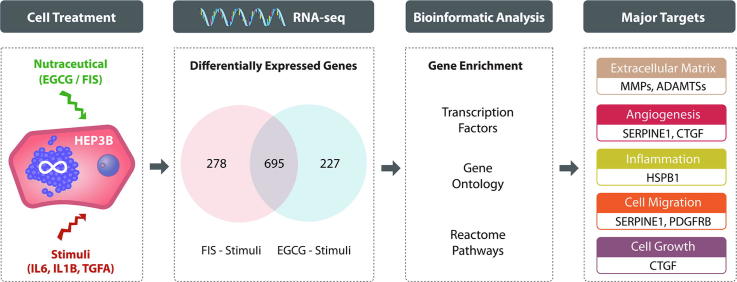
Graphical abstract
Abbreviations: HCC, hepatocellular carcinoma; EGCG, epigallocatechin gallate; FIS, fisetin; ECM, extracellular matrix; RT-qPCR, reverse transcription-quantitative real time PCR; DEGs, differentially expressed genes; DMSO, dimethyl sulfoxide; MEM, minimum essential medium; STIM, stimulated; SD, standard deviation; TF, transcription factor; GO, Gene Ontology; SEM, standard error of mean; EMT, epithelial to mesenchymal transition; ADAM, a disintegrin and metalloproteinase with thrombospondin motifs; MMPs, matrix metalloproteinases; ADAMTS9, ADAM metallopeptidase with thrombospondin type 1 motif 9; MMP11, Matrix Metallopeptidase 11; MMP9, Matrix Metallopeptidase 9; SERPINE1, Serpin Family E Member 1; CTGF, Connective Tissue Growth Factor; PDGFRB, Platelet Derived Growth Factor Receptor Beta; HSPB1, Heat Shock Protein Family B (Small) Member 1; HSPA2, Heat Shock Protein Family A (Hsp70) Member 2; CLIC3, Chloride Intracellular Channel 3
Keywords: RNA-sequencing, Hepatocellular carcinoma, Epigallocatechin gallate, Fisetin, Transcription factors, Gene Ontology, Reactome Pathways
Abstract
Hepatocellular carcinoma (HCC) is an essentially incurable inflammation-related cancer. We have previously shown by network analysis of proteomic data that the flavonoids epigallocatechin gallate (EGCG) and fisetin (FIS) efficiently downregulated pro-tumor cytokines released by HCC through inhibition of Akt/mTOR/RPS6 phospho-signaling. However, their mode of action at the global transcriptome level remains unclear. Herein, we endeavor to compare gene expression alterations mediated by these compounds through a comprehensive transcriptome analysis based on RNA-seq in HEP3B, a responsive HCC cell line, upon perturbation with a mixture of prototypical stimuli mimicking conditions of tumor microenvironment or under constitutive state.
Analysis of RNA-seq data revealed extended changes on HEP3B transcriptome imposed by test nutraceuticals. Under stimulated conditions, EGCG and FIS significantly modified, compared to the corresponding control, the expression of 922 and 973 genes, respectively, the large majority of which (695 genes), was affected by both compounds. Hierarchical clustering based on the expression data of shared genes demonstrated an almost identical profile in nutraceutical-treated stimulated cells which was virtually opposite in cells exposed to stimuli alone. Downstream enrichment analyses of the co-modified genes uncovered significant associations with cancer-related transcription factors as well as terms of Gene Ontology/Reactome Pathways and highlighted ECM dynamics as a nodal modulation point by nutraceuticals along with angiogenesis, inflammation, cell motility and growth. RNA-seq data for selected genes were independently confirmed by RT-qPCR.
Overall, the present systems approach provides novel evidence stepping up the mechanistic understanding of test nutraceuticals, thus rationalizing their clinical exploitation in new preventive/therapeutic modalities against HCC.
1. Introduction
Hepatocellular carcinoma (HCC) manifests as one of the most common and aggressive types of cancer on a global scale [1]. It develops under conditions of chronic inflammation [2] and liver cirrhosis [3], and is considered to be a highly vascularized tumor [4]. HCC incidence can be associated with a significant number of causative factors, the most important of which being viral hepatitis B and C, excessive consumption of alcohol, obesity, non-alcoholic fatty liver disease and dietary exposure to aflatoxin B1 [1], [3], [5], [6]. Underlying molecular traits of the disease, indicate that hepatocarcinogenesis is a particularly complex process, involving both genetic and epigenetic abnormalities [7]. Aberrant activation or inhibition of key signaling pathways that lead to excess cell proliferation and growth, along with an already established condition of oxidative stress, inflammatory tumor microenvironment, deregulated extracellular matrix (ECM) and rich angiogenic activity, shape the mosaic image of HCC incidence, progression and metastatic potential [8].
In most cases, HCC is diagnosed in advanced stages, resulting in the absence of many effective treatment regimes [9]. Therefore, it is urgent to identify reliable diagnostic biomarkers and develop promising drug candidates for disease prevention or therapy. To this end, a growing body of research has focused on natural dietary compounds known as nutraceuticals - a term coined from “nutrition” and “pharmaceuticals” [10] - due to their promising cancer chemopreventive/chemotherapeutic activities and limited side-effects [11]. Accordingly, several nutraceuticals have emerged, as inviting alternative candidates [12], [13] or complementary synergistic agents to conventional therapies [14], [15]. Their biological activities examined in several cancer types are associated with multiple effects on prominent cellular functions, including modulation of gene expression through epigenetic reprogramming or transcription factor alterations [11], [16], [17], [18], [19]. Flavonoids represent an important family of plant-derived nutraceuticals possessing potent anti-oxidative properties that render them attractive as inhibitors of oxidative stress, a cancer hallmark known to trigger signaling pathways of cell growth, survival, motility, inflammation, angiogenesis and metastasis, in both tumor and tumor-associated stroma cells [20].
Regardless of the growing attention to nutraceuticals there is not yet a solid mechanistic basis to support health claims in cancer patients. Thus, a more thorough understanding of their mode of action and molecular targets is strongly required. To this direction, we previously applied a systems biology approach combining data from high throughput Luminex immunoassays with network analysis in three human HCC cell lines exposed to selected nutraceuticals and/or distinct pro-tumor stimuli and explored the mechanisms underlying their modulatory effects on secreted cytokines at the level of phosphoproteins signaling [21]. Acquired results highlighted the polyphenol (-)-epigallocatechin gallate (EGCG) - the main catechin of green tea - and fisetin (FIS) - a flavonol widely found in many fruits and vegetables - as the most promising compounds against HCC [21], in agreement with other reports demonstrating their broad anti-cancer activities in numerous cancer preclinical models [22], [23], [24]. To pursue a further systems investigation of EGCG and FIS mechanisms at the transcriptome level, in the present work, HEP3B cells (a nutraceutical-responsive human HCC cell line [21]), are treated with test compounds under basal conditions or following induction with a mixture of prominent inflammatory/growth signaling stimuli, and the differential gene expression is monitored by RNA-seq analysis. Statistically significant differentially expressed genes (DEGs) are then searched for potentially co-modified targets and expression motifs, while thorough downstream data analyses are further conducted in order to explore significant connections of target genes to specific transcription factors, biological processes and pathways. Our results are expected to provide new mechanistic information about the modifications brought by EGCG and FIS on the global transcriptome landscape - especially under conditions simulating tumor inflammatory microenvironment - thus setting a base for their future usage in clinical chemopreventive and/or therapeutic interventions against HCC or other inflammation-related cancers.
2. Materials and methods
2.1. Nutraceutical acquisition and preparation
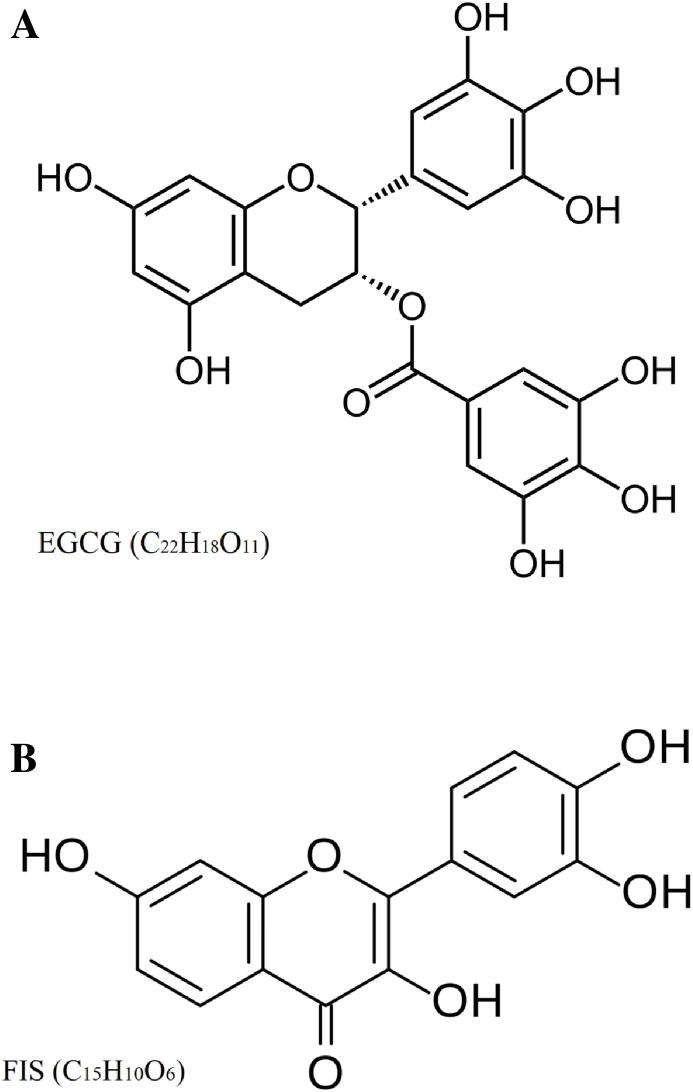
EGCG (Fig. 1A, purity ≥ 98%) was purchased from Sigma-Aldrich (St. Louis, USA) and STEMCELLTM Technologies (Vancouver, Canada), while FIS (Fig. 1B, HPLC purity ≥ 99) was provided by Carl Roth (Karlsruhe, Germany). Master stock solutions were prepared for both compounds in dimethyl sulfoxide (DMSO) and consequently were stored at −20 °C. Working dilutions contained up to 0.1% v/v DMSO.
Fig. 1.
Chemical structure of EGCG (A) and FIS (B).
2.2. Cell culture and treatment
HEP3B cells were maintained as described before [21]. HCC cells, seeded the day before in a 6-well plate (45 × 104 cells per well), were serum-starved for 4 h and then treated with EGCG (100 μM), FIS (10 μM) or DMSO (0.1% v/v) for 2 h. Cells were subsequently exposed to a mixture of stimuli consisting of recombinant interleukins IL-6 (0.1 μg/ml), IL-1B (0.01 μg/ml) and tumor growth factor A (TGFA) (0.2 μg/ml) (all purchased from PeproTech, USA) or vehicle (medium) and were further incubated for 22 h. Working concentrations for reagents and incubation length were chosen based on preliminary experiments performed to update conditions described in detail previously [21]. Cell viability under the used experimental conditions was evaluated by the MTT assay as previously described [25]. Samples of all possible treatment combinations of HEP3B cells i.e. EGCG, FIS, or DMSO at either constitutive (MEM) or stimulated (STIM) state were subjected to RNA extraction from two independent experiments.
2.3. RNA isolation and RNA-sequencing
Total RNA was isolated using the PureLink RNA Mini Kit (Invitrogen, USA) according to the manufacturer’s instructions. Quantification and quality control of isolated RNA was performed by measuring absorbance at 260 nm and 280 nm on a NANODROP ONEC spectrophotometer (Thermo Scientific, USA). The RNA-seq run was performed on biological duplicates from two independent experiments per treatment group on a NextSeq 500 Illumina platform that provided single-end reads of 85 bp length. Quality assessment, library preparation (TruSeqLT) and the actual sequencing run was conducted in the Biomedical Research Foundation of the Academy of Athens (BRFAA) sequencing facility. The entire dataset of RNA-seq FASTQ files has been deposited to the repository Mendeley Data and is accessible through https://doi.org/10.17632/n6bzf2nzj6.1 and https://doi.org/10.17632/kxrsf6cghn.1, for EGCG and FIS, respectively.
2.4. Read alignment and gene quantification
In the first step of the analysis we evaluated sequencing quality by implementing FastQC [26]. Quality control of sequenced reads indicated acceptable results, with the exception of two samples that were characterized by overall poor sequencing quality. Furthermore, we implemented the Trimmomatic read trimming tool [27] in an attempt to discard low quality fragments from the problematic samples. Sequenced reads of each sample were then mapped onto the reference human genome (hg19) using Tophat2 software [28] with default parameters. Apart from the two aforementioned samples, an overall mapping ratio of 82.9% ± 0.9 (mean, SD) was achieved. Low mapping rate (56.1% and 53.4%, respectively), along with poor sequencing quality, lead us to exclude the two samples from downstream analysis steps. Gene quantification for each sample was estimated by HTSeq-count [29] using the intersection-nonempty option as an overlap resolution mode.
2.5. Identification of differentially expressed genes
Raw counts produced by HTSeq-count were normalized based on the DESeq2 [30] normalization method that internally corrects for library size. DESeq2 linear models in R environment were implemented in order to identify statistically significant DEGs between nutraceutical-treated stimulated cells and DMSO-treated stimulated cells. Genes with an adjusted (Benjamini-Hochberg correction for multiple hypothesis testing) p-value < 0.05 were considered as differentially expressed. No log2(Fold Change) cut-off was implemented, as subtle-to-mild gene expression alterations are usually expected following nutraceutical treatment [31]. Lists of DEGs for each nutraceutical were then compared in order to highlight common targets. Co-modified genes were investigated by hierarchical clustering (Euclidean distance, average linkage) regarding their expression profile under the three examined conditions of stimulated cells i.e. DMSO-STIM, EGCG-STIM and FIS-STIM. Expression values used in the hierarchical clustering were normalized by DESeq2, transformed by the variance-stabilizing transformation [30] and finally mean-centered.
2.6. Transcription Factor, Gene Ontology and Reactome Pathway enrichment analyses
ChEA3 database web-server application was implemented on the co-modified genes, in order to perform transcription factor (TF) enrichment analysis [32]. ChEA3 covers 1632 site-specific TFs and offers a selection of six primary reference gene set libraries, generated from various sources of distinct data: a. GTEx and ARCHS4 libraries containing TF-gene co-expression RNA-seq data, b. ENCODE, Literature ChIP-seq and ReMap containing TF-target associations from ChIP-seq experiments and c. Enrichr Queries comprised of TF-gene co-occurrence from user-submitted lists. Individual enrichment results for each library are consequently integrated, thus, producing an improved composite rank of potentially implicated prioritized TFs. The MeanRank integration method was selected, as it generally offers a higher predictive performance [32].
Functional Gene Ontology (GO) and Reactome Pathway enrichment analysis of the co-modified DEGs was conducted using the Bioinfominer software, a bioinformatic tool for intelligent, automated interpretation of genomic data, which performs statistical and network analysis on various biological hierarchical vocabularies [33], [34]. Significance threshold for altered biological processes/pathways was set at a corrected hypergeometric p-value of 0.05.
2.7. Reverse transcription-quantitative real time PCR (RT-qPCR)
RNA (400 ng) was subjected to reverse transcription using the Takara PrimeScriptTM RT reagent Perfect Real Time Kit (Takara, Japan) and RT-qPCR was performed on a CFX Connect Real-Time System (Biorad, USA), using the SensiFASTTM SYBR Lo-ROX Kit (Bioline, UK) according to the manufacturer’s protocol i.e. polymerase activation at 95 °C for 2 min followed by 40 cycles of denaturation at 95 °C for 5sec and annealing/extension at 60 °C for 30sec, concluding with a melting curve construction from 65 to 95 °C with a 0.5 °C increment. Primers for selected genes were provided by Invitrogen (USA) and are presented in Table 1. Amplifications were performed in technical duplicates, using Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a reference gene for normalization purposes. RNA used was derived from the same samples subjected to RNA-seq and also from two independent biological replicates. Data analysis of the resulting Ct values was performed by using the 2-ΔΔCt method [35].
Table 1.
Primers used for RT-qPCR of selected genes.
| HGNC Gene Symbols | Forward Primer | Reverse Primer |
|---|---|---|
| ADAMTS9 | 5′-TCCGAGACTGCCGTAGAAAGA-3′ | 5′-CCGACAAAACCTGAAGCAAAA-3′ |
| MMP11 | 5′-CCGCCTCTACTGGAAGTTTG-3′ | 5′-GCACAGCCAAAGAAGTCAGG-3′ |
| MMP9 | 5′-TTGACAGCGACAAGAAGTGG-3′ | 5′-CAGTGAAGCGGTACATAGGG-3′ |
| SERPINE1 | 5′-CACAAATCAGACGGCAGCACT-3′ | 5′-CATCGGGCGTGGTGAACTC-3′ |
| CTGF | 5′-AGGAGTGGGTGTGTGACGA-3′ | 5′-CCAGGCAGTTGGCTCTAATC-3′ |
| PDGFRB | 5′-CGTCAAGATGCTTAAATCCACAGC-3′ | 5′-TGATGATATAGATGGGTCCTCCTTTG-3′ |
| HSPB1 | 5′-AAGGATGGCGTGGTGGAGATC-3′ | 5′-TCGTTGGACTGCGTGGCTAG-3′ |
| HSPA2 | 5′-AAAGGTCGTCTGAGCAAGGA-3′ | 5′-ATAGGACTCCAGGGCGTTTT-3′ |
| CLIC3 | 5′-TGTTTGTCAAGGCGAGTGAG-3′ | 5′-CGTGGTGAGGGTGAAAGGTA-3′ |
| GAPDH | 5′-CAGTCAGCCGCATCTTCTTTTG-3′ | 5′-AATCCGTTGACTCCGACCTTC-3′ |
2.8. Statistical analysis
RT-qPCR data were expressed as mean ± standard error of the mean (SEM) values, for each condition studied. Statistical analysis by Mann-Whitney test was performed using GraphPad Prism 6.0 software (GraphPad, San Diego, USA). A p-value < 0.05 indicates a statistically significant difference.
3. Results
3.1. Differential gene expression in EGCG and FIS-treated HEP3B cells
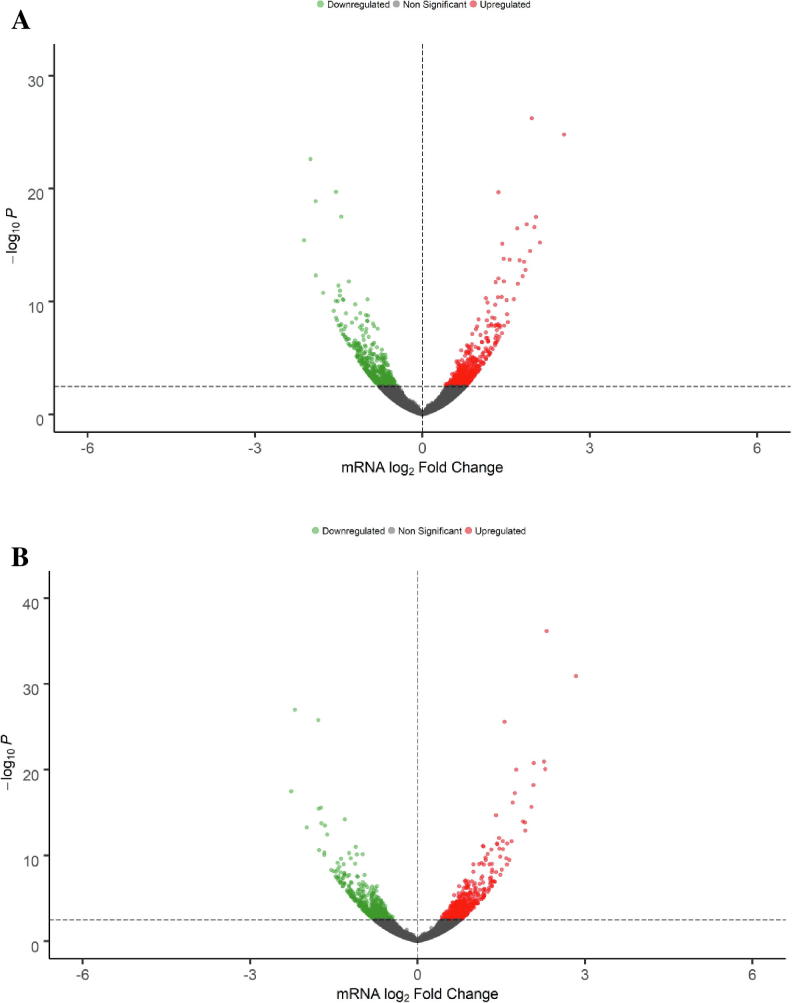
In order to identify potential transcriptome alterations that might be related with the previously published inhibitory effects of EGCG and FIS on HEP3B cells [21], we followed a similar treatment protocol with the modification that after pre-exposure of HEP3B cell cultures for 2 h with the test compounds or DMSO, a mixture of pro-inflammatory/growth signaling stimuli (IL-6, IL-1B and TGFA) or medium was added and the incubation further continued for 22 h. Gene expression was next monitored by RNA-seq in samples from all different treatment combinations (DMSO-MEM, DMSO-STIM, EGCG-MEM, EGCG-STIM, FIS-MEM, FIS-STIM) and acquired data were searched for statistically significant gene expression differences versus a. basal expression levels represented by DMSO-MEM and b. “stimulated” expression levels represented by DMSO-STIM. These comparisons revealed that gene expression alterations in EGCG-MEM and FIS-MEM versus DMSO-MEM did not reach (with few exceptions) the preset statistical criteria of significance, whereas a substantial number of statistically significant DEGs were identified in EGCG-STIM and FIS-STIM versus DMSO-STIM. Based on these results, in our subsequent downstream analysis we focused on the latter set of genes that were significantly modified by test compounds under stimulated conditions which eventually are more representative of live interactions between HCC tumor and its stroma. Volcano plots shown in Fig. 2 visualize the number of DEGs following exposure to EGCG (Fig. 2A) and FIS (Fig. 2B) compared to vehicle in stimulated cells. The horizontal dashed line indicates the preset statistical threshold corresponding to an adjusted p-value of < 0.05 whereas red and green dots represent up- and down-regulated genes, respectively. According to the aforementioned criterion the numbers of identified DEGs were 922 (458 over- and 464 under-expressed) after EGCG and 973 (498 over- and 475 under-expressed) after FIS treatment. It is worth noting that although no log2(Fold Change) cut-off was initially implemented, all determined DEGs are characterized by an observed |log2(Fold Change)| > 0.4. Differential analysis results for all investigated comparisons are provided in Supplementary File S1 (Tables S1-S4).
Fig. 2.
Volcano plots illustrating DEGs in EGCG-stimuli versus DMSO-stimuli (A) and FIS-stimuli versus DMSO-stimuli (B). Red and green dots represent up- and down-regulated genes, respectively; grey dots correspond to non-statistically significant altered genes. The horizontal dashed line indicates a statistical threshold corresponding to an adjusted p-value of < 0.05; x-axis: mRNA log2 Fold Change, y-axis: p-value in negative log10 scale. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
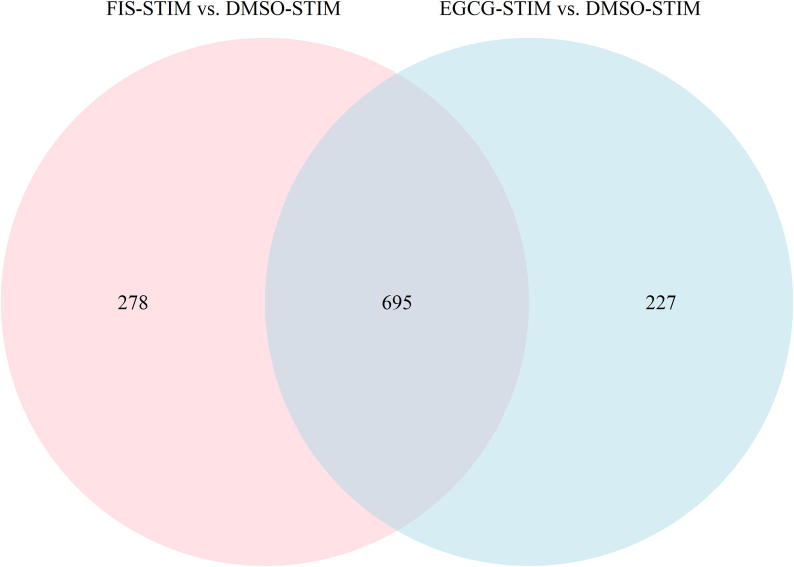
Venn diagram analysis on identified DEGs was next performed in order to detect possibly shared gene targets between EGCG and FIS. Notably, as shown in Fig. 3, the expression of the large majority of genes, i.e. 695 in total (Supplementary File S1 - Table S5) was modified by both nutraceuticals.
Fig. 3.
Venn diagram depicting common DEGs (adjusted p-value < 0.05) derived from nutraceuticals-treated versus DMSO-treated stimulated HEP3B cells. FIS-STIM: FIS-treated stimulated cells, EGCG-STIM: EGCG-treated stimulated cells, DMSO-STIM: DMSO-treated stimulated cells.
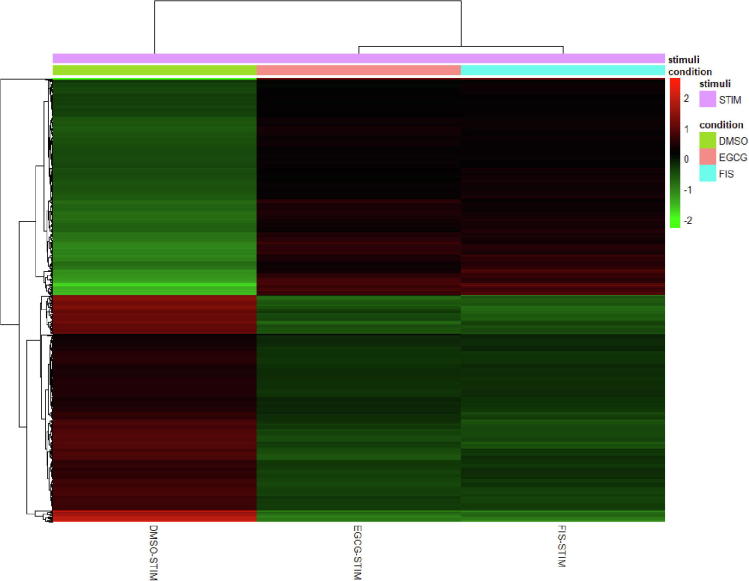
Furthermore, to compare the pattern of gene expression alterations, we next undertook hierarchical unsupervised clustering of the 695 co-modified genes (341 over- and 354 under-expressed) and their expression at all induced experimental settings (DMSO-STIM, EGCG-STIM, FIS-STIM). As illustrated in the heatmap representation (Fig. 4), cell treatment with EGCG and FIS in the presence of stimuli resulted in a quite identical gene expression profile, which was essentially opposite, compared to the one observed after exposure of cells to stimuli alone. These results indicate that test compounds might counteract the signaling aberrations mediated on HCC by paracrine pro-inflammatory and growth stimuli and reverse the imposed transcriptome changes in the majority of targeted genes.
Fig. 4.
Heatmap visualizing the hierarchical clustering of gene expression for the 695 common genes. Each row represents one of the common genes. Columns represent expression averages of replicates for each investigated group. Expression values are normalized counts from DESeq2, transformed by the variance-stabilizing transformation and mean-centered. Red color indicates relative over-expression, while green color indicates relative under-expression. DMSO-STIM: DMSO-treated stimulated cells, EGCG-STIM: EGCG-treated stimulated cells, FIS-STIM: FIS-treated stimulated cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Target genes association to transcription factors, biological processes and pathways
To gain evidence for potential mechanistic connection of transcriptome changes imposed by test nutraceuticals to specific TFs, biological processes and pathways, the list of the 695 co-modified genes was subjected to TF and functional enrichment analyses using established computational tools. ChEA3 TF analysis offers a better understanding of gene regulatory networks and illuminates associations between involved TFs and observed gene expression changes. Functional GO (category “Biological Process”) and Reactome Pathway enrichment analyses, provide an insight into the implicated biological terms and action mechanisms.
Table 2 presents the top 10 prioritized TFs according to their MeanRank integrated composite rank score, documented information about their major biological functions in the context of cancer (Supplementary File S2) and their acquired ranks for each individual library. As shown, crucial cancer-related processes -including epithelial-mesenchymal transition (EMT), ECM component expression, cell cycle and apoptosis control, DNA-damage, cell motility, invasion, metastasis, inflammation and angiogenesis- are known to be regulated by the majority of the indicated TFs. Table S6 in Supplementary File S1 provides a complete list of all 1632 site-specific TFs covered by ChEA3, prioritized based on their integrated MeanRank score, along with the overlapping genes found to be affected by test nutraceuticals for each TF entry. By crossing this list with the list of the 695 genes altered by FIS and EGCG, 45 TFs were found (highlighted by colour labeling at Table S6) whose gene expression is altered by test nutraceuticals in stimulated HEP3B cells.
Table 2.
Top 10 TFs derived from the ChEA3 enrichment analysis. TFs are ranked by an integrated composite score computed based on the individual library ranks.
| Final Rank | Transcription Factor | Biological Role1 | Individual Library Ranks |
|---|---|---|---|
| 1 | CREB3L2 | Cell Proliferation | ARCHS4 Coexpression,30;Enrichr Queries,19;GTEx Coexpression,2 |
| 2 | TWIST2 | EMT, Migration, Invasion | ARCHS4 Coexpression,21;GTEx Coexpression,14 |
| 3 | ELK3 | Invasion, Migration, Angiogenesis | Literature ChIP-seq,9;ARCHS4 Coexpression,43;Enrichr Queries,1;GTEx Coexpression,23 |
| 4 | ZNF469 | ECM | ARCHS4 Coexpression,7;GTEx Coexpression,32 |
| 5 | NR2F2 | Tumor Growth, Metastasis, Tumor Microenvironment | ARCHS4 Coexpression,20; ENCODE ChIP-seq,20; Enrichr Queries,5; ReMap ChIP-seq,47; GTEx Coexpression,8 |
| 6 | GLIS2 | EMT, Inflammation | ARCHS4 Coexpression,40;Enrichr Queries,10;GTEx Coexpression,13 |
| 7 | HIC1 | DNA-damage, Invasion, Metastasis | ARCHS4 Coexpression,24;Enrichr Queries,42;GTEx Coexpression,1 |
| 8 | NFATC4 | Cell Cycle and Apoptosis, Inflammation | ARCHS4 Coexpression,35;Enrichr Queries,27;GTEx Coexpression,19 |
| 9 | NPAS2 | Cell Survival, Cell Growth, DNA-damage | ARCHS4 Coexpression,28;Enrichr Queries,17;GTEx Coexpression,39 |
| 10 | TWIST1 | EMT, Angiogenesis, Invasion, Metastasis | ARCHS4 Coexpression,37;Enrichr Queries,53;ReMap ChIP-seq,19;GTEx Coexpression,7 |
Literature available in Supplementary File S2.
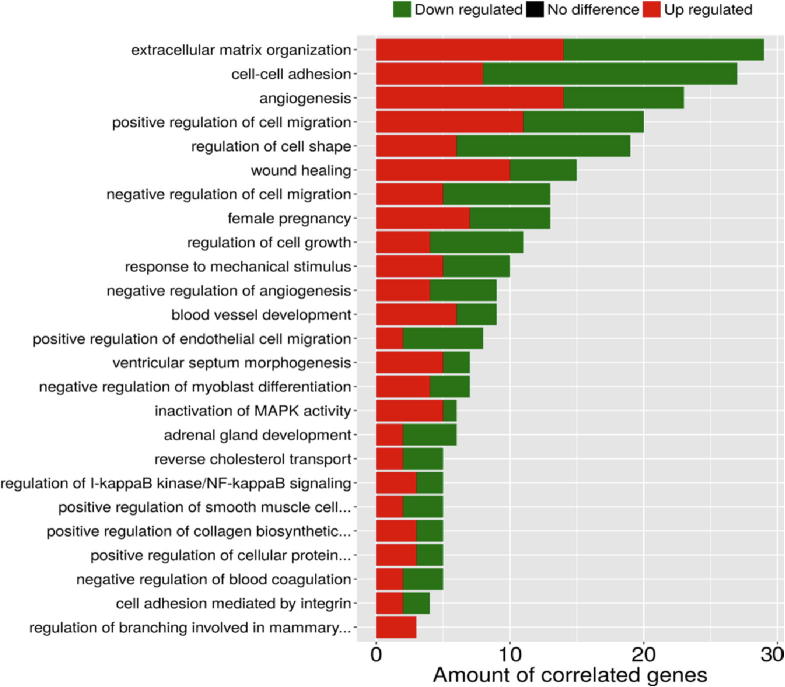
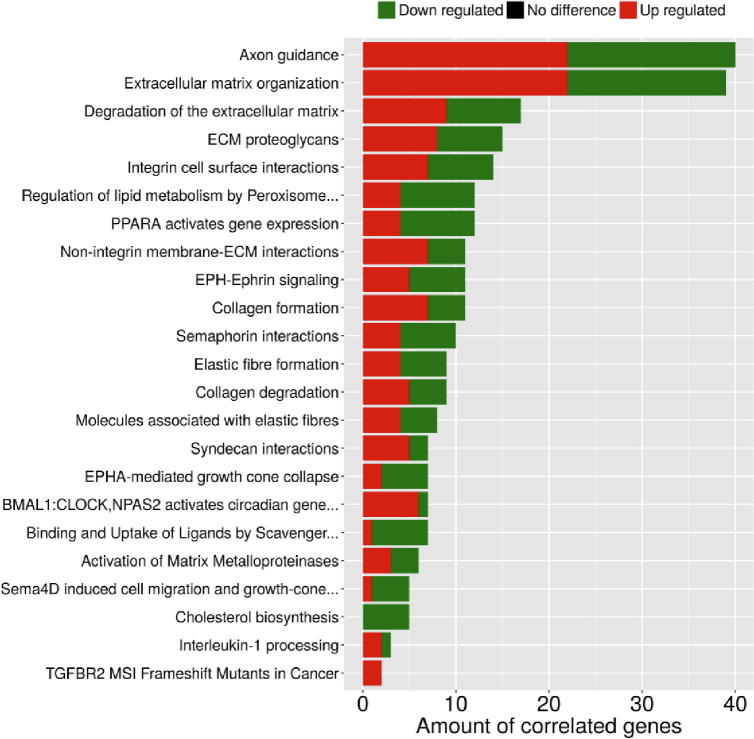
Fig. 5 summarizes the 25 over-represented GO terms which are significantly (corrected p-value < 0.05) associated with the co-modified genes. Altered biological processes involve a wide range of ontological terms related to ECM organization, wound healing, cell–cell adhesion, regulation of cell growth and cell shape, angiogenesis, regulation of cell migration including also specific endothelial cell migration related terms, inactivation of MAPK activity, EMT and regulation of I-kappaB kinase/NF-kappaB signaling. Furthermore, as shown in Fig. 6, common genes were linked by Reactome Pathways enrichment analysis to 23 statistically significant (corrected p-value < 0.05) pathways that were again predominately related to ECM dynamics (organization, degradation, interaction, activation of matrix metalloproteinases) but also to various cell–cell interaction signaling ontological pathways, regulation of lipid metabolism and cholesterol biosynthesis, interleukin-1 processing, RAF-independent MAPK1/3 activation. Detailed lists of all ontological terms - both processes and pathways - along with the implicated genes and significance statistics are provided in Supplementary Files S3& S4.
Fig. 5.
GO enrichment analysis associating the co-modified DEGs with statistically significant biological processes (corrected p-value < 0.05). Red color indicates upregulation, while green color indicates downregulation of gene expression. y-axis: ontological terms, x-axis: amount of correlated genes in each ontological term. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Reactome Pathway enrichment analysis associating the co-modified DEGs with statistically significant biological pathways (corrected p-value < 0.05). Red color indicates upregulation, while green color indicates downregulation of gene expression. y-axis: ontological terms, x-axis: amount of correlated genes in each ontological term. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
From the DEGs included in the prevalent ECM-related categories attention was paid in genes encoding members of ADAMTS and Matrix Metallopeptidases (MMPs) families due to their established key role in cancer progression [36], [37]. More specifically the expression of MMP11, MMP24 and ADAMTSL4 was found to be downregulated while the expression of MMP1, MMP9 and ADAMTS9 upregulated by test compounds. From the sets of genes belonging to terms related to regulation of cell growth, migration, angiogenesis, wound healing and blood coagulation we distinguished those encoding for Connective Tissue Growth Factor (CTGF) and Platelet Derived Growth Factor Receptor Beta (PDGFRB) - both under-expressed - along with a group of serpinE genes (SERPINED1, SERPINE1, SERPINE2) which were overexpressed after exposure to nutraceuticals. Notably, several genes encoding for members of the heat shock family of proteins, namely Heat Shock Protein Family B (Small) Member 1 (HSPB1), which is included in category “regulation of I-kappaB kinase/NF-kappaB signaling” as well as HSPA2, HSPA1A and HSP90AA1, were all found to be downregulated by test nutraceuticals.
3.3. Validation of RNA-seq data by RT-qPCR
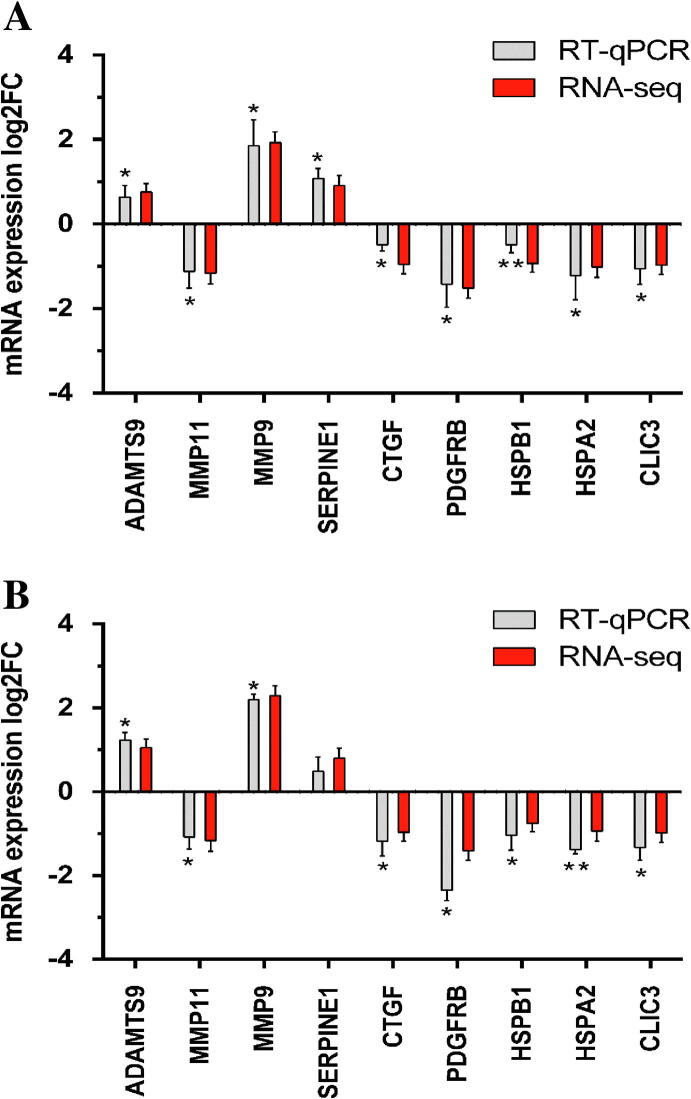
Experimental validation of RNA-seq data by RT-qPCR was performed on nine selected candidates included in the list of co-modified genes. More specifically, RT-qPCR was performed representatively for ADAMTS9, MMP11, MMP9, SERPINE1, CTGF, PDGFRB and HSPB1 since as mentioned above, these genes have been found in the functional enrichment analysis to significantly associate with one or more crucial ontological terms. Furthermore, two additional genes, namely HSPA2 and Chloride Intracellular Channel 3 (CLIC3), although not included into specific functional categories, were also selected for validation due to their importance in HCC or other cancer type advancement [38], [39]. As shown in Fig. 7A and 7B for EGCG and FIS respectively, the gene expression changes assessed by the two methodologies were in good correlation both in terms of direction and magnitude. It should be noted that repetitive RT-qPCR results were obtained with RNA templates derived from both the samples used in RNA-seq and from samples collected in two independent biological experiments, thus further supporting the reliability of the RNA-seq data and their subsequent computational analyses.
Fig. 7.
RT-real time qPCR validation of RNA-seq data for selected genes. RT-qPCR data represent mean ± SEM (n = 3–4, *p-value < 0.05, **p-value < 0.01). mRNA expression fold changes (log2 scale) derived from (A) EGCG-STIM and (B) FIS-STIM versus DMSO-STIM.
4. Discussion
Nutraceuticals have lately attracted a great attention in both the general public and the pharmaceutical market due to their potential in prevention, management or treatment of chronic diseases, including cancer. However, as governmental regulations vary widely by country, they are often sold without a precise scientific documentation of efficacy or safety [40]. Hence, studies attempting to investigate, at active but non-cytotoxic concentrations, their effects, involved molecular biomarkers and mode of action, are urgently needed to justify and expand their clinical use. In this work we have examined by RNA-seq the transcriptome alterations induced by two effective nutraceuticals, EGCG and FIS, on human HEP3B cells [21] in the presence (or not) of a mixture of inflammatory and growth stimuli (IL-6, IL-1B and TGFA), known to be actively involved in HCC advancement, thus reproducing to some extent the complex signaling crosstalk between tumor and the surrounding non-malignant stroma cells [41]. The major fraction of DEGs, i.e. 695 genes out of 922 and 973 altered in EGCG- and FIS-treated stimulated cells respectively, versus the stimuli-treated control cells, were found to be targeted by both compounds. Most importantly, comparison of the co-modified genes expression under the examined conditions in stimulated cells revealed that EGCG and FIS: a. modify gene expression in a near identical manner thus indicating involvement of common regulatory mechanisms with likely multiple consequences on cell system homeostasis and b. can essentially reverse the changes imposed by exposure to stimuli alone, thus underpinning their capability to “normalize” crucial pathogenetic dysfunctions sustained by the milieu of HCC tumors. Mechanistic associations to nutraceutical-mediated transcriptome differential expression were further explored by submitting the co-modified genes to TF, GO and Reactome Pathway enrichment analyses.
Accumulating evidence has demonstrated that cancer progression is strongly related with dysregulation of TFs as evidenced by the fact that they account for about 20% of all known oncogenes [42]. In the present work, integrated computational TF enrichment analysis based on co-modified DEGs revealed a prioritized list of potentially associated transcription regulators - activators or suppressors - implicated in a broad range of cellular processes related to malignant transformation and progression, thus supporting a promising modulatory action of test nutraceuticals. As a matter of fact, 45 of the associated TFs that are covered by the ChEA3 database, including ELK3/ELK1, NPAS2, FOSL1, HMGA2 and members of the FOX and SOX families, are presently found to be significantly modified by EGCG and FIS at the gene expression level. On the other hand, we and others have previously shown that test nutraceuticals can also act post-transcriptionally [19], [21], [23], for example by altering the phosphorylation and thus the activation status of their targets, as in cases of NFKB and CREB1 in HEP3B cells [21]. It would be interesting to further validate and investigate experimentally the potential effect of test compounds on “predicted” TFs at all possible levels (gene/protein expression, stability and function).
Concerning the functional GO and Reactome Pathways analyses, an interesting outcome is the prevalence of ECM-related terms thus indicating that EGCG and FIS may provide an effective modality to potentially “restore” ECM-dependent deregulated signaling that reinforces cellular transformation, invasion, metastasis, tumor-associated angiogenesis and inflammation [43]. Amongst the DEGs included in the ECM-related categories were those encoding distinct members of MMPs (MMP1, MMP9, MMP11 and MMP24) and ADAMTS (ADAMTS9 and ADAMTSL4) families of metalloproteinases. MMPs, which represent the most studied extracellular proteinases, have been proved to play a prominent role in ECM remodeling and cancer cell migration but also in regulation of molecular signaling within tumor microenvironment [37]. Although the contribution of ADAMTS to cancer progression is less understood, previous studies support aberrant expression of certain members of this family in malignant cells [36]. More specifically, in HCC, the promoter of ADAMTS9 is hypermethylated resulting in downregulated expression, which in turn is associated with poor survival [44]. Furthermore, in gastric cancers ADAMTS9 has been demonstrated to act as a functional tumor suppressor [45]. In the present work, test nutraceuticals downregulated the expression of MMP11, MMP24 and ADAMTSL4, while caused overexpression of MMP1, MMP9 and ADAMTS9 genes. These results only merely corroborate with published evidence supporting that the effect of EGCG and FIS on MMPs protein expression, in HCC and other cancers, is mostly inhibitory [17], [22], [25]. However, studies considering the failure of MMP inhibitors as potential anticancer therapeutics, rather indicate that MMPs, depending on the circumstances, may either suppress or promote cancer progression [37], [46]. Therefore, the biological relevance of the observed gene expression regulation pattern needs further investigation.
Apart from ECM dynamics, several other important GO terms related to regulation of cell growth, migration, angiogenesis, wound healing and blood coagulation have been demonstrated to significantly associate with a number of DEGs, including those encoding for CTGF, PDGFRB (downregulated) and a group of serpins (SERPINED1, SERPINE1, SERPINE2; upregulated) which are known to influence HCC progression. As a matter of fact, overexpressed CTGF has been found to promote cell growth and invasion in HCC [47] and play a keystone role in the transmission of growth signals between hepatoma and hepatic stellate cells [48]. High expression of PDGFRB is associated with poor prediction outcomes in advanced HCC [49] and with accelerated tumor growth in a mouse model of liver cancer [50]. The superfamily of serpins displays diverse biological functions, which in the context of cancer are mostly related with tumor angiogenesis, thrombolysis and blood coagulation [51]. Although, some serpins act as tumor suppressors limiting the invasive and metastatic potential of HCC cells [52], others, like SERPINE1, have been shown to positively correlate with increased tumor angiogenic activity [53]. Therefore, the demonstration of three serpin members as new EGCG and FIS targets poses interesting research questions for the potential consequences of their regulation on HCC. Furthermore, one of the most significantly altered (downregulated) genes in the DEGs dataset, CLIC3, a chloride channel protein encoding gene, emerged as well as a potential new target of EGCG and FIS. Chloride ion channels, being able to regulate ion homeostasis, pH levels and cell volume, appear to be important players in the regulation of cancer cell cycle, migration and infiltration [54]. For instance, CLIC3, a pro-invasive oxidoreductase, is secreted by cancer-associated fibroblasts and cancer cells, driving angiogenesis and cancer progression by promoting transglutaminase-2 (TGM2) - dependent invasion [39].
Finally, several DEGs are significantly associated with the term “regulation of I-kappaB kinase/NF-kappaB signaling” that refers to a key pathway orchestrating the transcription of a variety of genes controlling prominent pre-malignant processes such as cancer-related inflammatory response, cell proliferation/survival and angiogenesis [55]. HSPB1 (alternative symbol HSP27), a representative gene of this category encoding for heat-shock protein 27 is downregulated by EGCG and FIS. It has been previously shown that siRNA-mediated knockdown of HSPB1 in highly metastatic HCC cells significantly suppresses cell migration and invasion and induces cell apoptosis through regulation of NF-kappaB signal transduction [56]. Furthermore, extended literature supports the potential of both nutraceuticals to reduce the expression of HSPB1, HSP70 and HSP90 through inhibition of their promoter activity, thus attenuating cell proliferation and survival in various cancer models [57], [58]. In corroboration with these results we found that exposure to test nutraceutical downregulated the gene expression of three additional members of heat shock family of proteins: HSPA2, HSPA1A and HSP90AA1. Overexpression of HSPA2 in particular, correlates with aggressive HCC phenotypes and is an independent biomarker for poor prognosis of patients with HCC [38]. Overall current data further document heat shock proteins as key targets of EGCG and FIS in the context of HCC.
5. Conclusions
RNA-seq data analysis of stimulated HEP3B cells treated with EGCG and FIS provides novel evidence on their ability to efficiently reverse the transcriptome alterations mediated by micro-environmentally derived pro-inflammatory/growth signaling perturbations. Test nutraceuticals were found to act through expression regulation of common gene targets that significantly associate with prominent cancer-related TFs, biological processes and pathways. Although the innate molecular heterogeneity of HCC may restrict the generalization of current conclusions, our study sets a rational basis for application of systems biology methodologies in the global elucidation of molecular targets and action mode of promising anti-cancer nutraceuticals.
6. Availability of data and materials
Raw RNA-seq FASTQ data supporting the conclusions of this article have been deposited to Mendeley Data and are available at https://doi.org/10.17632/n6bzf2nzj6.1 (EGCG dataset) and https://doi.org/10.17632/kxrsf6cghn.1 (FIS dataset).
Funding
We acknowledge support of this work by the project “Synthetic Biology: from omics technologies to genomic engineering (OMIC-ENGINE)” (MIS 5002636) which is implemented under the Action Reinforcement of the Research and Innovation Infrastructure, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
CRediT authorship contribution statement
Panagiotis C. Agioutantis: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Vasilios Kotsikoris: Data curation, Formal analysis, Investigation, Methodology, Validation. Fragiskos N. Kolisis: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Validation, Writing - review & editing. Heleni Loutrari: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.03.006.
Contributor Information
Panagiotis C. Agioutantis, Email: panagiout@mail.ntua.gr.
Vasilios Kotsikoris, Email: vasiliskotsikoris@yahoo.gr.
Fragiskos N. Kolisis, Email: kolisis@chemeng.ntua.gr.
Heleni Loutrari, Email: elloutrar@med.uoa.gr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary File S1 - Tables S1 - S4: Differential expression analysis results for EGCG-MEM versus DMSO-MEM, FIS-MEM versus DMSO-MEM, EGCG-STIM versus DMSO-STIM and FIS-STIM versus DMSO-STIM. Table S5: List of 695 identified common genes. Table S6: List containing all 1632 unique TFs included in ChEA3 database, along with their ranks, integrated MeanRank score and overlapping genes.
Supplementary File S2 - Literature regarding the biological role of associated TFs, as supplementary information to Table 2.
Supplementary File S3 - Complete data from GO enrichment analysis.
Supplementary File S4 - Complete data from Reactome Pathway enrichment analysis.
References
- 1.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. The Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Berasain C., Castillo J., Perugorria M.J., Latasa M.U., Prieto J., Avila M.A. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci. 2009;1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag H.B. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Muto J., Shirabe K., Sugimachi K., Maehara Y. Review of angiogenesis in hepatocellular carcinoma. Hepatol Res. 2015;45(1):1–9. doi: 10.1111/hepr.12310. [DOI] [PubMed] [Google Scholar]
- 5.Marengo A., Rosso C., Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi L., Oliveira C.P., Carrilho F.J. Nonalcoholic fatty liver disease and hepatocellular carcinoma. BioMed Research International. 2014;2014 doi: 10.1155/2014/106247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravalli R.N., Steer C.J., Cressman E.N. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48(6):2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 8.Aravalli R.N., Cressman E.N., Steer C.J. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol. 2013;87(2):227–247. doi: 10.1007/s00204-012-0931-2. [DOI] [PubMed] [Google Scholar]
- 9.Llovet J., Ricci S., Mazzaferro V. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 10.Kalra E.K. Nutraceutical - definition and Introduction. AAPS PharmSci. 2003;5(3):27–28. doi: 10.1208/ps050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S.C., Kim J.H., Prasad S., Aggarwal B.B. Regulation of survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells through modulation of inflammatory pathways by nutraceuticals. Cancer Metastasis Rev. 2010;29(3):405–434. doi: 10.1007/s10555-010-9235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasri H., Baradaran A., Shirzad H., Rafieian-Kopaei M. New concepts in nutraceuticals as alternative for pharmaceuticals. Int J Prev Med. 2014;5(12):1487–1499. [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K.W., Bode A.M., Dong Z. Molecular targets of phytochemicals for cancer prevention. Nat Rev Cancer. 2011;11(3):211–218. doi: 10.1038/nrc3017. [DOI] [PubMed] [Google Scholar]
- 14.Pal H.C., Diamond A.C., Strickland L.R., Kappes J.C., Katiyar S.K., Elmets C.A. Fisetin, a dietary flavonoid, augments the anti-invasive and anti-metastatic potential of sorafenib in melanoma. Oncotarget. 2015;7(2):1227–1241. doi: 10.18632/oncotarget.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecumberri E., Dupertuis Y.M., Miralbell R., Pichard C. Green tea polyphenol epigallocatechin-3-gallate (EGCG) as adjuvant in cancer therapy. Clinical Nutr. 2013;32(6):894–903. doi: 10.1016/j.clnu.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Sung B., Prasad S., Yadav V.R., Aggarwal B.B. Cancer cell signaling pathways targeted by spice-derived nutraceuticals. Nutr Cancer. 2012;64(2):173–197. doi: 10.1080/01635581.2012.630551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darvesh A.S., Bishayee A. Chemopreventive and therapeutic potential of tea polyphenols in hepatocellular cancer. Nutr Cancer. 2013;65(3):329–344. doi: 10.1080/01635581.2013.767367. [DOI] [PubMed] [Google Scholar]
- 18.Shankar E., Kanwal R., Candamo M., Gupta S. Dietary phytochemicals as epigenetic modifiers in cancer: Promise and challenges. Semin Cancer Biol. 2016;40–41:82–99. doi: 10.1016/j.semcancer.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanmugam M.K., Lee J.H., Chai E.Z., Kanchi M.M., Kar S., Arfuso F. Cancer prevention and therapy through the modulation of transcription factors by bioactive natural compounds. Semin Cancer Biol. 2016;40–41:35–47. doi: 10.1016/j.semcancer.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Prasad S., Gupta S.C., Tyagi A.K. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett. 2017;387:95–105. doi: 10.1016/j.canlet.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Michailidou M., Melas I., Messinis D., Klamt S., Alexopoulos L., Kolisis F., Loutrari H. : Network-based analysis of nutraceuticals in human hepatocellular carcinomas reveals mechanisms of chemopreventive action. CPT: Pharmacomet Syst Pharmacol. 2015;4(6):350–361. doi: 10.1002/psp4.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negri A., Naponelli V., Rizzi F., Bettuzzi S. Molecular targets of epigallocatechin-gallate (EGCG): a special focus on signal transduction and cancer. Nutrients. 2018:10(12). doi: 10.3390/nu10121936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syed D.N., Adhami V.M., Khan N., Khan M.I., Mukhtar H. Exploring the molecular targets of dietary flavonoid fisetin in cancer. Semin Cancer Biol. 2016;40–41:130–140. doi: 10.1016/j.semcancer.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youns M., Abdel Halim Hegazy W. The natural flavonoid fisetin inhibits cellular proliferation of hepatic, colorectal, and pancreatic cancer cells through modulation of multiple signaling pathways. PLoS ONE. 2017;12(1) doi: 10.1371/journal.pone.0169335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loutrari H., Magkouta S., Papapetropoulos A., Roussos C. Mastic oil inhibits the metastatic phenotype of mouse lung adenocarcinoma cells. Cancers. 2011;3(1):789–801. doi: 10.3390/cancers3010789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S: FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. 2010.
- 27.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013:14(14). doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulos P., Papadodima O., Chatziioannou A., Loutrari H., Roussos C., Kolisis F.N. A transcriptomic computational analysis of mastic oil-treated Lewis lung carcinomas reveals molecular mechanisms targeting tumor cell growth and survival. BMC Med Genomics. 2009;2:68. doi: 10.1186/1755-8794-2-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keenan A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koutsandreas T., Binenbaum I., Pilalis E., Valavanis I., Papadodima O., Chatziioannou A. Analyzing and vizualizing genomic complexity for the derivation of the emergent molecular networks. Int J MonitSurveillTechnol. 2016;4(2):30–49. [Google Scholar]
- 34.Lhomond S., Avril T., Dejeans N., Voutetakis K., Doultsinos D., McMahon M. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol Med. 2018;10(3) doi: 10.15252/emmm.201707929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Rocks N., Paulissen G., El Hour M., Quesada F., Crahay C., Gueders M. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90(2):369–379. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Y., Zhao H., Li X.S., Kang H.R., Ma J.X., Yao F.F. Expression of HSPA2 in human hepatocellular carcinoma and its clinical significance. Tumour Biol J Int Soc Oncodev Biol Med. 2014;35(11):11283–11287. doi: 10.1007/s13277-014-2430-y. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez-Fernaud J.R., Ruengeler E., Casazza A., Neilson L.J., Pulleine E., Santi A. Secreted CLIC3 drives cancer progression through its glutathione-dependent oxidoreductase activity. Nat Commun. 2017;8:14206. doi: 10.1038/ncomms14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daliu P., Santini A., Novellino E. From pharmaceuticals to nutraceuticals: bridging disease prevention and management. Expert Rev Clinic Pharmacol. 2019;12(1):1–7. doi: 10.1080/17512433.2019.1552135. [DOI] [PubMed] [Google Scholar]
- 41.Novikova M.V., Khromova N.V., Kopnin P.B. Components of the hepatocellular carcinoma microenvironment and their role in tumor progression. Biochem. Biokhimiia. 2017;82(8):861–873. doi: 10.1134/S0006297917080016. [DOI] [PubMed] [Google Scholar]
- 42.Lambert M., Jambon S., Depauw S., David-Cordonnier M.H. Targeting transcription factors for cancer treatment. Molecules. 2018:23(6). doi: 10.3390/molecules23061479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu P., Weaver V.M., Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S.M., Wang L., Zhang B., Guo W. Promoter hypermethylation mediates the down-regulated expression of ADAMTS9 and associated with the prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2016;9(9):9646–9656. [Google Scholar]
- 45.Du W., Wang S., Zhou Q., Li X., Chu J., Chang Z. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32(28):3319–3328. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- 46.Coussens L.M., Fingleton B., Matrisian L.M. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 47.Xiu M., Liu Y.H., Brigstock D.R., He F.H., Zhang R.J., Gao R.P. Connective tissue growth factor is overexpressed in human hepatocellular carcinoma and promotes cell invasion and growth. World J Gastroenterol. 2012;18(47):7070–7078. doi: 10.3748/wjg.v18.i47.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makino Y., Hikita H., Kodama T., Shigekawa M., Yamada R., Sakamori R. CTGF mediates tumor-stroma interactions between hepatoma cells and hepatic stellate cells to accelerate HCC progression. Cancer Res. 2018;78(17):4902–4914. doi: 10.1158/0008-5472.CAN-17-3844. [DOI] [PubMed] [Google Scholar]
- 49.Chu J.S., Ge F.J., Zhang B., Wang Y., Silvestris N., Liu L.J. Expression and prognostic value of VEGFR-2, PDGFR-beta, and c-Met in advanced hepatocellular carcinoma. J. Exp. Clinical Cancer research: CR. 2013;32:16. doi: 10.1186/1756-9966-32-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maass T., Thieringer F.R., Mann A., Longerich T., Schirmacher P., Strand D. Liver specific overexpression of platelet-derived growth factor-B accelerates liver cancer development in chemically induced liver carcinogenesis. Int J Cancer. 2011;128(6):1259–1268. doi: 10.1002/ijc.25469. [DOI] [PubMed] [Google Scholar]
- 51.Law R.H., Zhang Q., McGowan S., Buckle A.M., Silverman G.A., Wong W. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui X., Liu Y., Wan C., Lu C., Cai J., He S. Decreased expression of SERPINB1 correlates with tumor invasion and poor prognosis in hepatocellular carcinoma. J Mol Histol. 2014;45(1):59–68. doi: 10.1007/s10735-013-9529-0. [DOI] [PubMed] [Google Scholar]
- 53.Geis T., Doring C., Popp R., Grossmann N., Fleming I., Hansmann M.L. HIF-2alpha-dependent PAI-1 induction contributes to angiogenesis in hepatocellular carcinoma. Exp Cell Res. 2015;331(1):46–57. doi: 10.1016/j.yexcr.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 54.Peretti M., Angelini M., Savalli N., Florio T., Yuspa S.H., Mazzanti M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochimica et Biophysica Acta. 2015;1848(10 Pt B):2523–2531. doi: 10.1016/j.bbamem.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto Y., Gaynor R.B. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem Sci. 2004;29(2):72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Guo K., Kang N.X., Li Y., Sun L., Gan L., Cui F.J. Regulation of HSP27 on NF-kappaB pathway activation may be involved in metastatic hepatocellular carcinoma cells apoptosis. BMC cancer. 2009;9:100. doi: 10.1186/1471-2407-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.A., Lee S., Kim D.E., Kim M., Kwon B.M., Han D.C. Fisetin, a dietary flavonoid, induces apoptosis of cancer cells by inhibiting HSF1 activity through blocking its binding to the hsp70 promoter. Carcinogenesis. 2015;36(6):696–706. doi: 10.1093/carcin/bgv045. [DOI] [PubMed] [Google Scholar]
- 58.Tran P.L.C.H.B., Kim S.A., Choi H.S., Yoon J.H., Ahn S.G. Epigallocatechin-3-gallate suppresses the expression of HSP70 and HSP90 and exhibits anti-tumor activity in vitro and in vivo. BMC Cancer. 2010;10:276. doi: 10.1186/1471-2407-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File S1 - Tables S1 - S4: Differential expression analysis results for EGCG-MEM versus DMSO-MEM, FIS-MEM versus DMSO-MEM, EGCG-STIM versus DMSO-STIM and FIS-STIM versus DMSO-STIM. Table S5: List of 695 identified common genes. Table S6: List containing all 1632 unique TFs included in ChEA3 database, along with their ranks, integrated MeanRank score and overlapping genes.
Supplementary File S2 - Literature regarding the biological role of associated TFs, as supplementary information to Table 2.
Supplementary File S3 - Complete data from GO enrichment analysis.
Supplementary File S4 - Complete data from Reactome Pathway enrichment analysis.
Data Availability Statement
Raw RNA-seq FASTQ data supporting the conclusions of this article have been deposited to Mendeley Data and are available at https://doi.org/10.17632/n6bzf2nzj6.1 (EGCG dataset) and https://doi.org/10.17632/kxrsf6cghn.1 (FIS dataset).