral Particles and Infectious Virions Derived from HBV Producer Cells
We next investigated whether the α-HBs SNR cells could sense infectious HBV particles (Dane particles). The α-HBs SNR cells with secNL reporter gene were exposed to purified virions derived from supernatant of HepG2.2.15.7 cells, which were stably expressing a complete HBV genome (Figure 2A). Increase in NanoLuc activity was observed in a dose-dependent manner with a half maximal effective concentration of 0.24 ng/mL (Figure 2B). We then proceeded to detect the functioning of α-HBs SNR cells exposed to stimuli from purified viral particles versus co-cultured infected cells at different time intervals. Following a single antigenic challenge with purified viral particles, a robust and significant output response was noted after 24 h (Figures 2C and S3A). These cells were then washed and re-suspended in fresh medium to remove the input stimuli, and the NanoLuc activity was measured again at 48 h; this process was again repeated at 72 h. We observed the output response to be annulled upon removing the stimulus (Figures 2C and S3A), denoting that the α-HBs SNR cells need continuous exposure to input signals in order to give a persistent output. To assess the correlation between continuity of input with persistence of output, the above experiment was carried out employing co-cultured HepG2.2.15.7 cells that provide continually increasing input signals by releasing actively replicating virions (Figure 2D). Similar to viral particle stimulation, input signals from infected cell co-culture elicited a brisk output response at 24 h. However unlike the former, the output response to co-culture stimulation persisted even at 48 and 72 h reiterating the need of constant exposure to input stimuli for obtaining a sustained output response. In the co-culture experiment, we also noted an increase in the magnitude of the output with increasing time, despite periodic washing free viruses from the medium (Figure 2D). The most probable explanation for this trend could be due to cell proliferation over time, causing the increase in cell numbers of both the input-signal-producing cells and the α-HBs SNR cells. To confirm this on individual cells, we performed a single-cell reporter assay using bioluminescence microscopy to measure luminescence signal intensity per cell, using ImageJ software. In the co-culture system, we observed not only higher numbers of luminescent cells (many cells being activated) but also highly luminescent cells (many receptors activated per cell) (Figure S3B).
Figure 2.
Sensing and Response to HepG2.2.15.7-Cell-Derived HBV Particles by the α-HBs SNR Cells
(A) Experimental design of α-HBs SNR cells expressing secNL in response to HBV particles and subviral particles derived from HepG2.2.15.7 cells. Purified HBV particles derived from cell supernatants of HepG2.2.15.7 cells were added to α-HBs SNR cells and incubated at 37°C for 48 h followed by measurement of NanoLuc signal.
(B) The NanoLuc activity of α-HBs SNR cells increased in a dose-dependent manner. α-HBs SNR cells were incubated with each amount of HBV particles for 48 h and NanoLuc activity in the cell supernatant measured. Control cells indicate Jurkat T cells harboring only secNL reporter gene without α-HBs SNR. The data shown represent the mean ± standard deviation (SD) of three independent experiments.
(C) Timecourse of NanoLuc activity in the cell supernatant of α-HBs SNR cells incubated with HBV particles. Upper panel indicates the time course of events in this experiment. α-HBs SNR cells were incubated with 0.5 ng/mL HBV particles. Cells were washed and re-suspended in fresh medium every 24 h and NanoLuc activity in the cell supernatant measured prior each medium change. α-HBs SNR cells without exposure to any input stimulus were used as controls. The data shown represent the mean ± SD of three independent experiments. ***p: <0.001. W/MC: wash and medium change.
(D) Timecourse of NanoLuc activity in the cell supernatant of α-HBs SNR cells co-cultured with HepG2.2.15.7 cells. Upper panel indicates the time course of events in this experiment. α-HBs SNR cells were co-cultured with HepG2.2.15.7 cells (HBV-producing cells) or HepG2 cells (uninfected negative control). Cells were washed and medium was refreshed every 24 h and NanoLuc activity in the cell supernatant measured prior each medium change. The data shown represent the mean ± SD of three independent experiments. **p: <0.01, ***p: <0.001. W/MC: wash and medium change.
(E) Response of α-HBs SNR cells incorporated with GFP reporter gene to HBV particles. GFP expressing α-HBs SNR cells were observed by fluorescence microscopy after 48 h of incubation in the presence or absence of 4 ng/mL HBV particles derived from HepG2.2.15.7 cells. Scale bars represent 40 μm.
(F) GFP expression of α-HBs SNR cells increased in a dose-dependent manner. GFP-expressing α-HBs SNR cells were observed by flow cytometry after 48 h incubation in the absence or presence of each amount of HBV particles derived from HepG2.2.15.7 cells. Control cells indicate Jurkat T cells harboring only GFP reporter gene without α-HBs SNR.
In order to characterize biosensor performance, we further explored the variations in receptor activity across entire populations of engineered cells by flow cytometry. α-HBs SNR cells with GFP reporter system challenged with HBV whole viral particles gave off fluorescence, which was visualized through fluorescence microscopy (Figure 2E). We next analyzed single cells with respect to population and output reporter intensities by flow cytometry. Both the fluorescence intensity and the population of fluorescing cells were found to be directly related to the concentration of input signals (Figure 2F).
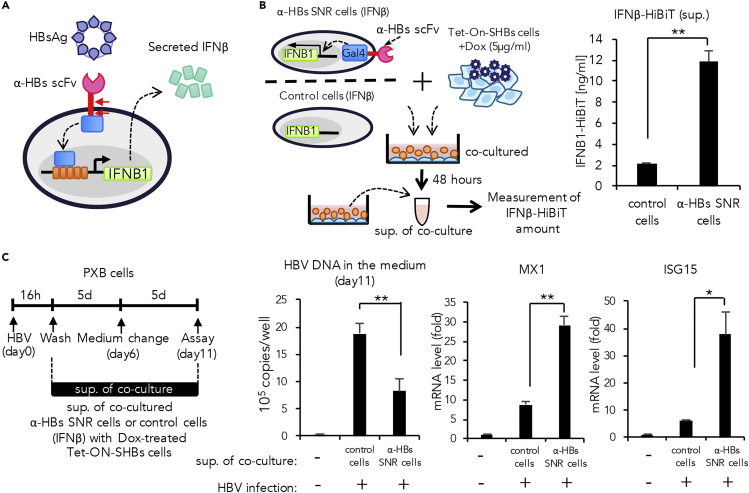
Generation of α-HBs SNR Cells Eliciting Innate Anti-viral Response
The innate immune system is essential for the initial detection of invading viruses and subsequent activation of anti-viral immunity. The innate immune response to viral pathogens originates from the antigen recognition by pattern recognition receptors (PRRs) that serve as important cellular sensors for detecting pathogen-associated molecular patterns (PAMPs). Activation of PRRs upregulates the expression of type I interferon and promotes clearance of the invading pathogens (Takeuchi and Akira, 2010, Thompson et al., 2011). We investigated whether the innate immune response against HBV could be duplicated with a simple logic function of antigen-input to interferon-output using our synNotch system.
We generated α-HBs SNR cells that could produce interferon β tagged at their carboxy terminals with the tiny 11-amino-acid luminescent tag HiBiT (Promega) (IFNβ-HiBiT) as a pre-defined output response (Figure 3A). The α-HBs SNR cells were co-cultured with Dox-treated Tet-ON-SHBs cells for 48 h, and secreted IFNβ-HiBiT was measured by the HiBiT-based protein quantitation system (Ranawakage et al., 2019, Sasaki et al., 2018). We observed a robust secretion of IFNβ-HiBiT by the α-HBs SNR cells upon the antigenic stimulation (Figure 3B).
Figure 3.
Direct Induction of the Innate Immune Molecule hIFNβ as Output Response in α-HBs SNR Cells
(A) α-HBs SNR cell design for induction of interferon-beta (IFNβ).
(B) Experimental design for induction of IFNβ in α-HBs SNR cells and quantification of induced IFNβ. α-HBs SNR cells were induced with HiBiT-tagged IFNβ by co-culture with Tet-ON-SHBs cells in presence of 5μg/mL doxycycline (Dox). After 48 h, IFNβ-HiBiT in the cell supernatant was quantified. Control cells indicate Jurkat T cells harboring only IFNβ-HiBiT gene without α-HBs SNR. The data shown represent the mean ± standard deviation (SD) of three independent experiments. **p: <0.01.
(C) Effect of IFNβ derived from the supernatant of α-HBs SNR cells stimulated by HBsAg from Dox-treated Tet-ON-SHBs cells. Primary human hepatocytes (PXB cells) were infected with HBV for 16 h and then were cultured in the presence or absence of supernatant containing α-HBs-SNR-cell-derived IFNβ (30%v/v). HBV DNA in the medium was extracted from these cell supernatants 11 days after infection and quantified using real-time PCR analysis. The expression of representative IFNβ-stimulated genes; MX1 and ISG15, were quantified using qPCR and normalized against the expression of β-actin in PXB cells. Control indicates the cell supernatant of Jurkat T cells harboring only IFNβ-HiBiT gene without α-HBs SNR, co-cultured with Dox-treated Tet-ON-SHBs cells. The data shown represent the mean ± SD of three independent experiments. *p: <0.05, **p: < 0.01.
We further proceeded to assess the antiviral activity of the IFNβ-HiBiT produced by α-HBs SNR cells. Primary human hepatocytes prepared from humanized mice with chimeric liver (PXB cells) were infected with wild-type HBV and then cultured in the presence or absence of cell supernatant containing IFNβ-HiBiT secreted from α-HBs SNR cells (30% v/v). Treatment with the cell supernatant significantly reduced HBV DNA in the medium reflecting the suppression of viral replication, with concomitant induction of interferon-stimulated genes MX1 and ISG15, which are known to inhibit HBV replication (Figure 3C). These results indicate that α-HBs SNR cells can initiate an effective synthetic anti-viral innate immune response following the recognition of HBs antigen.
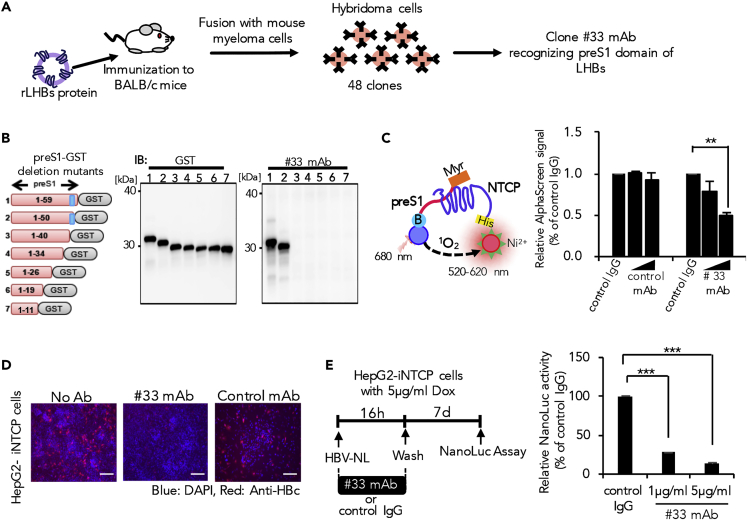
Generation of a Neutralizing Antibody Targeting HBV for Mounting into α-HBs SNR Cells
The humoral immune response is the aspect of acquired immunity mediated by antibody macromolecules in extracellular spaces secreted by B lymphocytes or plasma cells. Neutralizing antibodies (nAbs) generated during the humoral immune response form a highly effective defense in preventing or clearing viral infection (Burton, 2002, Hangartner et al., 2006). We examined the feasibility of simplifying the humoral immune response by directly producing nAbs upon antigen recognition using our newly developed synNotch system.
To achieve this, we attempted to create novel in-house neutralizing antibodies that could neutralize HBV and prevent infection. As it is known that large HBV surface protein (LHBs) binding to host NTCP mediates the early viral entry process, we attempted to develop monoclonal antibody (mAb) that specifically interfere with the LHBs-NTCP interaction (Yan et al., 2012). BALB/c mice were immunized with full-length LHBs recombinant protein derived from genotype C virus to subsequently establish 48 hybridoma clones, designated here as #1 to #48. We selected seven clones that were capable of detecting LHBs, of which only #33 mAb comprehensively recognized LHBs derived from all HBV genotypes tested (A, B, C, and D) (Figures 4A, S4A, and S4B). Epitope mapping analysis further revealed that #33 mAb could recognize the region between the 40th and 49th amino acid residues of preS1 domain that interacts with NTCP (Figure 4B). To examine the inhibitory function of this antibody in the preS1-NTCP interaction, we developed an AlphaScreen assay that gives off luminescence signals upon interaction of the two specific test proteins. This assay was designed such that binding of recombinant NTCP with only preS1 peptide, but not other non-specific peptides, yields a luminescence signal. Recombinant GST was used as the negative control protein to validate the specificity of the test system, as its binding with preS1 peptide did not produce a signal (Figure S4C). Using this assay, we tested the functional aspect of #33 mAb. Our results showed that this antibody could inhibit the preS1-NTCP interaction in a dose-dependent manner (Figure 4C). We further proceeded to explore the neutralizing activity of #33 mAb on HBV infection. HepG2 cells with tetracycline-inducible expression of NTCP (HepG2-iNTCP) cells (Miyakawa et al., 2018) were infected with NanoLuc luciferase-encoding HBV (HBV-NL) particles (Miyakawa et al., 2018, Nishitsuji et al., 2015) pre-treated with #33 mAb or control mAb. Our result demonstrated that the pre-treatment with #33 mAb, but not control mAb, significantly suppressed HBV infection (Figures 4D and 4E). Taken together, these results indicate that our newly developed anti-preS1 #33 mAb can efficiently neutralize HBV and inhibit infection by directly interfering with the preS1-NTCP interaction.
Figure 4.
Generation and Characterization of α-HBs Monoclonal Antibody
(A) Schematic diagram of hybridoma cell production and selection to generate α-HBs monoclonal antibodies (mAb). BALB/c mice were immunized with recombinant LHBs protein, and the splenocytes were fused with mouse myeloma cells. After producing hybridoma cells, one HBs mAb (# 33) that recognized preS1 domain of four HBV genotypes (A, B, C, and D) was selected.
(B) Epitope mapping of generated #33 mAb. Recombinant full-length or deletion-mutant preS1 proteins fused with GST protein were produced by wheat germ cell-free protein system and subjected to immunoblot using anti-GST antibody or #33 mAb. The predicted epitope of #33 mAb is shown in blue.
(C) Inhibition of preS1 domain and NTCP interaction by #33 mAb using AlphaScreen assay. AlphaScreen signal of biotinylated preS1 peptide and NTCP interaction was decreased in a dose-dependent manner by the addition of #33 mAb. Each value is normalized to those obtained from control IgG and represents the mean ± standard deviation (SD) of three independent experiments. **p: < 0.01.
(D and E) #33 mAb inhibits HBV infection. HepG2 Tet-ON-NTCP (HepG2-iNTCP) cells stimulated with 5μg/mL doxycycline (Dox) were infected with either HBV (D) or its reporter virus (HBV-NL) (E), in the presence of #33 mAb or negative control mAb. Viral infectivity was determined by intracellular HBcAg staining (D) or NanoLuc activity (E) of infected cell lysates. Each value is normalized to those obtained from control IgG and represents the mean ± SD of three independent experiments. ***p: < 0.001. Scale bars represent 100 μm.
α-HBs SNR Cells Incorporated with Neutralizing Antibody Gene Functionally Mimic Humoral Immune Cells
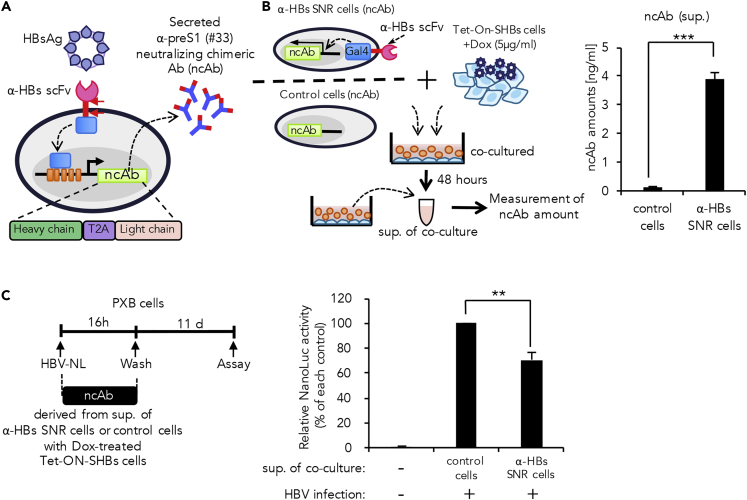
To introduce the mechanics of the humoral immune response into the α-HBs SNR cell system, we cloned the #33 antibody variable regions and constructed a mouse-human chimeric antibody (Figure S4D). The chimeric antibody was capable of inhibiting preS1 peptide-NTCP interaction (Figure S4E) and was also able to suppress HBV infection in PXB cells (Figure S4F). Hence we introduced this antibody into our synNotch system. Because antibody expression requires equivalent amounts of heavy chain and light chain, we designed a single gene cassette comprising of heavy and light chain genes connected with furin/T2A self-processing peptide and then inserted it downstream of UAS (Li et al., 2012). In this system, we expected the α-HBs SNR cells to secrete the neutralizing α-preS1 chimeric antibody (ncAb; originated from mouse #33 mAb) as an output response upon antigenic stimulation (Figure 5A).
Figure 5.
Direct Induction of a Neutralizing Chimeric Antibody (ncAb) as Output Response in α-HBs SNR Cells
(A) α-HBs SNR cell design for induction of α-preS1 (#33) ncAb and gene construct of chimeric antibody with heavy chain and light chain linked by T2A sequence.
(B) Experimental design for induction of ncAb in α-HBs SNR cells and quantification of induced ncAb derived from α-HBs SNR cells. ncAb was induced from α-HBs SNR cells by co-culture with Tet-ON-SHBs cells in the presence of 5μg/mL doxycycline (Dox). After 48 h, ncAb in the cell supernatant were quantified by AlphaScreen assay. Control cells indicate Jurkat T cells harboring only ncAb gene without α-HBs SNR. The data shown represent the mean ± standard deviation (SD) of three independent experiments. ***p: < 0.001.
(C) Secreted ncAb suppresses HBV-NL infection in PXB cells. Schematic diagram of HBV-NL infection experiment timeline. PXB cells were infected with HBV-NL in the presence or absence of secreted ncAb derived from α-HBs SNR cells. Infectivity was determined by measuring luciferase activity 11 days after infection. Control cells indicate the cell supernatant of Jurkat T cells harboring only ncAb gene without α-HBs SNR co-cultured with Dox-treated Tet-ON-SHBs cells. Values obtained from supernatant of control cells were set as 100%, and the presented data represent the mean ± SD of three independent experiments. **p: < 0.01.
We next investigated whether the α-HBs SNR cells could generate the pre-programmed output following the antigen stimulation. The α-HBs SNR cells were co-cultured with Dox-treated Tet-ON-SHBs cells. After 48 h, secreted ncAbs in the cell supernatant were measured by AlphaScreen using preS1 peptide as a probe. Our result showed that cells successfully secreted the ncAb upon antigen stimulation (Figure 5B).
Subsequently, we examined the neutralizing activity of the secreted antibody. The cell supernatant containing secreted ncAb was pre-incubated with HBV-NL and was then allowed to infect PXB cells. We found that the ncAb secreted from the α-HBs SNR cells significantly suppressed the infection of HBV-NL (Figure 5C). These results taken together highlight the capability of the α-HBs SNR cells to mimic the humoral immune cells and hints the possibility of in situ installation to protect from HBV infection.
Discussion
The availability of the modular chimeric receptor technology has given us the capability of rewiring cellular genetic circuits to provide a user-defined output for a specific input. We exploited the synthetic biology tool kit to engineer functional whole-cell biosensors for HBV surface antigen. For this purpose, we chose the synNotch technology as it senses extracellular membrane bound ligands and offers high input-output programmability (Morsut et al., 2016). In the conventional sense, canonical notch signaling is activated by receptor dissociation when the extracellular receptor domain engages with its corresponding membrane-bound ligand on adjacent cell, and the latter offers a mechanical shearing force by endocytosing the ligand-receptor complex (Parks et al., 2000). This is considered as one of the caveats of the synNotch system, as it could sense only the cell surface ligands and not the soluble ligands (Schwarz et al., 2017).
In our current study, we demonstrate that the synNotch receptor can also recognize and respond to virally encoded membrane antigens. The ligand analytes used for testing included recombinant HBs protein in liposomes (rHBs), enveloped subviral particles generated by SHBs expressing HepG2 cells, and the infectious whole viral Dane particles derived from HepG2.2.15.7 cells expressing a complete HBV genome. Interestingly, all these membrane anchored soluble ligands were capable of eliciting the activation of synNotch receptor. Stimulation of the α-HBs SNR cells under low cell density was able to trigger the output response despite spatial separation of the cells (Figure S2C). This raises the possibility that membrane-bound viral surface proteins generate sufficient mechanical force to cleave the notch domain to activate the canonical signaling cascade. Alternatively, signal activation could be initiated by less-known non-canonical pathways to activate notch signaling, which are yet to be elucidated in detail (Steinbuck and Winandy, 2018). Interestingly, α-HBs SNR cells were found to be more responsive to cell-derived than recombinant viral antigens. This was noticed when HBsAg secreted by Tet-On-SHBs cells activated the α-HBs SNR cells at nanomolar concentrations, whereas the rLHBs protein could achieve this effect only at micromolar concentrations (Figures 1B and 1C). The possible explanation for this could be the lesser antigen density and/or structural intactness of the rLHBs protein compared with the HBsAg (Bruss, 2004).
Yet another notable observation is the persistence of output with respect to the continuity of the input signals. The output response drastically dropped when the input signal was halted and similarly the output response increased and persisted with continual exposure to antigenic stimulation (Figures 2C and 2D). We also observed that increasing input signals activated more number of α-HBs SNR cells and also triggered more receptors on each cell (Figures 2F and S3B). Taken together, these findings suggest that the α-HBs SNR cells have a robust ON/OFF switch that is directly proportional to the concentration of the antigen encountered. This feature could possibly be exploited for monitoring reactivation of infection and viral blips in long-term infections and also for administering prompt in situ therapy during these incidents. One of the minor fallacies of this system was the constant low-level expression of the output genes even at the OFF state (Figures 2B and 2F). This is possibly due to the non-specific activation of the Gal4-VP64 transcription factor even at basal conditions and could be overcome either by further optimizing the existing system or by upgrading the construct with other more efficient trans-activating systems (Sevin-Pujol et al., 2017).
The α-HBs SNR cells could mimic the function of innate immune cells with PRRs that can recognize viral PAMPs and secrete type I interferons to limit further infection. HBV is known to evade innate immunity either by escaping recognition by PRRs or by dampening the production of interferons even after recognition by PRRs (Mutz et al., 2018). This creates an environmental niche with reduced immune pressure for the virus to survive and persist. Evasion from innate immunity is one of the key factors that helps HBV to establish a chronic infection with constant low-level viral replication (Suslov et al., 2018a). We have shown α-HBs SNR cells to secrete INFβ upon direct encounter with the viral antigen even without HBV invasion into cells (Figure 3B). Furthermore, the secreted IFNβ significantly reduced viral production in the infected cells (Figure 3C). This could be an interesting concept in immunotherapy of HBV as the α-HBs SNR cells overcome the stealth strategies used by the virus to evade the innate immunity mounted by natural cells (Suslov et al., 2018b).
In chronic infections, HBV evades humoral immune clearance either by producing excess of subviral decoys or by immune escape mutants (Rehermann and Nascimbeni, 2005). Moreover, chronic infection with the HBV is typically established neonatally and progresses during the phase of immune tolerance. The non-infectious subviral particles, which are produced in several fold excess to the infectious Dane particle, soak up the antibodies produced during natural humoral response and protects the virus from being neutralized (Rydell et al., 2017). These decoys are typically composed of small HBs that lacks the neutralizing epitope preS1 and therefore trigger the production of excess amounts non-PreS1 targeting antibodies. PreS1 is found only in the infectious viral particle and serves to bind with sodium taurocholate co-transporting polypeptide (NTCP) on hepatocytes, thereby favoring viral attachment and entry (Sankhyan et al., 2016). In this context, administration of specific neutralizing anti-preS1 antibodies has shown not only to neutralize virions to prevent infection of new cells but also to clear chronically infected cells by antibody-dependent cell cytotoxicity (ADCC) (Li et al., 2017). The novel anti-preS1 antibody designed in this study for synNotch output bound to NTCP had a broad neutralizing efficacy on the common HBV genotypes A, B, C, and D (Figures 4B and S3B) and is expected to be a promising next-generation therapeutic antibody against HBV infection.
Upon secretion by the α-HBs SNR cells, the anti-preS1 antibody caused a significant reduction in viral infectivity (Figure 5C). However, there were a considerable proportion of infected cells despite the presence of the secreted antibody when compared with the synthesized antibody. This could be due to two reasons; production of less quantity of output antibody and inadequate functional modification of the antibody after translation. The α-HBs SNR cells secreted only about ∼4 ng/mL of anti-preS1 antibody per well (Figure 5B), whereas marked reduction of infection was observed when we used 1 μg/mL per well (Figure 4E). The lower levels of output antibody generated could be due to the inherent flaw of the synNotch system where there is not much scope for signal amplification. Also, it is possible that the antibody secreted by α-HBs SNR cells (α-preS1 ncAb) had a diminished action compared with its synthesized counterpart (anti-preS1 #33 mAb). This is because we utilized the single frame system in which Ig heavy chain linked with light chain via furin/T2A self-processing peptide for expressing chimeric antibody under the control of the Gal4-VP64 transcription regulator. This method has been reported to achieve the well-balanced co-expression of heavy chain and light chain to generate functional antibodies in 293T cells or CHO cells (Li et al., 2012). However, the cleaving mechanism of the reportedly self-cleaving 2A peptide is not precisely understood, as it might also be dependent on endogenous cellular furin protease (Donnelly et al., 2001). The α-HBs SNR cells established from Jurkat T cell might not express adequate amounts of this enzyme to cleave heavy chain-light chain linker. To overcome these drawbacks, it is necessary to explore other output options such as single-chain- or heavy-chain-only antibodies capable of inhibiting HBV infection.
Because the antigen concentration in whole-blood or serum would be much lower than those in local environment of the lesions (tissue or organ) where viral proliferation is active, administration of the α-HBs SNR into systemic circulation may offer no significant advantage over existing monitoring methods and immunotherapeutic options. However, synNotch-mediated cell therapy may overcome the problems of existing biological treatments by producing therapeutic materials in situ, if they are locally installed to execute their predesigned function at the specific site. Desirably, the synNotch-expressing cells against HBV should be implanted into the liver to aid in the direct monitoring at the local environment and accentuate the immune response in the infective focus. This could be achieved by constructing the synNotch circuit in vitro in harvested autologous hepatocytes or mesenchymal cells that would engraft locally upon intra hepatic transfer (Nicolas et al., 2017). Alternatively, techniques facilitating in vivo gene delivery with viral or non-viral methods (hydrodynamic gene therapy, lipid-nanoparticles etc.) maybe used to create these sensors in situ in the liver (Yokoo et al., 2016). Further in vivo studies in these lines with suitable animal models will be able to provide more evidence on the added value of cell therapy for real-time monitoring and for overcoming the disadvantages of systemic administration of conventional immunotherapies for HBV infection (Roche and Samuel, 2011).
Although attractive, the use of engineered cells as agents of in vivo diagnostics and therapeutics is still at the nascent phase, and the safety and efficacy of this technology are the aspects that have to be addressed in the years to come (Fischbach et al., 2013). Apart from in vivo safety and biological efficacy, the use of genetically engineered biosensor cells needs to overcome other important issues such as cost, feasibility on commercial scale, and the overall risk-return balance. However, it could have several advantages such as early diagnosis during the initial phase of viral infection and/or monitoring re-activation before the disease condition progresses to severe stages. With ongoing basic research, it could be possible to develop this nascent technology into an innovative clinical intervention where living cells endowed with various biological functions practically sense clues of diseases in vivo, ultimately contributing to the advancement of medical technology via co-working with existing methods. Hence, we believe it is important to provide the evidence of such a proof-of-concept for future innovation through our current study.
Engineered cells are extensively being studied in the field of cancer. In vivo whole-cell biosensors have been shown to perform better than the detection of clinically used biomarkers of cancers (Aalipour et al., 2019). Also, in vivo sensing of cancers using engineered T cells that produce a reporter detectable by positron emission tomography (PET) is being studied (Keu et al., 2017). Cell-based therapies using engineered cells are a hotspot of current research and hold the promise of being the next pillar of medicine despite the gray areas that are expected to be addressed over time.
Limitations of the Study
Although the synNotch technology offers high customization of input-output response, the only significant hurdle this platform faces is the lack of signal amplification. This is one of the plausible reasons why the α-HBs SNR cells took over 24 h to produce a consistent output of the reporter system and secreted relatively lower quantities of neutralizing antibodies. Nevertheless, to the best of our knowledge, this is the first observation of viral envelope antigens activating synNotch signaling and the mechanism of which is yet to be elucidated. We speculate that the accessibility of the viral antigens to synNotch receptor might be dependent on thermo-fluid dynamics including temperature, solvent, virus concentration, and various other factors that are to be explored further. Although the α-HBs SNR cells have shown promising results in vitro, the in vivo efficacy of this platform should be further explored with suitable animal experiments.
However, if the drawbacks could be overcome by technological advancements in due course of time, this “first of its kind” platform offers a scope for engineering live cell biosensors that can be used for in vitro diagnostics or in vivo monitoring of viral infections. Also, the feasibility of programming these cells to directly mount the innate or humoral immune response against a viral infection opens a portal to revamp the landscape of infectious disease therapeutics, which we would like to call “Engineered Immunity.”
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported in part by an AMED Grant JP19fk0310103 and a Research Complex Program of Japan Science and Technology Agency (to AR) and by a GSK Japan Research Grant (to SM). We thank Dr. Naohito Nozaki for antibody production and Yutaro Yamaoka, Chizu Suzuki, Tatsuma Ban, Manami Tsunoi and Yuji Sato for their technical assistance. We also thank Kiho Tanaka for her inputs provided to make figure 360 videos.
Author Contributions
S.M., S.S.J., and K.M. designed and performed research, analyzed data, and wrote the manuscript. S.Y. performed research and analyzed data. K.W., H.N., H.K., N.Y., K.S., T.W., D.K., and T.T. contributed reagents and analyzed data. A.R. designed and supervised the research, analyzed the data, and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: February 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.100867.
Supplemental Information
References
- Aalipour A., Chuang H.-Y., Murty S., D’Souza A.L., Park S.-M., Gulati G.S., Patel C.B., Beinat C., Simonetta F., Martinić I. Engineered immune cells as highly sensitive cancer diagnostics. Nat. Biotechnol. 2019;37:531–539. doi: 10.1038/s41587-019-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeumler T.A., Ahmed A.A., Fulga T.A. Engineering synthetic signaling pathways with programmable dCas9-based chimeric receptors. Cell Rep. 2017;20:2639–2653. doi: 10.1016/j.celrep.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M., Cho J.H., Wong W.W. Synthetic biology: sensing with modular receptors. Nat. Chem. Biol. 2017;13:131–132. doi: 10.1038/nchembio.2290. [DOI] [PubMed] [Google Scholar]
- Bruss V. Envelopment of the hepatitis B virus nucleocapsid. Virus Res. 2004;106:199–209. doi: 10.1016/j.virusres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Burton D.R. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- Clark D.N., Hu J. Hepatitis B virus reverse transcriptase - Target of current antiviral therapy and future drug development. Antivir. Res. 2015;123:132–137. doi: 10.1016/j.antiviral.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M.L., Luke G., Mehrotra A., Li X., Hughes L.E., Gani D., Ryan M.D. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip”. J. Gen. Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Fischbach M.A., Bluestone J.A., Lim W.A. Cell-based therapeutics: the next pillar of medicine. Sci. Transl. Med. 2013;5:179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C., Wong R.J., Gish R.G. Global perspective on hepatitis B virus infections in the era of effective vaccines. Clin. Liver Dis. 2019;23:383–399. doi: 10.1016/j.cld.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Gui Q., Lawson T., Shan S., Yan L., Liu Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors. 2017;17:1623. doi: 10.3390/s17071623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangartner L., Zinkernagel R.M., Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol. 2006;6:231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
- Keu K.V., Witney T.H., Yaghoubi S., Rosenberg J., Kurien A., Magnusson R., Williams J., Habte F., Wagner J.R., Forman S. Reporter gene imaging of targeted T cell immunotherapy in recurrent glioma. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aag2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. Elife. 2017;6 doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wu Y., Qiu Y., Yao Z., Liu S., Liu Y., Shi J., Zheng D. 2A peptide-based, lentivirus-mediated anti-death receptor 5 chimeric antibody expression prevents tumor growth in nude mice. Mol. Ther. J. Am. Soc. Gene Ther. 2012;20:46–53. doi: 10.1038/mt.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jiang M., Xue J., Yan H., Liang X. Serum HBV RNA quantification: useful for monitoring natural history of chronic hepatitis B infection. BMC Gastroenterol. 2019;19:53. doi: 10.1186/s12876-019-0966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa K., Matsunaga S., Yamaoka Y., Dairaku M., Fukano K., Kimura H., Chimuro T., Nishitsuji H., Watashi K., Shimotohno K. Development of a cell-based assay to identify hepatitis B virus entry inhibitors targeting the sodium taurocholate cotransporting polypeptide. Oncotarget. 2018;9:23681–23694. doi: 10.18632/oncotarget.25348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., Lim W.A. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutz P., Metz P., Lempp F.A., Bender S., Qu B., Schöneweis K., Seitz S., Tu T., Restuccia A., Frankish J. HBV bypasses the innate immune response and does not protect HCV from antiviral activity of interferon. Gastroenterology. 2018;154:1791–1804.e22. doi: 10.1053/j.gastro.2018.01.044. [DOI] [PubMed] [Google Scholar]
- Nicolas C.T., Hickey R.D., Chen H.S., Mao S.A., Higuita M.L., Wang Y., Nyberg S.L. Liver regenerative medicine: from hepatocyte transplantation to bioartificial livers and bioengineered grafts. Stem Cells. 2017;35:42–50. doi: 10.1002/stem.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitsuji H., Ujino S., Shimizu Y., Harada K., Zhang J., Sugiyama M., Mizokami M., Shimotohno K. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci. 2015;106:1616–1624. doi: 10.1111/cas.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Klueg K.M., Stout J.R., Muskavitch M.A. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Dev. Camb. Engl. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Ranawakage D.C., Takada T., Kamachi Y. HiBiT-qIP, HiBiT-based quantitative immunoprecipitation, facilitates the determination of antibody affinity under immunoprecipitation conditions. Sci. Rep. 2019;9:6895. doi: 10.1038/s41598-019-43319-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann B., Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- Roche B., Samuel D. The difficulties of managing severe hepatitis B virus reactivation. Liver Int. 2011;31(Suppl 1):104–110. doi: 10.1111/j.1478-3231.2010.02396.x. [DOI] [PubMed] [Google Scholar]
- Rydell G.E., Prakash K., Norder H., Lindh M. Hepatitis B surface antigen on subviral particles reduces the neutralizing effect of anti-HBs antibodies on hepatitis B viral particles in vitro. Virology. 2017;509:67–70. doi: 10.1016/j.virol.2017.05.017. [DOI] [PubMed] [Google Scholar]
- Sankhyan A., Sharma C., Dutta D., Sharma T., Chosdol K., Wakita T., Watashi K., Awasthi A., Acharya S.K., Khanna N. Inhibition of preS1-hepatocyte interaction by an array of recombinant human antibodies from naturally recovered individuals. Sci. Rep. 2016;6:21240. doi: 10.1038/srep21240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Anindita P.D., Phongphaew W., Carr M., Kobayashi S., Orba Y., Sawa H. Development of a rapid and quantitative method for the analysis of viral entry and release using a NanoLuc luciferase complementation assay. Virus Res. 2018;243:69–74. doi: 10.1016/j.virusres.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Schwarz K.A., Daringer N.M., Dolberg T.B., Leonard J.N. Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol. 2017;13:202–209. doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin-Pujol A., Sicard M., Rosenberg C., Auriac M.-C., Lepage A., Niebel A., Gough C., Bensmihen S. Development of a GAL4-VP16/UAS trans-activation system for tissue specific expression in Medicago truncatula. PLoS One. 2017;12:e0188923. doi: 10.1371/journal.pone.0188923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbuck M.P., Winandy S. A review of notch processing with new insights into ligand-independent notch signaling in T-cells. Front. Immunol. 2018;9:1230. doi: 10.3389/fimmu.2018.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov A., Boldanova T., Wang X., Wieland S., Heim M.H. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology. 2018;154:1778–1790. doi: 10.1053/j.gastro.2018.01.034. [DOI] [PubMed] [Google Scholar]
- Suslov A., Wieland S., Menne S. Modulators of innate immunity as novel therapeutics for treatment of chronic hepatitis B. Curr. Opin. Virol. 2018;30:9–17. doi: 10.1016/j.coviro.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Kaminski J.J., Kurt-Jones E.A., Fitzgerald K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P.-F., Ling H., Foo J.L., Chang M.W. Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 2019;37:107393. doi: 10.1016/j.biotechadv.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo T., Kamimura K., Abe H., Kobayashi Y., Kanefuji T., Ogawa K., Goto R., Oda M., Suda T., Terai S. Liver-targeted hydrodynamic gene therapy: recent advances in the technique. World J. Gastroenterol. 2016;22:8862–8868. doi: 10.3748/wjg.v22.i40.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.