Abstract
Mycoplasma ovipneumoniae (Mo) is difficult to culture, resulting in many difficulties in related research and application. Since nucleotide metabolism is a basic metabolism affects growth, this study conducted a “point-to-point” comparison of the corresponding growth phases between the Mo NM151 strain and the Mycoplasma mycoides subsp. capri (Mmc) PG3 strain. The results showed that the largest difference in nucleotide metabolism was found in the stationary phase. Nucleotide synthesis in PG3 was mostly de novo, while nucleotide synthesis in NM151 was primarily based on salvage synthesis. Compared with PG3, the missing reactions of NM151 referred to the synthesis of deoxythymine monophosphate. We proposed and validated a culture medium with added serine to fill this gap and prolong the stationary phase of NM151. This solved the problem of the fast death of Mo, which is significant for related research and application.
Keywords: Mycoplasma ovipneumoniae, culture medium, transcriptome
INTRODUCTION
Mycoplasmal pneumonia of sheep and goats (MPSG) is a highly contagious infectious disease that is prevalent among sheep and goats [1]. In China, its main pathogens include Mycoplasma ovipneumoniae (Mo) and Mycoplasma mycoides subsp. capri (Mmc). Compared with Mmc, Mo is difficult to culture, resulting in many difficulties in related research and application. Mycoplasma is the smallest and simplest self-replicating bacterium [2]. Given its limited genome size, several nutrients are needed in its culture medium, and culturing it is time-consuming. In addition, different species of mycoplasma grow at different rates. Mmc is a species that is easy to culture. Its growth cycle is nearly 4 days, and includes a long stationary phase with a high survival rate through cell passages. Mo is a species that is difficult to culture, with a growth cycle of more than seven days. Cells die quickly after a short stationary phase, meaning that Mo cannot be passaged continuously. This limits conventional identification methods such as bacterial culturing, growth inhibition tests, growth precipitation tests, agglutination tests and antigen preparation. Growing these organisms needs complex media, reflecting the loss of many genes that encode anabolic enzymes and the retention of genes that encode catabolic pathways [3]. Mycoplasma's metabolic network is simple compared to that of more complex organisms such as Escherichia coli [4,5]. Therefore, comparing genomes from mycoplasma and from E. coli will provide little useful information. Thus, this paper compared nucleotide metabolism between Mmc and Mo. Since nucleotides are precursors to nucleic acid synthesis, they are critical to basic life activities and growth.
The entire genome sequences of these 2 strains were sequenced in our laboratory. The Mmc PG3 strain has a genome of 1.01 Mb, encoding 846 genes, while the Mo NM2010 strain has a genome of 864 kb, encoding approximately 888 genes [6]. These served as the reference genomes for this study. RNA-seq detects which transcripts are over- and under-represented in a certain condition. This in turn can serve as a basis on which functions are active/inactive in a cell. Moreover, it helps in detecting over-/unexpressed genes and thus finding missing reactions and predicting which enzymes, upstream and downstream products, or activators can fill these gaps in theory.
In this paper, a point-to-point comparison between the corresponding phases of PG3 and NM151 strains was performed. We focused on the key enzymes that only expressed in PG3 or NM151 and hypothesized that key enzymes could affected Mo growth. Then found an approach to fill the metabolic gap, which is significant for Mo culture and application.
MATERIALS AND METHODS
Strain and culture
Mmc PG3 strain, Mo NM151 strain, Mo NM130 strain, Mo NM85 strain and Mo NM811 strain used were isolated from Huhhot in China. They were recovered and cultured in a modified Hayflick's broth (1.0 g/L glucose, 21.0 g/L PPLO, 0.4 g/L sodium pyruvate, 10 mL of 25% (w/v) yeast extract, 200 mL donor equine serum (HyClone, USA), 10 mL 10,000 IU/mL penicillin-G, 2.5 mL of 0.4% (w/v) phenol red, and 20 mM HEPES at pH 7.6–7.8, adjusted with 0.1 M sodium hydroxide at 37°C, in a 5% CO2 atmosphere[7]. Solid medium was prepared by adding 1% (w/v) agar to the modified Hayflick's broth.
RNA isolation, library construction, and sequencing
According to growth curves of PG3 and NM151 (Supplementary Fig. 1). The sampling times of PG3 were 18 h, 35 h, 58 h and 83 h and the sampling times of NM151 were 24 h, 48 h, 72 h and 96 h. These correspond to the logarithmic phase (PG3/NM151-log), stationary phase (PG3/NM151-sta), decline phase (PG3/NM151-dec), and late-decline phase (PG3/NM151-latedec), respectively. All culturing was performed in triplicate. The cells were stored at −80°C for a maximum of 24 h before RNA isolation.
The total RNA of samples was isolated using the RNAprep Pure Cell/Bacteria Kit (Tiangen, China) per the manufacturer's instructions. RNA quality was checked using RNase-free agarosegel electrophoresis and determined with a Bioanalyzer 2100 (Agilent, USA). The total RNA concentration was measured using an ND1000 spectrophotometer (Thermo Fisher Scientific, USA) at 260nm and 280nm. Samples were sent to LC Bio Tech (China) for further processing.
Total RNA samples (10 μg) were subjected to further purification with a Ribo-Zero Magnetic kit (G+/G− Bacteria) (Illumina, USA) to enrich mRNA and remove rRNA. Following purification, the fraction was fragmented into smaller pieces using a Truseq RNA sample preparation kit (Illumina). Next, cleaved RNA fragments were reverse-transcribed to create the cDNA, which was next used to synthesize U-labelled second-stranded DNAs with DNA polymerase I, RNase H, and deoxyuridine triphosphate (dUTP). An A-base was then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contained a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. Single- or dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme treatment of the U-labelled second-stranded DNAs, the ligated products were amplified with PCR under the following conditions: initial denaturation at 95°C for 3 min; eight cycles of denaturation at 98°C for 15 sec, annealing at 60°C for 15 sec, and extension at 72°C for 30 sec; and then final extension at 72°C for 5 min. The average insert size for the final cDNA library was 300 bp (± 50 bp). Paired-end sequencing (2 × 150 bp) was performed on the Illumina Hiseq 4000 platform (Illumina).
Strand-specific libraries were prepared using a dUTP second-strand marking protocol with reagents from Invitrogen (Thermo Fisher Scientific). The first-strand cDNA was synthesized from 400 ng of precipitated fragmented RNA using 3 μg of random primers, 4 μg of actinomycin D, and SuperScript III Reverse Transcriptase. After extracting and precipitating the first-strand, the second-strand cDNA was synthesized as previously described, using dUTP rather than dTTP. Paired-end sequencing (2 × 150 bp) was performed on the Illumina HiSeq-2000 platform (Illumina). Clean data were obtained from the raw data by removing the sequences of the adapters and low-quality reads (reads containing sequencing adaptors or primers and nucleotides with a Q quality score below 20). Clean reads were aligned with the Mo and Mmc using Bowtie 2 [8]. The mapped reads of each sample were assembled using Rockhopper [9]. The relative abundance of the transcripts was calculated as the reads per kilobase per million mapped (RPKM).
Differentially expressed genes analysis
Differentially expressed genes (DEGs) among the different samples were detected using FunPat in the R package [10]. The DEGs were defined with a false discovery rate (FDR) of 1% or less, while a fold change ≥ 2 was set as the threshold for significant differential expression. DEGs were identified as enriched in metabolic pathways (p < 0.01) by searching against the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database [11,12,13]
Validation of RNA-sequencing analysis via real time quantitative polymerase chain reaction (RT/q-PCR)
To validate genes that were differentially expressed, an independent experiment was conducted using real time quantitative polymerase chain reaction (RT/q-PCR). Total RNA was used to synthesize cDNA using the PrimeScript RT reagent kit (Takara, China) according to the manufacturer's instructions. The primers that are designed for each gene are given in Supplementary Table 1. Primer specificity was determined using melting curve analysis and agarose gel electrophoresis of PCR products. RT/q-PCR was performed using an Applied Biosystems 7500 with SYBR Premix Ex Taq (Tli RNaseH Plus) and ROX plus (Takara) according to the manufacturer's protocol. The PCR cycles were as follows: one cycle of 30 sec at 95°C, followed by 40 cycles at 95°C for 5 sec, 54°C for 30 sec, and 72°C for 34 sec. After amplification, fluorescent data were converted to threshold cycle (Ct) values for analysis. TheCTvalues were determined at the threshold fluorescence value of 0.2 (ΔCt = CtTarget gene − Ctreference gene). The data were then statistically analysed using a t-test in the SPSS statistical package (Version 22.00). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference gene.
Modified medium detection by the colony forming unit (CFU) method
Cells were counted by the CFU method. To start, 200 μL of NM151 suspension was inoculated into 1.8 mL of medium and cultured for 240 h at 12-h collection intervals. Bacterial suspensions were serially diluted 10-fold up to 10−12. For each dilution, 0.1 mL of suspension was seeded in solid medium and cultured at 37°C for five days. At the end of incubation, the colonies were counted microscopically.
RESULTS
Transcriptomic data for PG3 and NM151
To obtain a global view of the transcriptome, total RNAs were isolated from PG3 and NM151 in the logarithmic (log), stationary (sta), decline (dec), and late-decline phases (latedec), then subjected to high-throughput Illumina sequencing. After filtering out low-quality reads and primer contamination, the number of properly paired reads among the 24 samples was between 12.8 and 37.7 million. The results of these reads mapped back to the reference genome (Mmc PG3 strain, GenBank: ANIV00000000.1 and Mo NM2010 strain, GenBank: JAKV00000000.1) were between 50% and 90% (Supplementary Table 2).
RNA-sequencing analysis validation via RT/q-PCR
RNA samples were isolated, as previously described, reverse-transcribed, and analysed. Four genes were randomly chosen from each comparable group, including pnp, nusA, atpA, gapdh and an unnamed gene. The RT/q-PCR results were consistent with the RNA-sequencing data (Supplementary Fig. 2).
DEGs enriched to nucleotide metabolism
There were 848 (PG3/NM151, 408/515), 462 (369/167), 730 (485/319) and 652(473/253) DEGs in PG3-log vs. NM151-log, PG3-sta vs. NM151-sta, PG3-dec vs. NM151-dec and PG3-latedec vs. NM151-latedec, respectively. The number of DEGs mapped to KEGG pathways was 320, 196, 299 and 266. The number of DEGs enriched to nucleotide metabolism is shown in Fig. 1.
Fig. 1. Statistics of DEGs per group (PG3 vs. NM151).
The first line is the number of DEGs and core genes of PG3 and NM151 in the four comparison groups, respectively. Same as above, the second line is the number of DEGs and core genes enriched in the KEGG pathway. The third and fourth lines are the specific genes of PG3 and NM151 enriched in purine nucleotides and pymidine nucleotides metabolism.
DEG, differentially expressed gene; KEGG, Kyoto Encyclopedia of Genes and Genomes
Comparison of PG3 and NM151 nucleic acid metabolism
In this study, the enzymes, whose differentially expressed genes were coded, and which were involved in synthesizing purine and pyrimidine nucleotides between PG3 and NM151 in the four growth stages, were compared.
Enzymes of PG3 and NM151 in purine nucleotide metabolism
The enzymes from PG3 and NM151 in the four stages are shown in Supplementary Table 3; there were 22 enzymes in PG3 and 14 in NM151. The number of enzymes in PG3 was larger than NM151, particularly in the stationary stage. The reactions in which these enzymes were involved in are given in Fig. 2.
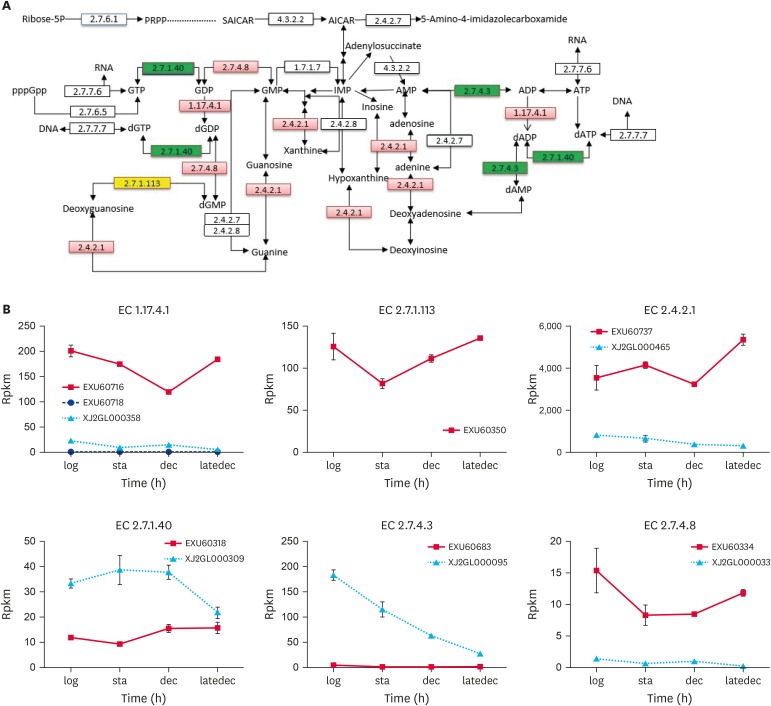
Fig. 2. Specific enzymes in purine metabolism.
(A) was a pathway part of Ko00230, only the enzymes, whose differentially expressed genes were coded between the two species, were listed. The enzyme boxes were divided into four equal parts, one for each growth phase. If the gene were upregulaed in PG3 compared with NM151, this enzyme rectangle in red. And the opposite (downregulation) could be green. If the gene is not expressed in NM151, it will be yellow. (B) were the RPKM value of genes in PG3 and NM151 during the four growth stages.
RPKM, reads per kilobase per million mapped; PRPP, Phosphoribosyl pyrophosphate; SAICAR, phosphoribosylaminoimidazolesuccinocarboxamide; AICAR, aminoimidazole carboxamide ribonucleotide; GTP, guanosine triphosphate; GDP, guanosine diphosphate; GMP, guanosine monophosphate; IMP, inosine monophosphate; AMP, adenosine monophosphate; ADP, adenosine diphosphate; ATP, adenosine triphosphate; d, deoxy-.
In the logarithmic phase, there were four enzymes in PG3. Two of them were ribonucleoside-diphosphate reductase (EC 1.17.4.1) and deoxyguanosine kinase (EC 2.7.1.113), which were also found in the decline and late-decline phases. They catalyse the reduction of guanosine diphosphate and adenosine diphosphate and enhance the reaction to dGTP synthesis. At this time, NM151 had 7 enzymes. Of these, purine nucleoside phosphorylase (EC 2.4.2.1) was a key enzyme in the purine salvage synthesis pathway. They were also found in the stationary and decline phases. The other two enzymes were pyruvate kinase (EC 2.7.1.40) and adenylate kinase (EC 2.7.4.3). Pyruvate kinase was extensively involved in glycolysis and nucleic acid metabolism. Adenylate kinase catalyses (d) ATP and (d) AMP conversion. During the stationary phase, PG3 had six enzymes, while only purine nucleoside phosphorylase remained in NM151. In the decline and late-decline phase, the number of enzymes in NM51 increased, but was still less than the number in PG3. This indirectly affected the metabolic capacity of NM151. Guanylate kinase (EC 2.7.4.8) plays the same role as deoxyguanosine kinase by strengthening DNA synthesis. This occurred in PG3. Guanosine triphosphate (GTP) diphosphokinase (EC 2.7.6.5), which catalyses pppGpp-generated GTP and synthesizes RNA, was not annotated in the NM151 genome.
Enzymes of PG3 and NM151 pyrimidine nucleotide metabolism
Enzyme trends involved in pyrimidine nucleotide metabolism during the four growth phases are similar to those of purine nucleotide metabolism (Supplementary Table 4). The reactions of these enzymes are given in Fig. 3.
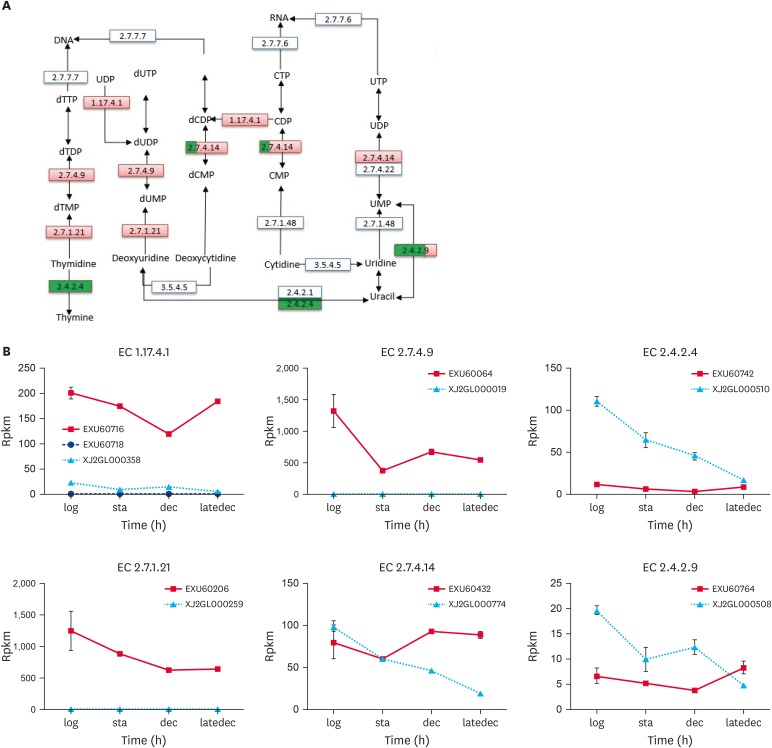
Fig. 3. Specific enzymes in pyrimidine metabolism.
(A) was a pathway part of Ko00240, only the enzymes, whose differentially expressed genes were coded between the 2 species, were listed. The enzyme boxes were divided into 4 equal parts, one for each growth phase. If the gene were upregulaed in PG3 compared with NM151, this enzyme rectangle in red. And the opposite (downregulation) could be green. If the gene is not expressed in NM151, it will be yellow. (B) were the RPKM value of genes in PG3 and NM151 during the 4 growth stages.
TTP, thiamine triphosphate; TDP, thiamine diphosphate; TMP, thiamine monophosphate; UDP, uridine diphosphate; UTP, uridine triphosphate; UMP, uridine monophosphate; CTP, cytidine triphosphate; CMP, cytidine monophosphate; CDP, cytidine diphosphate; d, deoxy-.
During the logarithmic phase, NM151 had 7 enzymes, whereas PG3 only had ribonucleoside-diphosphate reductase (EC 1.17.4.1). Ribonucleoside-diphosphate reductase was responsible for reducing cytosine and uracil nucleotides in the corresponding deoxy-cytosine and uracil. In the stationary phase, the enzymes in NM151 decreased sharply, with only 2 remaining enzymes. Among them, thymidylate kinase (EC 2.7.4.9) catalyses the conversion of deoxyuridine diphosphate (dUDP) and deoxyuridine monophosphate (dUMP). But the enzyme that converts dUMP to deoxythymine monophosphate (dTMP) was unnoticed, indicating that this step was its gap reaction. In addition, thymidine phosphorylase (EC 2.4.2.4), which was involved in the salvage synthesis of thymidylate, was not enriched during this stage. This indirectly affected DNA synthesis. Conversely, the number of enzymes in PG3 was seven. Thymidine kinase (EC 2.7.1.21) and thymidine phosphorylase (EC 2.4.2.4), involved in the salvage synthesis of thymidylate, were absent in NM151.
Detection of Mo in the modified medium
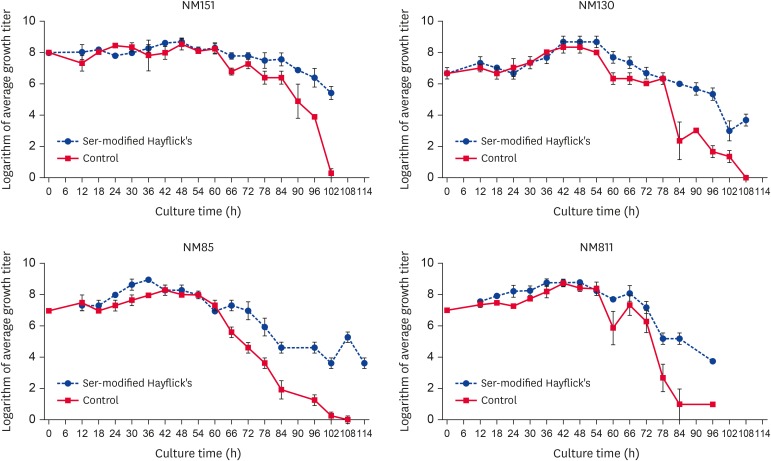
To further examine our conclusions, experiments on growth for 4 strains of Mo are desirable. These 4 strains are NM151, NM85, NM130, and NM811. We improved the culture medium by the following methods: i) serine (30 mg/L) was added to modified Hayflick's broth; ii) the modified Hayflick's broth was used as a control. The NM151 growth curves were constructed by the CFU method. The result was that, by adding serine (ser-modified Hayflick's) significantly prolonged the stationary phase after being cultured for 66 h (p < 0.05) (Fig. 4).
Fig. 4. Growth curves in ser-modified Hayflick's mediums.
All cultures were performed three times, and colonies were counted in duplicate for each dilution. The growth curve of the Mo NM151 strain was constructed by plotting the logarithm of the colony counts for each dilution on the Y-axis and the sampling time on the X-axis.
DISCUSSION
Mo is difficult to culture, resulting in many difficulties in related research and application. Since nucleotide metabolism is a basic metabolism affects growth, this study compared metabolic characteristics between Mo and Mmc.
The results showed that nucleotide synthesis in PG3 was mostly de novo, while nucleotide synthesis in NM151 was primarily based on salvage synthesis. Regarding the number of enzymes involved in purine nucleotide metabolism, there were more enzymes in PG3, especially in the stationary phase. Therefore, compared with PG3, the decrease in enzymes involved in purine nucleotide metabolism during the stationary phase of NM151 may have inhibited its metabolism and growth. In the decline and late-decline phases, the number of enzymes in NM151 increased again. Of these enzymes, UMP/CMP kinase (2.7.4.14), uracil phosphoribosyltransferase (EC 2.4.2.9) and thymidine phosphorylase showed that pyrimidine nucleotide formation in NM151 was strong in this phase, and its main route was salvage synthesis. Therefore, the major period limiting the NM151 pyrimidine nucleotide metabolism was the stationary phase, while the missing reactions are the synthesis of thymidylate in this paper. This inhibits growth and metabolism indirectly. Compared to NM151, more enzymes are linked to DNA synthesis and purine nucleotide de novo synthesis in PG3. This indicates that PG3 possessed the relative integrity of metabolic pathways and strong metabolism ability. Purine nucleoside phosphorylase was the only enzyme in purine metabolism in the NM151 strain. It modulates salvage synthesis effectively. In addition, it was a guarantee of its normal catabolism and anabolism.
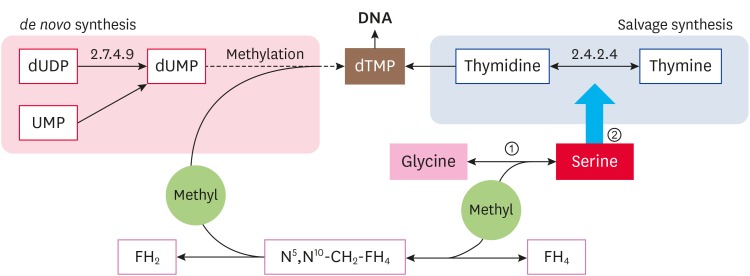
In pyrimidine nucleotide metabolism, dTMP synthesis was noteworthy. There are 2 pathways for dTMP synthesis. One is the de novo synthesis of dTMP, which requires 2 steps: i) UMP was reduced to dUMP, and ii) dUMP was transformed into dTMP by methylation. The other way is the salvage pathway. In the stationary phase, the remaining enzymes in NM151 did not encounter the de novo synthesis of dTMP. Of these 2 enzymes, thymidylate kinase catalyses the transformation of dUDP and dUMP. However, generating dUMP did not translate to dTMP efficiently [14]. Thymidine phosphorylase (EC 2.4.2.4), which was responsible for dTMP salvage synthesis was not an enzyme at this stage. It has been reported that serine can provide methyl for converting dUMP to dTMP and contributes to the de novo synthesis of thymine nucleotides [15]. Mycoplasma can produce thymidine phosphorylase when consuming serine in the cell culture medium supernatant and can cleave the exogenous thymidine into thymine in order to synthesize genetic material itself [16]. Therefore, we proposed that serine will promote dTMP synthesis in NM151 and indirectly promote its metabolism and growth (Fig. 5).
Fig. 5. The pathway of serine participates in the metabolism of nucleotides.
① serine can provide methyl for dUMP converting to dTMP and contributes to the de novo synthesis of thymine nucleotides; ② mycoplasma can produce thymidine phosphorylase (EC 2.4.2.4) when consuming serine in the cell culture medium supernatant.
Researchers have been studying and screening mycoplasma culture components. The most common cultures use pleuropneumonia-like organisms (PPLO) as their base, then add serum, yeast extract, growth factors and other nutrients, as well as drugs to inhibit bacterial growth. In the UK, mycoplasma medium is usually rich in PPLO and being widely used. Here, we retained the modified Hayflick's broth, which was PPLO-enriched, while the modified Hayflick's broth with added serine prolonged the Mo stationary phase.
In conclusion, this paper proposed and confirmed that a culture medium with added serine can prolong the stationary phase of NM151. In addition, we found that positive rate of Mo separation was improved when the programme was applied. Lastly, one of the reasons for the difficulties posed by Mo culture and passage is that the stationary phase of Mo was too short, and cells died quickly. It is difficult to master the accurate passage of time. Through the improvements offered by this experiment, the stationary phase was prolonged.
Footnotes
Funding: This work was supported by Science and Technology program of Inner Mongolia (201802066), Science and Technology Transformation of Inner Mongolia Agricultural University (YZGC2017027), Science and Technology project of Inner Mongolia (201502070).
Conflict of Interest: The authors declare no competing financial interest.
- Conceptualization: Wang X, Hao Y.
- Formal analysis: Wang X.
- Funding acquisition: Hao Y.
- Investigation: Wang X.
- Methodology: Wang X, Hao Y, Zhang W.
- Project administration: Hao Y.
- Resources: Hao Y.
- Software: Wang X, Zhang W.
- Supervision: Hao Y, Zhang W.
- Validation: Wang X.
- Visualization: Wang X, Hao Y.
- Writing - original draft: Wang X, Hao Y, Zhang W.
- Writing - review & editing: Wang X, Hao Y, Zhang W.
SUPPLEMENTARY MATERIALS
Primers for RT/q-PCR
Transcriptomic data for PG3 and NM151
Specific enzymes involved in purine nucleotide metabolism of PG3 and NM151 during the 4 growth stages
Enzymes involved in PG3 and NM151 pyrimidine nucleotide metabolism during the 4 growth stages
Growth curve and sampling time for (A) RNA-seq of PG3 and RNA-seq of NM151.
Results of RT/q-PCR.
References
- 1.Wang XH, Wang YF, Huang HB, Bai F, Shi X, Ma C, Gao Y, Zhang J, Zhang W, Hao Y. Understanding the metabolism of Mycoplasma mycoides subsp. capri in vitro by a transcriptomic analysis. J Integr Agric. 2018;17:428–435. [Google Scholar]
- 2.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miles RJ. Catabolism in mollicutes. J Gen Microbiol. 1992;138:1773–1783. doi: 10.1099/00221287-138-9-1773. [DOI] [PubMed] [Google Scholar]
- 4.Wodke JA, Puchałka J, Lluch-Senar M, Marcos J, Yus E, Godinho M, Gutiérrez-Gallego R, dos Santos VA, Serrano L, Klipp E, Maier T. Dissecting the energy metabolism in Mycoplasma pneumoniae through genome-scale metabolic modeling. Mol Syst Biol. 2013;9:653. doi: 10.1038/msb.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yus E, Maier T, Michalodimitrakis K, van Noort V, Yamada T, Chen WH, Wodke JA, Güell M, Martínez S, Bourgeois R, Kühner S, Raineri E, Letunic I, Kalinina OV, Rode M, Herrmann R, Gutiérrez-Gallego R, Russell RB, Gavin AC, Bork P, Serrano L. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 6.Wang XH, Huang HB, Cheng C, Wang RC, Zheng JQ, Hao YQ, Zhang W. Complete genome sequence of Mycoplasma ovipneumoniae strain NM2010, which was isolated from a sheep in China. J Integr Agric. 2014;13:2562–2563. [Google Scholar]
- 7.Xu CG, Hao YQ, Zhang L, Hao RX, Liu XL, Huang ZY. Molecular cloning and immune response analysis of putative variable lipoproteins from Mycoplasma mycoides subsp capri. Genet Mol Res. 2014;13:1527–1539. doi: 10.4238/2014.March.12.5. [DOI] [PubMed] [Google Scholar]
- 8.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanavia T, Finotello F, Di Camillo B. FunPat: function-based pattern analysis on RNA-seq time series data. BMC Genomics. 2015;16:S2. doi: 10.1186/1471-2164-16-S6-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun S, Xuan F, Ge X, Fu H, Zhu J, Zhang S. Identification of differentially expressed genes in hepatopancreas of oriental river prawn, Macrobrachium nipponense exposed to environmental hypoxia. Gene. 2014;534:298–306. [PubMed] [Google Scholar]
- 14.Ziemkowski P, Felczak K, Poznański J, Kulikowski T, Zieliński Z, Cieśla J, Rode W. Interactions of 2′-fluoro-substituted dUMP analogues with thymidylate synthase. Biochem Biophys Res Commun. 2007;362:37–43. doi: 10.1016/j.bbrc.2007.07.097. [DOI] [PubMed] [Google Scholar]
- 15.Wang JY. Biochemistry. Beijing: Higher Education Press; 2002. p. 400. [Google Scholar]
- 16.Sinigaglia F, Scheidegger D, Talmadge K, Garotta G. A sensitive and quantitative microassay for the detection of mycoplasma contamination: inhibition of IL-2 dependent cell line proliferation. J Immunol Methods. 1985;76:85–92. doi: 10.1016/0022-1759(85)90483-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for RT/q-PCR
Transcriptomic data for PG3 and NM151
Specific enzymes involved in purine nucleotide metabolism of PG3 and NM151 during the 4 growth stages
Enzymes involved in PG3 and NM151 pyrimidine nucleotide metabolism during the 4 growth stages
Growth curve and sampling time for (A) RNA-seq of PG3 and RNA-seq of NM151.
Results of RT/q-PCR.