Abstract
Modification on nucleic acid plays a pivotal role in controlling gene expression. Various kinds of modifications greatly increase the information-encoding capacity of DNA and RNA by introducing extra chemical group to existing bases instead of altering the genetic sequences. As a marker on DNA or RNA, nucleic acid modification can be recognized by specific proteins, leading to versatile regulation of gene expression. However, modified and regular bases are often indistinguishable by most conventional molecular methods, impeding detailed functional studies that require the information of genomic location. Recently, new technologies are emerging to resolve the positions of varied modifications on both DNA and RNA. Intriguingly, by integrating regional targeting tools and effector proteins, researchers begin to actively control the modification status of desired gene in vivo. In this review, we summarize the characteristics of DNA and RNA modifications, the available mapping and editing tools, and the potential application as well as deficiency of these technologies in basic and translational researches.
1. Introduction
Genetic information flows from DNA to RNA, and then express as proteins. However, nucleic acid has other tricks to expand its information-encoding capacity. Beyond the genetic code, there is another hidden layer of complexity, usually mediated by chemical modifications [1]. These so-called epigenetic markers play a pivotal role in numerous biological processes by tuning gene expression network [2]. Instead of altering the genetic code, DNA and RNA modifications regulate gene expression via ‘reader’ proteins and relevant effectors (‘writer’ and ‘eraser’ proteins) [3]. To date, dozens of distinct DNA modifications and even more RNA modifications have been characterized (the main types are illustrated in Fig. 1) [4], [5]. These modifications are installed or removed by writer or eraser proteins, and serve as functional markers specifically recognized by reader proteins [6]. Recent studies revealed variety of DNA and RNA modifications actively participate in diverse biological processes, including embryonic development, organ differentiation, or even human diseases if dysregulated [7]. Along with the detailed characterizing of relevant effector proteins such as writer or eraser, researchers attempt to establish or eliminate modifications at desired loci through novel molecular tools, and study the biological function and related regulatory mechanisms of these modifications [8].
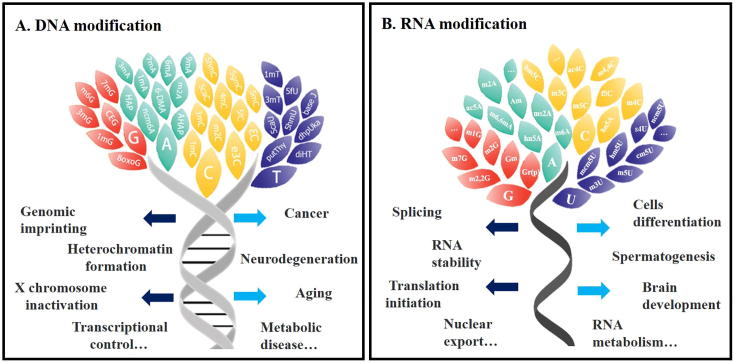
Fig. 1.
Diverse modifications on nucleic acids and their possible functions. Panel A, DNA modifications. Panel B, RNA modifications. Leaves of different colors represent different types of modifications on DNA or RNA bases.
For years, the studying of nucleotide modification had been impeded by several reasons. Given the relatively low abundance, most modifications are difficult to detect and study. For instance, as the most abundant DNA modification in mammals, 5mC accounts for ~1% of the total DNA bases [9], while m6A, the most abundant modification on mRNA, accounts for 0.1–0.4% in all adenosine bases [10]. The proportion of other types of modifications are even lower [5]. Moreover, these minority bases are highly dynamic and sometimes reversable, varying from different physiological conditions or developmental stages [11]. As the first step to study these elusive gadgets, researchers dedicated to develop innovating methods to detect and locate the precise loci across the genome [12]. However, each type of modification has its unique properties, preventing the invent of a universal method. With the fast development of advanced technologies such as high-throughput sequencing, the positions of each type of modification had been mapped to genome or transcriptome one by one [13]. Meanwhile, another group of researchers began to look for novel tools which can alter the modification status of desired location, although the effectors for each modification vary and lack detailed characterization [14], [15]. To better understand the diversity and complexity of nucleic acid modifications, we summarize the current knowledge of representative type of modifications on nucleic acid and relevant technologies, highlighting the importance of mapping and editing tools for this research field.
2. Nucleotide modifications and effector proteins
2.1. DNA modifications
DNA modification mediated biological processes had been extensively studied over the past decades [16]. As indicated in Fig. 1, each of the four bases may have corresponding modifications, among which 5-methylcytosine (5mC) is best characterized as the most abundant one in mammals. As an epigenetic mark, 5mC tightly associates with other chromatin elements, exerting widespread effects on gene expression during multiple developmental and physiological processes, or even human diseases [17]. The enzymes responsible for 5mC deposition and removal have been well defined in mammals, including DNMT1/3A/3B proteins as methyltransferases, and TET1/2/3 as demethylases [17]. These effectors so-called “writer” and “eraser” provide effective tools to manipulate the modification status of DNA, and result in potential targets for therapeutic purpose. Other modifications such as 5-hydroxymethylcytosine (5hmC), 5-formyl cytosine (5fC) and 5-carboxyl cytosine (5caC), though much less abundant, have also been considered as functional regulators but need further investigation [18].
Another kind of DNA modification, N6-methyladenine (6mA), had been originally known as the most prevalent DNA modification in bacteria [19], [20]. It was also found recently in certain kinds of eukaryotes with much less abundance [21]. Besides 6mA, N4-methylcytosine (4mC) is exclusively present in bacteria [22]. Most DNA methyltransferases in bacteria belong to the restriction-modification (RM) systems which defense invading nucleic acid such as virus [23], while certain ‘solitary’ DNA methyltransferases are probably derived from ancestral RM systems but lose their restriction enzyme partner. An example of solitary DNA methyltransferase is the Dam enzyme, which methylates adenosine to 6mA on the 5′-GATC-3′ motif and can be recognized as the writer protein in bacteria [24]. Besides Dam, there is a large class of DNA methyltransferases working on specific sequence motifs, whereas demethylase is supposed not to exist in bacteria [22].
2.2. RNA modifications
Though it has been determined decades earlier that RNA especially noncoding RNAs need various chemical modifications to be fully functional, modifications on mRNA are spotlighted only recently and discovered to be involved in key processes of RNA metabolism [3]. Among the ~150 types of RNA modifications, m6A is of the most abundant one on mRNA with important roles in the regulation of gene expression, thus affecting multiple biological processes such as embryonic development, neuronal regulation and oncogenesis [7], [25]. The proportion of m6A is estimated to be 0.1%–0.4% of total adenosine residues in cellular mRNA, accounting for 2–3 sites per transcript [10]. Although the presence of m6A in eukaryotes has been discovered for years, determining the locations in mRNA was delayed due to the absence of accurate mapping methods until recently [10], [26], [27]. With the development of m6A-specific antibody, researchers started to interrogate the distribution and relative effector proteins in the genome-wide view [28]. To date, the key participants involved in m6A pathway have been well-characterized in mammals, including METTL3/14 and supporting proteins as the “writer” to deposit methyl group at desired positions, FTO/ALKBH5 as the “eraser” to remove modification, and YTH family proteins and associated proteins as the “reader” to decode modification [3]. However, in bacteria, mRNA seems not to contain modification though sporadic studies reported trace level in certain strains [29]. Stimulated by the progress of m6A studies, other modifications also began to catch much attention and research interests. For example, m6Am, a companion modification to m6A, was reported to destabilize mRNA and controlled by the writer protein PCIF1 and eraser protein FTO [30].
3. Methods to map nucleotide modifications
Resolving the genomic localization of nucleotide modifications is essential to study their functions. Owing to the different chemical properties of modified bases, the methods used to map each type of modification diverged significantly [31]. Take m6A for example, it is not distinguishable from regular A base by base-pairing strategy. Thus, new methods are proliferating in recent years to map variety of modifications across the genome or transcriptome. We generally attribute these methods to 3 classes and will highlight 5mC in DNA and m6A in RNA as exemplified objectives (Fig. 2).
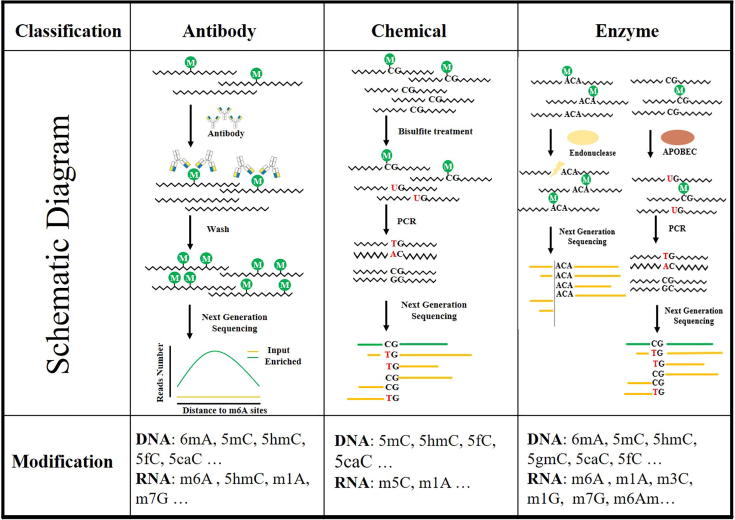
Fig. 2.
Schematic diagram of mapping methods for nucleic acid modifications. Various methods could be roughly divided into three types according to different mechanisms: antibody enrichment [28], chemical reaction [36], [37], [50] and enzyme recognition [43], [44]. The modification signal can be captured by preprocessing using one or combined strategies then resolved by sequencing.
Strategy 1: antibody enrichment. Well-characterized antibody usually has high specificity and affinity to bind DNA or RNA fragments with specific modification, thus will be suitable for the following pull-down experiment. After immunoprecipitation, the enriched fragments are supposed to have much more modifications than the background sequences (input). By high-throughput sequencing and bioinformatic comparing of fragments from the enriched portion versus the background portion, the modification sites can be estimated to locate in the “peak” region, where the enriched sequence reads are significantly more than the background sequence reads. These enrichment-based methods have been widely used for the initial profiling of various modification, including 5mC in DNA (MeDIP-seq) [32] and m6A in RNA (MeRIP-seq) [33]. However, these methods are limited by the low resolution that can only locate the modification to a range of ~100 nt length, but not the exact site. More complicated, sometimes it is the stoichiometry but not the existence determines the function [10], [27], however, the IP-based methods are powerless to detect the modification intensity in different conditions. To solve these problems, researchers further optimized the IP-based detection methods, trying to achieve better precision. For example, PA‐m6A‐seq increases the resolution of m6A signature from ~100 nt to ~30 nt by introducing exonuclease trimming [34]; the miCLIP and m6ACE-seq exploit photo-crosslinking to generate mutation or truncation near m6A site during reverse transcription [34], [35].
Strategy 2: chemical reaction. According to some unique characteristics of modification, certain chemical reaction can be specifically applied to modified or unmodified bases. During PCR or reverse transcription, the chemically reacted/labeled positions will cause polymerase or reverse-transcriptase (RT) stop, or introduce nucleotide transition, thus can be derived by the following sequencing. The most successful and extensive application of this strategy in modification mapping might be bisulfite sequencing (BS-seq) for 5mC [36], [37]. After treatment by the chemical reagent bisulfite, the cytosine residues will be converted to uracil that read as T, but leave 5mC unaffected. Another example is the PSI-seq for pseudouridine (Ψ), which uses N-Cyclohexyl-N′-(2-morpholinoethyl)carbodiimide (CMC) to selectively modify pseudouridines and generate RT stop [38]. Similar to antibody enrichment strategy, chemistry-mediated pull-down can also be applied to enrich the modified DNA or RNA fragments. Take m7G-seq method for example, it first converts m7G into abasic site, then label the site with biotin to facilitate pulldown enrichment [39]. The disadvantage of chemical reaction is also obvious: the chemical treatment is usually harsh and tend to destroy the chain of nucleic acid. And the specificity is sometimes a problem since the difference between modified and regular bases is not sufficient enough for the chemical reagent to distinguish and react specifically.
Strategy 3: enzyme recognition. Some naturally occurring enzymes recognize modification to function in vivo. For instance, restriction enzymes belonging to the restriction-modification system (RM system) exploit modification as the signature to distinguish native DNA and foreign invaders. Taking this advantage, methods to map 5mC and 6mA have been well established based on certain modification-sensitive restriction enzyme [40], [41]. Similarly, by screening the endonucleases, researchers found the MazF and ChpBK enzyme belonging to the toxin-antitoxin (TA) system, can be blocked by m6A, but cut the RNA with unmodified sequence motif [42]. The m6A-REF-seq/MASTER-seq method leveraged this feature to resolve the m6A map in single-base resolution [43], [44]. Recently developed bisulfite-free method, EM-seq, utilizes TET and βGT to oxidize 5mC and 5hmC to 5gmC, then uses deaminase APOBEC next to deaminate unmodified C to U [45]. TET and βGT enzymes are also used in TAB-seq method which maps 5hmC in single-base resolution across the genome [46]. Another method for 5hmC mapping, ACE-seq, uses deaminase APOBEC to convert unmodified C and 5mC to U, leaving 5hmC intact by βGT treatment [47]. After comparing converted versus unconverted sequences, the 5hmC signature can be derived by bioinformatic analysis. Comparing to chemical-based methods, enzymes usually have higher specificity and sensitivity, but depend on the searching of appropriate enzyme in nature and the innate characters of the tool protein. Directed evolution may accelerate the searching process or even creates novel enzyme with enhanced activity.
It is worth noting that the 3rd-generaton sequencing provides an alternative but promising strategy to map DNA and RNA modifications that modified bases could be directly detected while base calling. Nevertheless, due to high error rate and limited discrimination of modified bases, this method is waiting to be practicable in future research [48].
These above-mentioned methods are usually joint used or integrated to supplement their respective advances and achieve more reliable results. For example, the newly developed method TAPS utilize enzyme TET to oxidize 5mC to 5caC first, then chemically reduce 5caC to dihydrouracil (DHU) with pyridine borane. The DHU will be readout as T by DNA polymerase [49]. Ideally, researchers expect a method can map the modifications in single-base resolution, and quantify the stoichiometry of the modified base versus unmodified at each site. In addition, this method should detect every modification locus across the genome, but needs minimal input material, or even works on single cell. To achieve this, innovative method is still needed.
4. Tools to edit nucleotide modifications
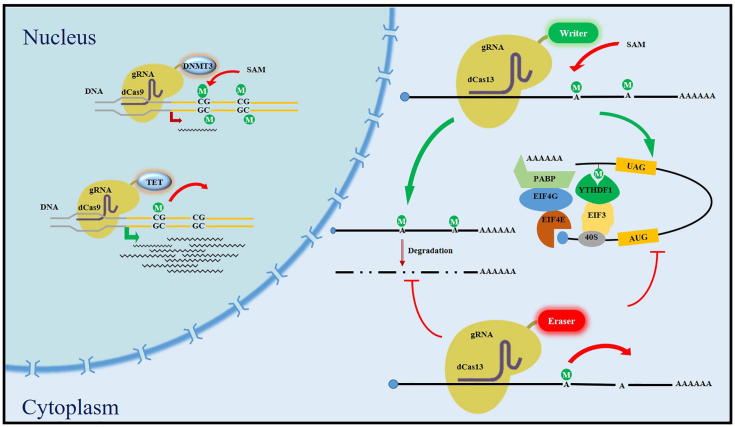
Though massive researches have been conducted to show the significance of DNA or RNA modifications by perturbing the effector proteins which elevate or suppress the global modification level, direct evidence elucidating the function of site-specific modification is still lacking. Meanwhile, safe and effective tools for modification editing are considered of great promise in clinical applications. Therefore, new tools and technologies targeting specific genomic region with the capacity to edit the modification status are urgently needed [14], [51], [52]. Along with the advancement of CRISPR-Cas system in genetic editing, researches attempted to transform the Cas protein for epigenetic editing purpose. Shortly after the application of Cas9 in human genome editing, several groups succeeded to edit the DNA methylation by adapting Cas9 protein in various systems such as cancers and embryonic stem cells [14], [53]. They combined the 5mC methyltransferase DNMT3A with dCas9 (a mutated Cas9 without endonuclease activity), making it a reality to add 5mC modification to the target site directed by guide RNA [53]. They proved that targeted de novo methylation of a CTCF loop anchor site by DNMT3A-dCas9 blocked CTCF binding and interfered with DNA looping, resulting in regulation of gene expression in the neighboring region. In order to enhance the editing efficiency, dCas9 was fused to repetitive peptide epitopes (SunTag), which recruited multiple copies of antibody-fused DNMT3A to amplify the local concentration of effector and methylate the target region [51]. The targeted demethylation was carried out by the effector protein TET, which oxidized 5mC and then the products were recognized by the endogenous DNA-repair mechanism and substituted with regular cytosine [54], [55]. It is worth noting that the DNA methylation is tightly associated with other epigenetic signals, such as histone modifications and chromatin architecture, thus editing the DNA modification will ultimately remodel the transcription environment and potentially control gene expression [56].
Comparing to DNA, RNA is more dynamic and structured. To efficiently adding or removing modifications to RNA, advanced tools are needed. With the continuous understanding and improvement of dCas9 system, an antisense oligonucleotide (PAMer) was supplied to assist dCas9 protein targeting RNA and edited the related modifications by the similar strategy working on DNA [57], [58]. By fusing the dCas9 to m6A writer proteins, Liu et al. developed a molecular tool to target specific sequence on transcript and modulate m6A modification to desired site. They showed that the directed modification on the 5′ UTR of Hsp70 mRNA promotes translation efficiency under stress conditions [59]. The guide RNA directed site-specific demethylation was also realized by tethering dCas9 to the m6A eraser FTO [57], [60]. Meanwhile, the discovery of Cas13 family proteins intrinsically targeting RNA provided new opportunity. Tethering catalytically inactive Cas13b (dCas13b) and effector protein, Li et al. had successfully edit the m6A modification on specific genes and tune the mRNA stability through demethylation (Fig. 3) [61].
Fig. 3.
Schematic diagram of CRISPR-based DNA and RNA modification editing system. Inside the nucleus, DNA modification 5mC can be added or removed by dCas9 fused effectors DNMT3 (writer) or TET (eraser), resulting in transcription regulation. In the cytoplasm, RNA modification m6A can be deposited or removed by dCas13 fused effector proteins, leading to modification mediated post-transcriptional regulation, such as mRNA degradation or translational repression via the reader proteins.
Though the CRISPR-Cas system is currently the most widely used tool to target specific DNA or RNA sequence, other programable tools are also being developed. Cpf1 is another RNA-guided endonuclease which had been widely used in genome editing but yet to be accommodated for modification editing [62]. Engineered zinc finger (ZF) and transcription activator-like effectors (TALEs) system can be linked to epigenetic effectors to exert the modification manipulation purpose as well [63]. Taking advantage of endogenous RNA binding proteins, the CRISPR-Cas-inspired RNA targeting system (CIRTS) uses engineered fusion protein composed of ssRNA binding protein, RNA hairpin binding protein and effector protein to target specific RNA sequence by the assistant of a guide RNA [64]. This tool is built entirely from human protein parts, and potentially avoids immune issues when applied to diseases therapies. In addition, PUF protein is another choice to target RNA, which can be modulated to bind specific nucleotides by substituting the amino acids in its particular structural repeats [65].
5. Prospective and deficiency
As an additional layer of information beyond sequences, DNA and RNA modifications emerge to revolutionize our comprehension of gene regulation, while determining the causal relationship remain a big challenge. To interrogate the mechanism behind, mapping the exact position of each type of modifications across the genome would be the first step. With the rapid development of novel technologies, especially the high-throughput sequencing facilitated methods combined with other biological or chemical approaches, mapping the main types of modifications such as 5mC and m6A is not a barrier any more, however, given that ~150 RNA modifications and ~100 DNA modifications in total, it is still a great challenge to map them precisely in the genome-wide [34], [35]. As our understanding of the modifications deepens, editing tools are desired to alter the modification status of a given target, correcting the falsely modified base in disease or interfering the regular modification in basic research. In turn, the directed targeting of DNA and RNA modifications provides new tool to study the mechanism and function of regional modification on a specific gene. For example, Liu et al. demonstrated that artificially depositing m6A on the 3′ UTR of Actb mRNA resulted in fast degradation, whereas editing the 5′ UTR of the same gene did not show such effect [57]. Although previous studies based on transcriptome-wide bioinformatic analysis had already proposed that the function of m6A modification was region-dependent, this work provided direct evidence for the first time supporting the hypothesis. In this review, we mainly take 5mC m6A as examples to introduce the relevant tools and technologies, since these two modifications are most well-documented in DNA and RNA. Nevertheless, the relevant methods described above can be easily adapted and applied to other modifications, as long as the effector proteins responsible to that specific modification had been characterized.
The advancement of these tools also provides brand new choices for basic and clinical researches. For example, numerous studies using epigenetic editing tools have been conducted in the complex diseases, especially cancers [14]. A dCas9-TET1CD fusion protein was created to modulate the activity of tumor-suppressor genes by demethylating the target regions [54]. In addition, these kinds of tools had been used to explore the mechanisms by which tumor cells hijack enhancers by dysregulating MYC, upregulate homeobox oncogenes via site-specific hypomethylation, or facilitate tumor initiation by preventing senescence entry via hit-and-run epigenetic events [66]. With fast development of these manipulating technologies, the application scope has largely expanded to explore novel treatment strategy for more diverse diseases. For example, Kantoret et al. proposed a new approach to treat Parkinson’s disease through dCas9-DNMT3A system, which established the hypermethylation pattern in first intron of SNCA, with the goal of repressing its expression [67]. Similar studies focusing on the treatment of neurodegenerative diseases and muscular dystrophy had been reported as well [68].
On the other hand, it is undeniable that these tools may inherit the innate shortcomings of CRISPR system. The greatest concern is off-target effect, which introduces unintended genetic or epigenetic changes at inappropriate locations [69], [70]. Substantial efforts had been undertaken to estimate and minimize the mistakes in different systems. According to recent studies, off-target effect varies in different species in which the tool applied and the subtype of CRISPR system [71], [72]. These studies proposed several possible ways to avoid this problem: 1. Modulate the effective concentration and delivery of sgRNA-Cas9 complexes; 2. Rational design and modify the sgRNA molecules; 3. Screen for new variants of CRISPR proteins or engineer the current version [73]. Benefit from the fast development of genome editing technologies, the DNA/RNA modification editing tools based on reconstructed CRISPR-Cas systems would undoubtedly keep improving in parallel. However, the introduction of effector protein might generate extra off-targeting. In a single-base editor system, the main source of the off-target effect does not come from CRISPR system, but from the effector protein deaminase: high activity of the enzyme results in excessive modification all through the genome, failing to resolve the signals of the intended loci from background [74]. Therefore, careful evaluation of the effector protein activity is highly recommended before designing an editing tool for DNA/RNA modifications. Rational adjustment or even protein evolution of the effector would be necessary in some cases.
The new technologies have armed us to better depict and understand the appearance, mechanics and functions of nucleic acids modifications. Intriguingly, the advancement of this field will very likely to make great contribution to both fundamental and translational researches in the near future.
Author contribution
Li-Qian Chen, Wen-Shuo Zhao and Guan-Zheng Luo wrote the manuscript. All authors reviewed the writing and approved the final version of the manuscript. Guan-Zheng Luo provided funding.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant nos. 31922015, 31622031, 91753129) and Natural Science Foundation of Guangdong Province (Grant no. 2018B030306044).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.03.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fu Y., He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol. 2012;16:516–524. doi: 10.1016/j.cbpa.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K., Zhao B.S., He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 5.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2012;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas S., Rao C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur J Pharmacol. 2018 doi: 10.1016/j.ejphar.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 7.Barber B.A., Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat – Anat Anzeiger. 2010;192:261–274. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Chinnusamy V., Mohapatra T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front Genet. 2018:9. doi: 10.3389/fgene.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 12.Chen K., Luo G.Z., He C. 1st ed. Elsevier Inc.; 2015. High-Resolution Mapping of N6-Methyladenosine in Transcriptome and Genome Using a Photo-Crosslinking-Assisted Strategy; p. 560. [DOI] [PubMed] [Google Scholar]
- 13.Churko J.M., Mantalas G.L., Snyder M.P., Wu J.C. Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ Res. 2013;112:1613–1623. doi: 10.1161/CIRCRESAHA.113.300939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urbano, Smith, Weeks, Chatterjee Gene-specific targeting of DNA methylation in the mammalian genome. Cancers (Basel) 2019;11:1515. doi: 10.3390/cancers11101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Wang L., Zou X., Duan S., Li Z., Deng Z. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol Adv. 2019;37:708–729. doi: 10.1016/j.biotechadv.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Robertson K.D. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 17.Li E., Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a019133. a019133–a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breiling A., Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenet Chromatin. 2015;8:24. doi: 10.1186/s13072-015-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell J.L., Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 20.Messer W., Noyer-Weidner M. Timing and targeting: The biological functions of Dam methylation in E. coli. Cell. 1988;54:735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 21.Ratel D., Ravanat J.L., Berger F., Wion D. N6-methyladenine: The other methylated base of DNA. BioEssays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casadesus J., Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray N.E. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- 24.Wion D., Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olynik B.M., Rastegar M. The genetic and epigenetic journey of embryonic stem cells into mature neural cells. Front Genet. 2012:3. doi: 10.3389/fgene.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 28.Feederle R., Schepers A. Antibodies specific for nucleic acid modifications. RNA Biol. 2017;14:1089–1098. doi: 10.1080/15476286.2017.1295905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty B.K., Kushner S.R. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019;363:eaav0080. doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 31.Mongan N.P., Emes R.D., Archer N. Detection and analysis of RNA methylation. F1000Research. 2019;8:559. doi: 10.12688/f1000research.17956.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohn F., Weber M., Schübeler D., Roloff T.-C. Methylated DNA Immunoprecipitation (MeDIP) Methods Mol Biol. 2009:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 33.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 34.Chen K., Lu Z., Wang X., Fu Y., Luo G.-Z., Liu N. High-resolution N6-Methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew Chem Int Ed. 2015;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu Y., Luo G.Z., Chen K., Deng X., Yu M., Han D. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Tollefsbol T.O. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol. 2011:11–21. doi: 10.1007/978-1-61779-316-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovejoy A.F., Riordan D.P., Brown P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enroth C., Poulsen L.D., Iversen S., Kirpekar F., Albrechtsen A., Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz736. e126-e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin S.-G., Kadam S., Pfeifer G.P. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq223. e125-e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G.-Z., Wang F., Weng X., Chen K., Hao Z., Yu M. Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat Commun. 2016;7:11301. doi: 10.1038/ncomms11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imanishi M., Tsuji S., Suda A., Futaki S. Detection of N 6 -methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem Commun. 2017;53:12930–12933. doi: 10.1039/c7cc07699a. [DOI] [PubMed] [Google Scholar]
- 43.Badian N.A. Reading disability in an epidemiological context incidence and environmental correlates. J Learn Disabil. 1984;17:129–136. doi: 10.1177/002221948401700301. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Campos M.A., Edelheit S., Toth U., Safra M., Shachar R., Viukov S. Deciphering the “m6A Code” via antibody-independent quantitative profiling. Cell. 2019:1–17. doi: 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Vaisvila R. EM-seq: detection of DNA methylation at single base resolution from picograms of DNA. J Chem Inf Model. 2019;53:1689–1699. [Google Scholar]
- 46.Yu M., Han D., Hon G.C., He C. Tet-assisted bisulfite sequencing (TAB-seq) Methods Mol Biol. 2018:645–663. doi: 10.1007/978-1-4939-7481-8_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutsky E.K., Denizio J.E., Hu P., Liu M.Y., Nabel C.S., Fabyanic E.B. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat Biotechnol. 2018;36:1083–1090. doi: 10.1038/nbt.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardui S., Ameur A., Vermeesch J.R., Hestand M.S. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46:2159–2168. doi: 10.1093/nar/gky066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Siejka-Zielińska P., Velikova G., Bi Y., Yuan F., Tomkova M. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat Biotechnol. 2019;37:424–429. doi: 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- 50.Boschi-Muller S., Motorin Y. Chemistry enters nucleic acids biology: enzymatic mechanisms of RNA modification. Biochem. 2013;78:1392–1404. doi: 10.1134/S0006297913130026. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y.-H., Su J., Lei Y., Brunetti L., Gundry M.C., Zhang X. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 2017;18:176. doi: 10.1186/s13059-017-1306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molla K.A., Yang Y. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019 doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choudhury S.R., Cui Y., Lubecka K., Stefanska B., Irudayaraj J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget. 2016;7. doi: 10.18632/oncotarget.10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhutani N., Burns D.M., Blau H.M. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillemer E., Whitlock C., Weissman I.L. Transformation-associated proteins in murine B-cell lymphomas that are distinct from Abelson virus gene products. Proc Natl Acad Sci U S A. 1984;81:4434–4438. doi: 10.1073/pnas.81.14.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X.-M., Zhou J., Mao Y., Ji Q., Qian S.-B. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. 2019 doi: 10.1038/s41589-019-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei J., He C. Site-specific m6A editing. Nat Chem Biol. 2019;15:848–849. doi: 10.1038/s41589-019-0349-8. [DOI] [PubMed] [Google Scholar]
- 59.Tsuchida Y., Kaneko Y., Kanda N., Makino S., Utakoji T., Saito S. Possible relationship of chromosome abnormalities and gene amplification with effects of chemotherapy: a neuroblastoma xenograft study. Prog Clin Biol Res. 1985;175:171–180. [PubMed] [Google Scholar]
- 60.Rau K., Rösner L., Rentmeister A. Sequence-specific m 6 A demethylation in RNA by FTO fused to RCas9. RNA. 2019;25:1311–1323. doi: 10.1261/rna.070706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Chen Z., Chen F., Ling Y., Peng Y., Luo N. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. BioRxiv. 2019 doi: 10.1093/nar/gkaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015 doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konermann S., Brigham M.D., Trevino A.E., Hsu P.D., Heidenreich M., Cong Le. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rauch S., He E., Srienc M., Zhou H., Zhang Z., Dickinson B.C. Programmable RNA-guided RNA effector proteins built from human parts. Cell. 2019;178 doi: 10.1016/j.cell.2019.05.049. 122–134.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinoda K., Suda A., Otonari K., Futaki S., Imanishi M. Programmable RNA methylation and demethylation using PUF RNA binding proteins. Chem Commun. 2020;56:1365–1368. doi: 10.1039/c9cc09298f. [DOI] [PubMed] [Google Scholar]
- 66.Schuijers J., Manteiga J.C., Weintraub A.S., Day D.S., Zamudio A.V., Hnisz D. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep. 2018;23:349–360. doi: 10.1016/j.celrep.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kantor B., Tagliafierro L., Gu J., Zamora M.E., Ilich E., Grenier C. Downregulation of SNCA expression by targeted editing of DNA methylation: a potential strategy for precision therapy in PD. Mol Ther. 2018;26:2638–2649. doi: 10.1016/j.ymthe.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young A.B. Four decades of neurodegenerative disease research: how far we have come! J Neurosci. 2009;29:12722–12728. doi: 10.1523/JNEUROSCI.3767-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 70.Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther – Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J., Manghwar H., Sun L., Wang P., Wang G., Sheng H. Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR Cas9-edited cotton plants. Plant Biotechnol J. 2019;17:858–868. doi: 10.1111/pbi.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tycko J., Myer V.E., Hsu P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell. 2016;63:355–370. doi: 10.1016/j.molcel.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019:eaaw7166. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.