Highlights
-
•
The ferret model is used to study human disease and physiology.
-
•
TaqMan realtime RT-PCR assays for ferret cytokine and chemokine mRNA were developed.
-
•
Cytokine and chemokine patterns in ferret cells were similar to other mammals.
-
•
A comprehensive panel of mRNAs can be measured in samples of limited quantity.
Keywords: Ferret, Real time RT-PCR, Cytokine, Gene, TaqMan
Abstract
The ferret is an excellent model for many human infectious diseases including influenza, SARS-CoV, henipavirus and pneumococcal infections. The ferret is also used to study cystic fibrosis and various cancers, as well as reproductive biology and physiology. However, the range of reagents available to measure the ferret immune response is very limited. To address this deficiency, high-throughput real time RT-PCR TaqMan assays were developed to measure the expression of fifteen immune mediators associated with the innate and adaptive immune responses (IFNα, IFNβ, IFNγ, IL1α, IL1β, IL2, IL4, IL6, IL8, IL10, IL12p40, IL17, Granzyme A, MCP1, TNFα), as well as four endogenous housekeeping genes (ATF4, HPRT, GAPDH, L32). These assays have been optimized to maximize reaction efficiency, reduce the amount of sample required (down to 1 ng RNA per real time RT-PCR reaction) and to select the most appropriate housekeeping genes. Using these assays, the expression of each of the tested genes could be detected in ferret lymph node cells stimulated with mitogens or infected with influenza virus in vitro. These new tools will allow a more comprehensive analysis of the ferret immune responses following infection or in other disease states.
1. Introduction
Ferrets are an outbred population widely used to study influenza virus infection (Belser et al., 2011, Laurie et al., 2010, Rockman et al., 2012, Hurt et al., 2010) as well as a range of other diseases, including SARS-coronavirus (CoV) (reviewed in (Roberts et al., 2008)) and henipaviruses, such as infection with Hendra virus and Nipah virus (Bossart et al., 2009, Pallister et al., 2009, Pallister et al., 2011, Geisbert et al., 2012). The anatomical and physiological similarity between human and ferret lungs also enables ferrets to be used as a model to study lung carcinomas (reviewed in Baric et al., 2013). Recently, absence of the cystic fibrosis transmembrane conductance regulator (CFTR) was associated with spontaneous disease induction in the lung and pancreas in ferrets, showing similar pathology to that of cystic fibrosis in humans (Sun et al., 2010, reviewed in Keiser and Engelhardt, 2011). The broad utility of this model is highlighted by the use of ferrets to study pneumococcal transmission, reproductive biology and human fetal brain development (reviewed in Baric et al., 2013).
While the ferret is a good model for human respiratory virus infections, reagents to identify ferret leukocytes and immune mediators are limited. Studies have identified cross-reactive antibodies that recognize populations of ferret leukocytes, such as CD8, CD11β, CD44 and CD25, and cytokines IFNγ, TNFα, IL4 and IL8 (Rutigliano et al., 2008, Martel and Aasted, 2009, Pillet et al., 2011). Cloning and sequencing of ferret cytokine genes have enabled molecular approaches targeting the corresponding mRNAs (von Messling et al., 2006, Danesh et al., 2008, Nakata et al., 2008, Ochi et al., 2008, Qin et al., 2013). Expression of cytokine and chemokine genes has been assessed in ex vivo samples following infection of naïve or vaccinated ferrets with influenza virus or SARS-CoV by microarray analysis (Cameron et al., 2008, Fang et al., 2010, Rowe et al., 2010). Cytokine and chemokine gene profiles have also been assessed ex vivo and in in vitro epithelial cultures using SYBR green real time RT-PCR assays (Svitek and von Messling, 2007, Cameron et al., 2008, Danesh et al., 2008, Danesh et al., 2011, Svitek et al., 2008, Kim et al., 2009, Fang et al., 2010, Hamelin et al., 2010, Kobinger et al., 2010, Rowe et al., 2010, Kang et al., 2011, Meunier and von Messling, 2011, Meunier and von Messling, 2012, Pillet et al., 2011, Huang et al., 2012, Maines et al., 2012, Meunier et al., 2012, Belser et al., 2013, Zeng et al., 2013). TaqMan chemistry incorporates target-specific fluorescent labeled probes enabling multiple genes can be assessed in a single real time PCR reaction (Giulietti et al., 2001). To date, TaqMan real time RT-PCR assays have only been developed for a smaller number of ferret-specific gene targets (Nakata et al., 2009, Suguitan et al., 2012).
To enable a broader characterization of the immune response in the ferret model, we developed a panel of TaqMan assays to detect mRNA of fifteen ferret cytokines, chemokines and immune mediators (IFNα, IFNβ, IFNγ, IL1α, IL1β, IL2, IL4, IL6, IL8, IL10, IL12p40, IL17, granzyme A, MCP-1, TNFα) and four housekeeping genes (ATF4, GAPDH, L32 and HPRT). The cytokine and chemokine profile induced by stimulation of ferret leukocytes with mitogens or influenza virus was also assessed to investigate the relevance of the ferret immune response to human infection studies.
2. Materials and methods
2.1. Design of ferret cytokine and housekeeping gene primers and probes
Sequences for cytokine, chemokine and housekeeping genes of multiple species were obtained from Genbank (http://www.ncbi.nlm.nih.gov/genbank) and aligned. Regions of conservation were identified and primers were designed using PrimerSelect (DNASTAR Lasergene8, Madison, USA) or PrimerExpress (Applied Biosystems, California, USA) to amplify the region from ferret cDNA. Cloned genes were sequenced and TaqMan real time PCR primers and probes designed using PrimerExpress. All oligonucleotide primers and probes used in this study, including those previously published, are listed in Table 1 . Primers for IFNα were designed to amplify multiple subtypes (1–12) (Easlick et al., 2010, Hillyer et al., 2012).
Table 1.
Oligonucleotide primer and probe sequences used in this study. Primers and probes developed for the TaqMan real time RT-PCR assay in this study are highlighted in bold italics. Primers (and probes) from other published real time RT-PCR studies, TaqMan and SYBR Green, are indicated. Primers used to clone inserts for plasmid controls are also indicated, and referenced as appropriate.
| Gene target | Forward primer 5′ → 3′ | Reverse primer 5′ → 3′ | Probe | Use | Reference |
|---|---|---|---|---|---|
| IL1α | CTGAAACCTCAAAGACATCTCATCTT | GCTGGCTGCCACCATCA | FAM-CCTTCAAGGAGGATGTG- MGBNFQ | TaqMan | This study |
| IL1α | TTCTCTGAATCAGAAATCCTTCTATGAT | CTCTTACAAAGAGAGTAAACATTCATTTGG | Cloning | This study | |
| IL1β | CCTGGTGCTGTATAACTCGTATGAG | TTGGTTCACACTAGTTCCGTTGA | VIC-TCGGGCGCTCCAC- MGBNFQ | TaqMan | Reverse primer matches (Fang et al., 2010, Rowe et al., 2010) |
| IL1β | AGATGAAGTGCTGCTTCCAAGAC | GTGCTGATGTACCAGTTGGGAAA | Cloning | This study | |
| IL1β | GGACTGCAAATTCCAGGACATAA | TTGGTTCACACTAGTTCCGTTGA | SYBR green | Fang et al. (2010) and Rowe et al. (2010) | |
| IL2 | GTTAAAAATTATGAGAGCCCCAGGA | TTGAGTTCTTCTGCTAGACATTGAAGA | FAM-CTACATGCCCAAGAAG-MGBNFQ | TaqMan | This study |
| IL2 | GCAACTCTTGTCTTGCATCGTAC | TCAAGTCAGTGTTGAGAAGATGC | Cloning | This study | |
| IL2 | TGCTGCTGGACTTACAGTTGCTCT | CAATTCTGTGGCCTTCTTGGGCAT | SYBR green | Svitek and von Messling (2007) | |
| IL4 | CCAACAGATTGCTCAGAGGACTT | CACCGAACAGGTCATGTTTGC | 6FAM-CAGGAACCTCAGGAACAT-MGBNFQ | TaqMan assay | This study |
| IL4 | GATCTATTAATGGGTCTCACCTC | CAGCTTCAATGCCTGTAGTATTTCTG | Cloning | This study | |
| IL4 | CGTTGAACATCCTCACAGCGAGAAAC | TTGCCATGTTCCTGAGGTTCCTGTGA | SYBR green | Svitek and von Messling (2007) | |
| IL4 | TCACCGGCACTTTCATCCACGGACATAACTT | GAGCTGCTGAAGCACAGTTGCAGCTCTGC | SYBR green | Maines et al. (2012) | |
| IL4 | TCACCGGCACTTTCATCCA | TTCTCGCTGTGAGGATGTTCA | SYBR green | Fang et al. (2010) | |
| IL6 | GCAGAGAACAACCTAAATCTTCCAA | TGATTGAATTGAGACTGGAAGCA | 6FAM-CTGGCAGAAGAGGAC-MGBNFQ | TaqMan | This study |
| IL6 | CAAATGTGAAGACAGCAAGGAGGCA | TCTGAAACTCCTGAAGACCGGTAGTG | SYBR green, cloning | Svitek and von Messling (2007) | |
| IL6 | CAAGTGGCTGAAACACGTAACAA | GGCTGAACTGCAGGAAATCC | FAM-TCACCTCATCCTACGGAGCCTTG-TAMRA | TaqMan | Nakata et al. (2009) |
| IL8 | GGCACCTTGCATCAACATGA | AAGCAGGAAAACTGCCAAGAGA | 6FAM-TTCCAAGCTGGCTGTTG-MGBNFQ | TaqMan | This study |
| IL8 | CAAGAGCCAGGAAGAAACCAGACC | TGATTCTTGGATACCACAGAGAATG | Cloning | This study | |
| IL8 | AACCCACTCCACGCCTTTCCATC | GGCACACCTCTTTTCCATTGAC | SYBR green | Maines et al. (2012) | |
| IL10 | GCTGCGGCGCTGTCA | CTCCACCGCCTTGCTCTTAT | VIC-CGATTTCTGCCCTGTGAG-MGBNFQ | TaqMan | This study |
| IL10 | CCTTCGGCAGGGTGAAGACTTTC | ATGTCAAACTCACTCATGGCTTTGTAG | Cloning | This study | |
| IL10 | TCCTTGCTGGAGGACTTTAAGGGT | TCCACCGCCTTGCTCTTATTCTCA | SYBR green | Svitek and von Messling (2007) | |
| IL10 | CGAGAACCACGACCCAGAA | CCGCAGGGTCTTCAGCTTT | FAM-TCAAGGAGCACGTGAACTCGCTGG-TAMRA | TaqMan | Nakata et al. (2009) |
| IL12p40 | GGTGCTATTCACAAGCTCAAGTATG | GGTTTGATGATGTCCCTGATGA | VIC- TACACCAGCAGCTTC-MGBNFQ | TaqMan, cloning | This study |
| IL12p40 | ATCGAGGTTGTGGTGGGTGCTATT | TAGGTTCATGGGTGGGTCTGGTTT | SYBR green | Svitek and von Messling (2007) | |
| IL17 | GGACGGTAAACTACCACATGAACTC | AGACTCCCTTCGCAGAACCA | VIC-TCCCCATCCAGCAAGA-MGBNFQ | TaqMan | This study |
| IL17 | CGCAATGAGGACCCTGAGAGA | ggtgacacaggtgcagcccac | Cloning | This study | |
| Granzyme A | GGATCCTCCCTCTCCCTAAGAA | CCCAGCCTGCAACTTGACA | VIC-ATGATGTCAAACCCGAAAC-MGBNFQ | TaqMan | This study |
| Granzyme A | ACTGGGTGTTGACTGCAGCTC | ACATATCAGAGGGCTTCCAGAATC | Cloning | This study | |
| MCP1 | GCAGCAAGTGTCCCAAAGAAG | GACTGGGGTCAGCGCAGAT | FAM-ATCCTCAAGACATTCCT-MGBNFQ | TaqMan | This study |
| MCP1 | GCAGCAAGTGTCCCAAAGAAG | gctttgcagtttgggtttgg | Cloning b | This study | |
| CCL2 (MCP1) | GCTCCCTATTCACTTGCTGTTTC | GATTCGATAGCCCTCCAGCTT | SYBR green | Rowe et al. (2010) | |
| TNFα | TGCCATCAGACGGGCTGTA | ACATCCTCGGCCCTTGAAG | VIC-CTTATCTACTCGCAGGTCC-MGBNFQ | TaqMan | This study |
| TNFα | TGGAGCTGACAGACAACCAGCTAA | TGATGGTGTGGGTAAGGAGCACAT | SYBR green, cloning | Svitek and von Messling (2007) | |
| TNFα | CCAGATGGCCTCCAACTAATCA | GGCTTGTCACTTGGAGTTCGA | SYBR green | Fang et al. (2010) and Rowe et al. (2010) | |
| TNFα | ATGTTGTAGCAAACCCTGAAGCT | ATTGGCCAGGAGGGCATT | FAM-ACTCCAATGGCTGAGCCGACGTG-TAMRA | TaqMan | Nakata et al. (2009) |
| IFNαa | TCCATCCTGAGGAACTACTTCCAG | AGGCACAAGGGCTGTATTGC | 6FAM-GAATCTCCCTCTATCTGC-MGBNFQ | TaqMan | This study |
| IFNαa | CTCAGCWGCCACTMCCTC | CATGATCTCTGCTCGGACCAT | Cloning | This study | |
| IFNα | ATGCTCCTGCGACAAATGAGGAGA | TTCTGCAGCTGCTTGCTGTCAAAC | SYBR green | Svitek and von Messling (2007) | |
| IFNβ | ATATTTCTCCACCACGGTTCTTG | ACTCCACACTGCTGCTGCTTAG | VIC-AACTATAACTTACTTCGATTCCA-MGBNFQ | TaqMan | This study |
| IFNβ | ATGACCAGCAGGTGTATCCTCC | AAAAGCTCCTGAGGATTTCTGCT | Cloning | This study | |
| IFNβ | GGTGTATCCTCCAAACTGCTCTCC | CACTCCACACTGCTGCTGCTTAG | SYBR green | Maines et al. (2012) | |
| IFNγ | AACTGGAGAGAGGAGAGTGACAAAA | GTCTTCCTTGATGGTATCCATGC | VIC-TCTCCTTCTACTTGAAACTGT-MGBNFQ | TaqMan | This study |
| IFNγ | ATGAATTATACAACTATATGCTTAG | TTATTTCGATGCTCTGCGGCCTCG | Cloning | This study | |
| IFNγ | CCATCAAGGAAGACATGCTTGTCAGG | CTGGACCTGCAGATCATTCACAGGAA | SYBR green | Svitek and von Messling (2007) | |
| IFNγ | TGGTGGGCCTCTTTTCTTAGATAT | AGAAGGAGACAATTTGGCTTTGA | FAM-TTGAAGAACTGGAGAGAGGAGAGTGACAAAAAAA-TAMRA | TaqMan | Nakata et al. (2009) |
| Matrix | GACCRATCCTGTCACCTCTGAC | GGGCATTYTGGACAAAKCGTCTACG | 6FAM-TGCAGTCCTCGCTCACTGGGCACG-MGBNFQ | TaqMan | CDCc |
| Matrixd | TGTAAAACGACGGCCAGTAGCAAAAGCAGGTAG | CAGGAAACAGCTATGACCAGTAGAAACAAGGTAGT | Cloning | CDCc | |
| ATF4 | TTTACCTTCCTGCAACCACTTC | TCATGGTAATGTAAGCAGTAGAGTC | NED-CTGTCCTCCACTCCAGATCATTCCT-MGBNFQ | TaqMan, cloning | Bruder et al. (2010) |
| HPRT | CACTGGGAAAACAATGCAGA | ACAAAGTCAGGTTTATAGCCAACA | NED-TGCTGGTGAAgAGGACCCCTCG-MGBNFQ | TaqMan, cloning | Peters et al. (2007) e |
| L32 | TGGTTATAGGAGCAACAAGAAA | GCACATCAGCAGCACTTCA | NED-TGTTGCCCAGTGGCTTCTGG-MGBNFQ | TaqMan, cloning | Adapted from Peters et al. (2007) e |
| GAPDH | TGCGGCCAAGGCAGTAG | AGGCCATGCCAGTGAGCTT | VIC-CTGAGCTGAATGGGAAG-MGBNFQ | TaqMan | This study |
| GAPDH | AACATCATCCCTGCTTCCACTGGT | TGTTGAAGTCGCAGGAGACAACCT | SYBR green, cloning | Svitek and von Messling (2007) | |
| GAPDH | TTGCTGACAATCTTGAGGGAGTT | CTGCTGATGCCCCCATGT | FAM-TCATACTTCTCATGGTTCACACCCATCACG-TAMRA | TaqMan | Nakata et al. (2009) |
Targeted to all IFNα subtypes (Easlick et al., 2010, Hillyer et al., 2012).
Note same forward primer as TaqMan assay.
Primers obtained from the Centers for Disease Control, Atlanta, USA through WHO (http://www.who.int/csr/resources/publications/swineflu/sequencing_primers/en/index.html).
M13 Tag underlined.
Nucleotide changes to sequences underlined.
2.2. Oligonucleotide primer and probe generation
Lyophilized oligonucleotide primers were synthesized by Geneworks (Adelaide, Australia) and dissolved in nuclease-free water (Promega, Madison, USA) at 100 μM. All TaqMan® MGB™ probes were synthesized by Applied Biosystems with a 5′reporter dye (either FAM, NED or VIC) and a 3′non-fluorescent quencher (NFQ).
2.3. Ferrets
Adult male and female ferrets (weight 500–1500 g) were purchased from independent breeders and housed at CSL Limited (Victoria, Australia) using services provided under a Support Services Agreement. Serum samples were tested by hemagglutination inhibition assay to ensure seronegativity (titer <10) to currently circulating influenza strains before use. Experiments using ferrets were conducted with approval from the CSL Limited/Zoetis Australia Animal Ethics Committee, in accordance with the Australian Government National Health and Medical Research Council Australian code of practice for the care and use of animals for scientific purposes (NHMRC, 2013).
2.4. Influenza virus
A/Tasmania/2004/2009 (A(H1N1)pdm09) influenza virus was passaged in the allantoic cavity of embryonated hen's eggs and stored in aliquots at −80 °C. To heat inactivate, virus was incubated at 60 °C for 30 min.
2.5. In vitro culture of ferret lymph node cells with mitogens or influenza virus
Retropharyngeal lymph nodes were collected from naïve ferrets and placed in RPMI-1640 AQ media™ (Sigma–Aldrich, New South Wales, Australia) supplemented with 10% (v/v) fetal calf serum (Interpath Services, Victoria, Australia), 2 mM L-glutamine (SAFC Biosciences, USA), 50 U/ml penicillin/50 μg/ml streptomycin (Sigma–Aldrich) (complete-RPMI). Single cell suspensions were made by mashing the tissue and passing through a sterile 40 μM cell strainer (BD, San Jose, USA). Cell suspensions were washed twice then resuspended in complete-RPMI. Lymph node cells from each ferret (5 × 106 per well) were plated in a 24-well plate in 1 ml complete-RPMI with or without 10 μl of live or heat-inactivated virus (104 TCID50) or 5 μg/ml Concanavalin A (ConA), Phytohaemagglutinin (PHA-P), Lipopolysaccharide (LPS), Ionomycin (Iono) or Phorbol 12-myristate 13-acetate (PMA) (all from Sigma–Aldrich) in duplicate or triplicate. Cell cultures were incubated at 37 °C in 5% CO2 in a humidified incubator for the indicated periods.
2.6. RNA extraction
Total RNA was extracted from cultured cells using the RNeasy® Mini kit (Qiagen, Victoria, Australia) according to the manufacturer's instructions. Briefly, cells from a single well were pelleted and resuspended in 600 μl RLT buffer. The sample was vortexed and then run through a QIAshredder column. RNA was extracted from the supernatant using the Animal Cells Spin protocol, without on-column DNase digestion and eluted with a 30 μl volume. RNA purity was assessed (A 260/A 280) using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Massachusetts, USA). RNA was stored at −80 °C.
2.7. Reverse transcription
For removal of genomic DNA, 800 ng RNA was incubated with 2 units DNase I (RNase-free) (New England Biolabs, Massachusetts, USA) in DNase reaction buffer (final volume 10 μ1) at 37 °C for 10 min. The reaction was terminated by the addition of EDTA (final concentration 5 mM, Sigma–Aldrich) and incubation at 75 °C for 10 min. cDNA was generated using the Superscript III First Strand synthesis System for RT-PCR (Invitrogen, California, USA) with random hexamer primers, according to the manufacturer's instructions. RNaseH treatment was performed. A simultaneous reaction without the reverse transcription enzyme was performed in parallel to generate a ‘-RT’ control. The reaction volume resulted in 10 ng initial RNA/μl final cDNA preparation for culture samples, except at points indicated in the text, where 1 ng initial RNA/μl final cDNA preparation was used. cDNA standards were generated in parallel with cDNA test samples for each experiment. cDNA standards were prepared using RNA pooled from a range of test samples within each experiment, with at least 80 μl of 100 ng initial RNA/μl final cDNA standard prepared. Ten-fold and two-fold serial dilutions (five dilutions of each) were prepared and used for standard curve generation for efficiency calculations. cDNA was stored at −20 °C.
2.8. Polymerase chain reaction (PCR)
Amplification of gene-specific products was performed using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen) according to the manufacturer's instructions. Reactions were run on a S1000™Thermal Cycler (Bio-Rad Laboratories Pty Ltd., New South Wales, Australia).
2.9. Cloning of ferret genes
Total RNA was extracted from stimulated ferret cell cultures, reverse transcribed and ferret genes amplified by conventional and TaqMan real time PCR, using primers indicated in Table 1. Both cDNA and ‘no RT’ control samples were run to ensure the specificity of the amplification. The matrix influenza gene was amplified from RNA extracted from virus isolate A/California/7/2009 (A(H1N1)pdm09). The products were agarose gel-purified using the QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions. Purified DNA was quantified and ligated into pGEM®-T Easy vector (Promega) (IFNγ, IL2, IL4, IL6, IL8, IL10, IL12p40, IL17, MCP1, TNFα, ATF4, HPRT, GAPDH, L32, Matrix) or pCR™4Blunt-TOPO® vector (Invitrogen) (IFNα, IFNβ, IL1α, IL1β, Granzyme A) according to manufacturer's instructions. Ligation reactions were transformed into One Shot® TOP10 chemically competent E. coli (Invitrogen) by heat shock according to the manufacturer's protocol. Positive transformants were identified by PCR using M13 primers (Promega) and gene-specific primers. Plasmid DNA was isolated from bacterial cultures using the QIAprep Spin miniprep kit (Qiagen) according to the manufacturer's instructions.
2.10. Real time PCR assay
The generation of oligonucleotide dimers for each TaqMan primer pair was assessed using Power SYBR® Green PCR MasterMix (Applied Biosystems) with melting curve analysis, according to the manufacturer's instructions. Primers which resulted in oligonucleotide dimer generation were redesigned and retested. A comparison between primer pairs was also performed using Power SYBR® Green PCR MasterMix without a melting curve, according to the manufacturer's instructions. All other real time PCR assays were performed using the TaqMan® Fast Universal PCR Master Mix (2×), No AmpErase UNG (Applied Biosystems), according to the manufacturer's instructions. One microliter cDNA sample was assayed per reaction. Each reaction consisted of 1 cycle of 95 °C for 20 s, followed by 50 cycles of 95 °C for 3 s and 60 °C for 30 s. Real time PCR runs for each gene included cDNA standards (10-fold and 2-fold dilutions, in duplicate), DNA plasmid controls (10-fold dilutions; 5 dilutions), no template control and test samples. When test samples were run over multiple plates for a single gene, DNA plasmid controls were included on each plate and the same mastermix preparation was used for all PCR plates. DNA plasmid controls were reproducible (<1 Ct difference) between plates. All real time PCR reactions were run on an Applied Biosystems 7500 Fast Real-Time PCR system using the 7500 Fast System Software, Version 1.4.0.25 (Applied Biosystems). Data were analyzed using 7500 Fast System Software, Version 1.4.0.25 except for plasmid DNA efficiency calculations which were determined using 7500 Fast System Software Version 2.0.5.
2.11. Calculations of reaction efficiency, housekeeping gene stability and fold change of gene expression
The efficiency of each gene amplification was calculated by plotting the average Ct (y-axis) against the logarithm of the input amount of RNA/μl cDNA (x-axis). Both a 10-fold dilution series and a 2-fold dilution series were used for each gene and RNA set. Real time PCR efficiency (E) = (10−1/slope) for 10-fold dilution series (Pfaffl, 2004) and (2−1/slope) for 2-fold dilution series. % real time PCR efficiency = (E − 1) × 100. If the standard deviation for the efficiencies determined using 10-fold and 2-fold dilution series fell within 8%, the average efficiency was used in all calculations. If the standard deviation was >8%, the efficiency calculated using 2-fold dilutions was used. The geometric mean of the efficiencies for the indicated genes was used for the housekeeping gene efficiency. The gene stability of housekeeping genes was calculated using geNorm in qbase+ Version 2.5 (Biogazelle) (Vandesompele et al., 2002). The fold change of expression of a gene was calculated using relative quantitation with kinetic PCR correction (Pfaffl, 2004). Fold change = (E target)ΔCt target (control−test)/(E HKP)ΔCt HKP (control−test) where ‘HKP’ was the geometric mean of all housekeeping genes for each data point as indicated in figure legends and ‘ΔCt’ = Ct control – Ct test. “Undetermined’ data points in cytokine gene expression were assigned a Ct of “40” to enable calculation of fold change and are indicated in figure legends. The control sample for cultures was unstimulated cells cultured in complete-RPMI.
2.12. Statistics
The ability to perform duplex real time PCR was assessed using two tailed t-test. Data were analyzed using R software (Version 2.15.3 (2013-03-01)) (Team, 2013). Expression of target genes in cultured cells was compared using one-way ANOVA and Tukey's post hoc test (only differences to unstimulated cells are indicated). The mean of duplicate or triplicate wells was calculated and used as a single value for each ferret. *p < 0.05, **p < 0.01, ***p < 0.001.
3. Results
3.1. Development of a two-step TaqMan real time PCR assay to measure expression of ferret immune mediator and housekeeping genes
Cytokine, chemokine and housekeeping genes were amplified from total mRNA generated from cultures of ferret lymph node cells using primers targeting conserved regions of each gene. All genes cloned in this study were sequenced and the sequences run in BLASTn whereby they matched other published and predicted sequences for the corresponding ferret gene on GenBank, especially those derived from the ferret genome project (Di Palma et al., 2013) (Table 2 ). Sequences generated in this study were also translated and all sequences aligned to known sequences of the corresponding proteins from other species in GenBank (data not shown). All primer/probe sets for the TaqMan real time PCR assay (Table 1) were tested to ensure specificity and optimum efficiency using plasmid DNA standards. The absence of non-specific amplicons was verified for all primer/probe sets (Fig. 1A). The % efficiency of each primer/probe set was consistent and between 94 and 103% which is equivalent to an efficiency of 1.94–2.03 (Fig. 1B), indicating that the template doubled after each cycle during exponential amplification. The correlation co-efficient for all samples was between 0.980 and 0.999 (data not shown). The sensitivity of the primer/probe sets for each gene was consistent across the different TaqMan assays, with all genes detected down to 0.0001 pg DNA plasmid (average 25.2 RNA copies), except GAPDH, which was detected down to 0.00001 pg DNA (2.5 RNA copies) (Table 3 ). The limits of detection were consistent for previously published TaqMan primer/probes sets (Nakata et al., 2009) and all tested SYBR Green primer sets (Svitek and von Messling, 2007, Fang et al., 2010, Rowe et al., 2010, Maines et al., 2012) when analyses were performed using the same samples (Table 3). There was no change in the sensitivity of the assay when primer/probes were mixed in the indicated duplex real time PCR reactions (Fig. 1C), or when plasmid samples were prepared in pools (Fig. 1D), or a combination of both duplex real time PCR reactions and sample pools (Fig. 1E), except for L32, which was less sensitive when assayed with other genes (Fig. 1E).
Table 2.
Ferret genes amplified in this study and sequence similarly to other published ferret experimental and predicted sequences by BLASTn. Features of the BLASTn alignments are indicated. Note that the aligned sequences are divided into sequences submitted from laboratory-derived experimental data as well as sequences from the ferret genome which have been predicted and designated using the gene prediction tool.
BLASTn sequences that aligned with the ferret sequence generated in this study.
Relationship between the length the of the BLASTn sequence and the ferret sequence generated in this study. <100% indicates that the BLASTn sequence is shorter. Range for all BLASTn sequences is indicated.
Range for all BLASTn sequences is indicated.
Predicted using the NCBI eukaryotic gene prediction tool, Gnomon.
Fig. 1.
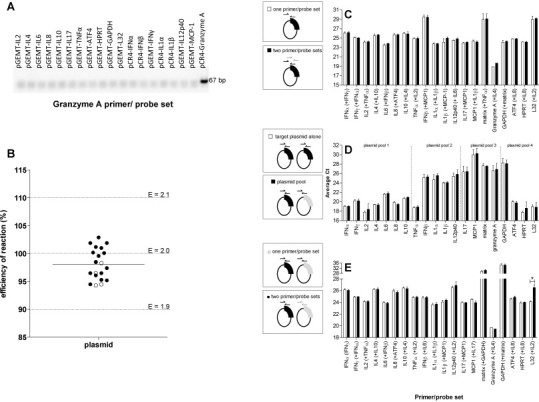
Development of TaqMan assay to detect expression of ferret immune mediators and housekeeping genes using plasmid DNA standards. (A) The specificity of all primer/probe sets was tested against all DNA plasmid standards (1 pg) by TaqMan real time PCR and assessed by gel electrophoresis. An example is shown. (B) The efficiency of each reaction was determined using a 10-fold dilution standard curve. Cytokine and chemokine (black circles) and housekeeping (white circles) genes are shown. The mean efficiency for all genes is indicated by the horizontal line. The reaction efficiency (E) is indicated with the corresponding % efficiency, with the ideal value ‘E = 2’ and the acceptable range (1.9–2.1), indicated. (C) One or two primer/probe sets were combined in a real time PCR reaction and assayed against each of the single target genes. (D) One primer/probe set was assayed against the target gene in a pool (four to seven plasmids) or alone. (E) One or two primer/probe sets were combined in a real time PCR reaction and assayed against the target gene in a paired pool. (C–E) All samples were run in triplicate, with mean and standard deviation indicated. *p < 0.05.
Table 3.
Comparison of the sensitivity of TaqMan and SYBR Green real time PCR assays. All reactions were performed with the same plasmid standard, except in cases where the appropriate plasmid standard encompassing both primer sets was not available, and a cDNA sample was used instead. The same samples were used to compare the TaqMan assays in this study with either previously published TaqMan assays or previously published SYBR Green assays for an individual gene. Individual lines indicate different experiments.
| Gene target | Sensitivity of real time reactiona |
References for other published studies | |||||
|---|---|---|---|---|---|---|---|
| TaqMan primer/probes from this study | TaqMan primer/probes from other published studies | SYBR Green primers from other published studies | |||||
| TaqMan assay |
TaqMan Assay |
SYBR Green assay |
|||||
| Copy number/RNA amount | Average Ct ± std dev | Copy number/RNA amount | Average Ct ± std dev | Copy number/RNA amount | Average Ct ± std dev | ||
| IL1α | 20.8b | 34.8 ± 0.1 | n.a. | n.a. | |||
| IL1β | 21.9b | 33.8 ± 0.3 | 21.9b | 35.3 ± 0.2 | Fang et al. (2010) and Rowe et al. (2010) | ||
| IL2 | 26.3b | 34.6 ± 0.3 | 26.3b | 29.9 ± 0.8 | Svitek and von Messling (2007) | ||
| IL4 | 26.6b | 34.7 ± 0.5 | 26.6b | 35.3 ± 0.4 | Maines et al. (2012) | ||
| IL4 | 26.6b | 34.7 ± 0.5 | 26.6b | 33.2 ± 0.1. | Svitek and von Messling (2007) | ||
| IL6 | 28.9b | 36.8 ± 0.3 | 28.9b | 33.6 ± 0.9 | Svitek and von Messling (2007) | ||
| IL6 | 0.1 ng | 38.2 ± 0.6 | 0.1 ng | 38.7 ± 0.7 | Nakata et al. (2009) | ||
| IL8 | 26.9b | 37.1 ± 0.6 | 26.9b | 34.0 ± 0.4 | Maines et al. (2012) | ||
| IL10 | 27.1b | 35.0 ± 0.3 | 27.1b | 33.9 ± 0.9 | Svitek and von Messling (2007) | ||
| IL10 | 27.1b | 33.3 ± 0.5 | 27.1b | 34.6 ± 0.5 | Nakata et al. (2009) | ||
| IL12p40 | 29.6b | 33.2 ± 0.4 | n.a.d | n.a.d | |||
| IL12p40d | 1 ng | 33.9 ± 0.4 | 1 ng | 34.6 ± 1.2 | Svitek and von Messling (2007) | ||
| IL17 | 28.2b | 33.9 ± 0.4 | n.a. | n.a. | |||
| Granzyme A | 22.8b | 35.3 ± 0.7 | n.a. | n.a. | |||
| MCP1 | 29.0b | 34.9 ± 0.2 | n.a.d | n.a.d | |||
| MCP1d | 0.1 ng | 33.3 ± 0.3 | 1 ng | 34 ± 0.9 | Rowe et al. (2010) | ||
| TNFα | 29.1b | 34.1 ± 0.6 | 29.1b | 31.1 ± 0.6 | Svitek and von Messling (2007) | ||
| TNFαd | 0.1 ng | 33.7 ± 0.6 | 0.1 ng | 34.5 ± 0.7 | Fang et al. (2010) and Rowe et al. (2010) | ||
| TNFα d | 0.1 ng | 34.6 ± 0.5 | 0.1 ng | 35.0 ± 0.6 | Nakata et al. (2009) | ||
| IFNα | 22.9b | 34.7 ± 0.8 | 22.9b | 34.5 ± 0.8 | Svitek and von Messling (2007) | ||
| IFNβ | 22.5b | 35.7 ± 0.7 | 22.5b | 31.6 ± 0.4 | Maines et al. (2012) | ||
| IFNγ | 26.3b | 35.1 ± 0.5 | 26.3b | 33.6 ± 1.1 | Svitek and von Messling (2007) | ||
| IFNγ | 26.3b | 34.8 ± 0.2 | 26.3b | 35.9 ± 0.1 | Nakata et al. (2009) | ||
| Matrix | 23.3b | 37.2 ± 0.8 | n.a. | n.a. | |||
| ATF4 | 28.9b | 33.9 ± 0.5 | n.a. | n.a. | |||
| HPRT | 29.1b | 35.9 ± 0.3 | n.a. | n.a. | |||
| GAPDH | <10c | 37.6 ± 0.9 | 2.8c | 37.8 ± 1 | Svitek and von Messling (2007) | ||
| GAPDHd | 0.3 ng | 33.4 ± 1.5 | 0.03 ng | 31.7 ± 0.6 | Nakata et al. (2009) | ||
| L32 | 29.3b | 36.6 ± 0.7 | n.a. | n.a. | |||
n.a.: not applicable.
Sensitivity refers to the minimum number of copies detected after serial ten-fold dilutions of DNA plasmid, in all triplicates. All three replicates must have a detectable Ct value for inclusion.
Copy number is equivalent to 0.0001 pg DNA plasmid standard.
Copy number is equivalent to 0.00001 pg DNA plasmid standard.
Plasmid did not overlap with previously published primer sequence.
3.2. Optimization of two-step real time PCR assay using RNA from cultured lymphoid cells
To enable comparison between samples from multiple ferrets, the expression of all genes was determined as a fold change, using the calculation for relative quantification to housekeeping gene(s), with kinetic PCR efficiency adjustment (Pfaffl, 2004). As this equation relies on consistency between samples in the RNA quantity assayed, and the optimum reaction efficiency, as well as minimal fluctuation in the expression of housekeeping genes (endogenous controls) (Peters et al., 2007, Mane et al., 2008, Bruder et al., 2010), these parameters were optimized using RNA extracted from cultured ferret lymph node cells stimulated with mitogens or with influenza virus.
As the RNA quantity varied between samples (range 1.39–11.17 μg/well, Fig. 2A), we first determined the minimum amount of RNA for use in the TaqMan real time RT-PCR assay. The reproducibility of both reverse transcription (Fig. 2B) and real time PCR (Fig. 2C) steps was assessed separately for samples with different amounts of RNA. A minimum of 1 ng total RNA produced highly reproducible results for both reverse transcription and real time PCR steps (standard deviation <0.4Ct) and the Ct value was able to detect gene expression (Fig. 2B and C) where Ct (cycle threshold) is the cycle number at which the fluorescent signal of the reaction crosses the threshold to exceed background level. A set of cDNA standards was generated from the mitogen- and influenza virus-stimulated ferret lymph node cells and the reaction efficiency for all genes determined (examples of graphs and calculations for efficiency are shown in Fig. 3A, compilation of efficiencies for all genes is shown in Fig. 3B and C). Although the reaction efficiencies clustered for each of the ferret lymph node cDNA preparations (mitogen or virus stimulation in Fig. 3B and C, respectively), the efficiencies using the 10-fold dilutions of cDNA samples were more broadly spread than for the plasmid preparations (compare 10-fold dilution in Fig. 3B and C with Fig. 1B). Furthermore, analysis of the 10-fold dilutions indicated some variability due to low level cytokine mRNA expression (such as for IL1α, IFNα). Use of 2-fold dilutions of the cDNA standards enabled the efficiency to be determined and data points overlaid with the data points generated by the 10-fold dilution series, suggesting this approach is acceptable to determine cytokine levels (Fig. 3A). To minimize the potential effect of initial sampling variability, a calculation of gene stability was performed for housekeeping genes (Vandesompele et al., 2002). Assessment of the stability of expression of housekeeping genes following stimulation with different mitogens (Fig. 4A) or influenza virus (Fig. 4B) demonstrated that expression of some housekeeping genes, such as GAPDH, was affected more than others and this effect was specific to different stimuli (compare Fig. 4A to B). As GAPDH was more variable in expression than the other housekeeping genes; ATF4, L32 and HPRT following mitogen stimulation, GAPDH was excluded and a combination housekeeping genes was used as a reference (Fig. 4C). In contrast, following virus stimulation, all housekeeping genes were relatively stable and thus all housekeeping genes were able to be used as references (Fig. 4D). These data suggests that reaction efficiencies and the most stable housekeeping genes should be determined for a type of sample set of cDNA under analysis to be able to accurately calculate gene expression.
Fig. 2.
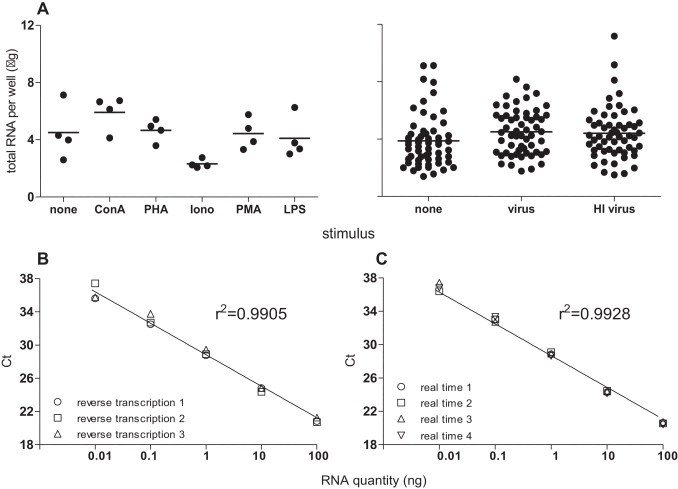
Quantification of total RNA isolated from cultured ferret lymphoid cells and assessment of variability in reverse transcription and real time PCR assay steps. (A) Cells from naïve ferret lymph nodes were cultured with or without the indicated mitogens or with influenza virus for 24 or 48 h, respectively. Each point indicates the average amount of RNA isolated from duplicate or triplicate culture wells of cells from a single ferret. Total RNA was extracted and quantified by spectrophotometry. The mean for each group is indicated by horizontal bar. (B) cDNA was reverse transcribed from the same RNA samples on three separate occasions. All samples were assessed in the same real time PCR assay detecting the housekeeping gene ATF4. (C) cDNA was reverse transcribed in bulk and assayed detecting ATF4, in four separate real time PCR reactions. (B, C) Individual data points represent each experiment. The line of best fit was calculated using linear regression.
Fig. 3.
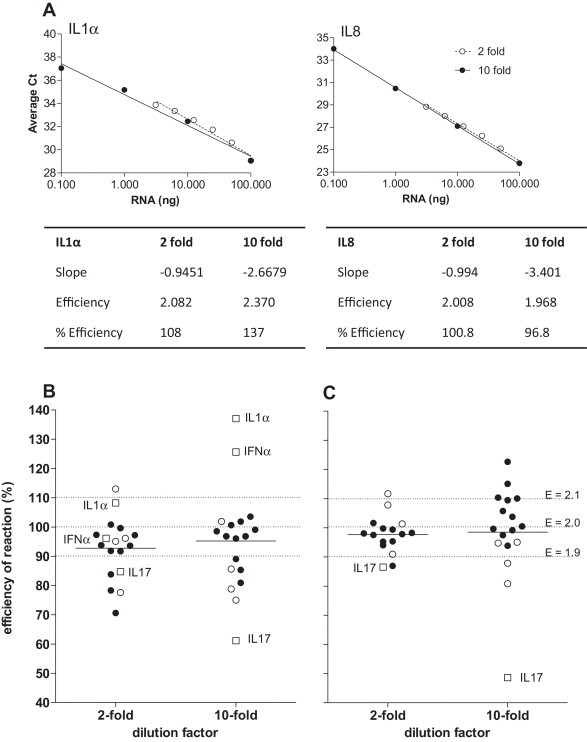
Assessment of efficiency of TaqMan assay for ferret cytokine mRNAs using cDNA. The efficiency of each reaction was determined using standard curves generated from cDNA pools in 10-fold or 2-fold dilution series (average of duplicate values are shown). (A) Examples of calculations for efficiency using cDNA standards from mitogen-stimulated cells to detect IL1α and IL8 expression. (B, C) The reaction efficiencies for each gene for a set of cDNA standards (10-fold and 2-fold dilutions) were calculated. The cDNA standards were generated using mitogen (B)- or influenza virus (C)-stimulated cells. Housekeeping genes (ATF4, HPRT, GAPDH, L32) are indicated by white circles, outliers are indicated by white squares, all other genes are indicated by black circles. Mean is indicated for each group by horizontal line. The % efficiency and corresponding efficiency (E) are indicated.
Fig. 4.
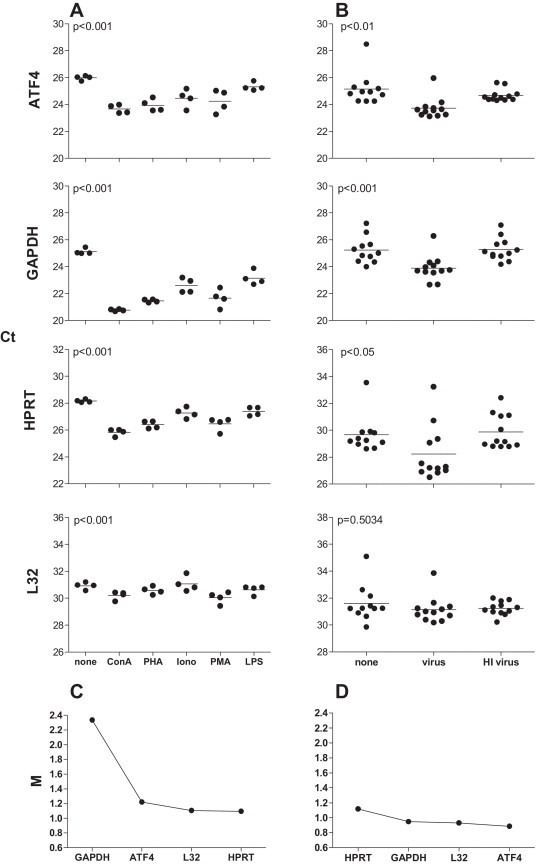
Assessment of housekeeping gene expression in cultured ferret lymphoid cells. Expression of housekeeping genes was determined in lymph node cells from naïve ferrets stimulated with mitogens for 24 h (A) or with influenza virus for 48 h (B). All stimulations were performed in duplicate or triplicate wells for each ferret and each data point represents the mean Ct of the culture wells for a single ferret. (A, B) Mean for each group is indicated by horizontal line and p value indicates overall differences between groups. (C, D) The ‘average pairwise variation of a gene with all other housekeeping genes’ (M) after stepwise exclusion of the more variable genes as calculated using geNorm (Vandesompele et al., 2002). The least stable gene is shown on the left, the most stable gene on the right.
3.3. Stimulation of ferret lymph node cells with mitogens or influenza virus differentially modulates expression of innate and adaptive immune mediators
To test the ability of ferret leukocytes to produce mRNA cytokines and chemokines, lymph node cells from naïve ferrets were cultured with various mitogens known to activate T and B lymphocyte and macrophage/monocyte responses (Fig. 5 ) and with live or heat inactivated influenza virus (Fig. 6 ) and the cytokine and chemokine expression profiles were determined (summarized in Table 4 ). Culture with the lectins Concanavalin A (ConA) or Phytohaemagglutinin (PHA), which act by cross linking T cell receptors via sugars on the surface of human T lymphocytes (Chilson and Kelly-Chilson, 1989), induced similar cytokine profiles, increasing expression of IL2, IL4, IL6, IL10, IL17, Granzyme A, TNFα and IFNγ, most with high fold changes, consistent with effective stimulation of T lymphocytes (Fig. 5). PHA also induced expression of IL12p40 mRNA. The addition of Con A or PHA significantly reduced the expression of MCP-1 and IFNα in their respective cultures (Fig. 5). Ionomycin and Phorbol 12-myristate 13-acetate (PMA) are mitogenic for lymphocytes as they bypass surface receptors and activate cellular responses by increasing intracellular calcium and directly activating protein kinase C, respectively (Nishezuka, 1984, Al Wabel et al., 1993). Culture with ionomycin significantly increased mRNA expression of IL2, IL10 and IFNγ. PMA significantly increased levels of TNFα and IL1β. Expression of IFNα and IL12p40 mRNAs was significantly reduced upon culture with Ionomycin or PMA (Fig. 5).
Fig. 5.
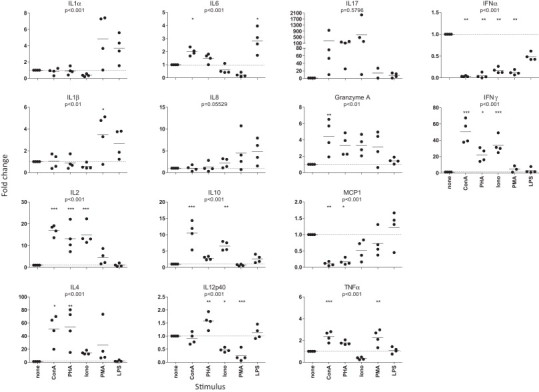
Gene expression in mitogen-stimulated ferret lymph node cells. Lymph node cells from four naïve ferrets were stimulated with the indicated mitogens for 24 h, then RNA was isolated, reverse transcribed and assayed by real time PCR using 10 ng initial RNA/reaction. Each data point represents the average fold change of duplicate or triplicate culture wells compared to the geometric mean of L32, HPRT and ATF4 housekeeping genes. IFNβ was not detected. Horizontal bars indicate the mean for each group; dotted line indicates no fold change. Statistical difference in fold-change compared to cells with no stimulation is indicated.
Fig. 6.
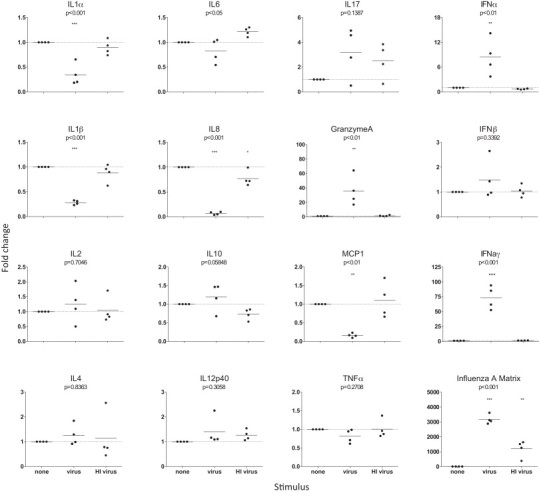
Gene expression in influenza virus-stimulated ferret lymph node cells. Lymph node cells from four naive ferrets were stimulated with the live or heat inactivated (HI) A (H1N1)pdm09 virus for 48 h, then RNA was isolated, reverse transcribed and assayed by real time PCR using 10 ng initial RNA/reaction. Each data point represents the average fold change of duplicate or triplicate culture wells compared to the geometric mean of GAPDH, L32, HPRT and ATF4 housekeeping genes. The influenza A matrix gene was not detected in the ‘none’ sample and was substituted with a value of 40; all other genes had a detectable Ct in all samples. Horizontal bars indicate the mean for each group; dotted line indicates no fold change. Statistical difference in fold-change compared to cells with no stimulation is indicated.
Table 4.
Changes in cytokine and chemokine expression following stimulation of ferret lymph node cells with mitogens or influenza virus.
| Gene | Stimulus |
|||||
|---|---|---|---|---|---|---|
| ConA | PHA | Iono | PMA | LPS | Influenza virus | |
| IL1α | nca | nc | nc | +b | + | ↓*** |
| IL1β | nc | nc | nc | +*d | + | ↓*** |
| IL2 | +*** | +*** | +*** | + | nc | nc |
| IL4 | +* | +** | + | + | nc | nc |
| IL6 | +* | + | nc | ↓c | +* | nc |
| IL8 | nc | nc | nc | + | + | ↓*** |
| IL10 | +*** | + | +** | nc | + | nc |
| IL12p40 | nc | +** | ↓* | ↓*** | nc | nc |
| IL17 | + | + | + | nc | nc | nc |
| Granzyme A | +** | + | + | + | nc | +** |
| MCP1 | ↓** | ↓* | ↓ | nc | nc | ↓** |
| TNFα | +*** | + | ↓ | +** | nc | nc |
| IFNα | ↓** | ↓** | ↓** | ↓** | ↓ | +** |
| IFNβ | n.d.e | n.d. | n.d. | n.d. | n.d. | nc |
| IFNγ | +*** | +* | +*** | nc | nc | +*** |
No change in expression compared to unstimulated cells.
Upregulation in expression compared to unstimulated cells.
Down-regulation in expression compared to unstimulated cells.
Significant change *p < 0.05, **p < 0.01, ***p < 0.001.
Not detected.
Lipopolysaccharide (LPS) is a potent activator of naïve and mature B lymphocytes (Andersson et al., 1973, Smith et al., 1979), monocytes and macrophages. Stimulation with LPS induced significant levels of IL6 and a small increase in expression of IL8. No expression of IFNβ was detected in any culture stimulated with mitogen. Real time PCR assays performed as duplex PCRs with the same primer pairs as shown in Fig. 1E detected up- and down-regulation of the same cytokines as assays using single primer/probe sets (data not shown). Real time assays run with 10-fold less cDNA (1 ng RNA/μl cDNA) for IL6, IFNγ, L32 and ATF4 detected the same fold differences (data not shown).
Culture of ferret leukocytes with live or heat inactivated A(H1N1)pdm09 virus for 48 h induced different cytokine profiles from those stimulated by mitogens (Fig. 6). Live A(H1N1)pdm09 virus induced a significant reduction in expression of IL1α, IL1β, IL8 and MCP1, as well as large increases in expression of Granzyme A, IFNα and IFNγ, whereas heat inactivated virus induced negligible effect on cytokine expression. Note that cultures assayed at 24 and 72 h also showed similar results (data not shown).
4. Discussion
In this study, we describe a series of real time TaqMan RT-PCR assays that can be used to characterize the expression of cytokines and chemokines of the innate and adaptive immune response in ferrets. The sequences of the ferret genes cloned in this study and used for design of the primers and probes, aligned with other previously published sequences as well as with the predicted sequences from the Ferret Genome Project (Di Palma et al., 2013). Previous studies have predominantly utilized SYBR green real time RT-PCR and required a minimum quantity of 10 ng initial RNA per reaction for detection of a single ferret cytokine or chemokine (Svitek et al., 2008, Meunier et al., 2012, Meunier and von Messling, 2012). Here we have demonstrated that 1 ng total RNA is sufficient in a TaqMan real time RT-PCR assay to detect most cytokine and chemokine mRNAs. By multiplexing reactions with probes using different fluorochromes, samples with low amounts of RNA, such as ferret respiratory samples (Suguitan et al., 2012), may still be analyzed using this assay without loss of sensitivity, providing a significant sample and cost reduction. In our study we showed that a minimum of duplex assays could be used, mixing primers and probes specific for a housekeeping gene and a cytokine/chemokine gene or two cytokine/chemokines genes. With further optimization, a higher number of targets may be able to be multiplexed.
Real time PCR results can be reported in copy number or as fold change both compared to a control, depending on whether absolute or relative quantification is required (Giulietti et al., 2001, Pfaffl, 2004). Absolute quantification requires DNA or in vitro transcribed RNA standards to be included in each reaction, whereas relative quantification can be achieved by including a set of cDNA standards generated from the sample of interest, or no standards at all, if the efficiency of each reaction is identical (Pfaffl, 2004). As it is difficult to maintain the stability of a large number of RNA standards (Giulietti et al., 2001), absolute quantification was not used in this study. Furthermore, the necessity for a standard curve on each assay plate would increase the number of plates required for an experiment, which may become impractical when assaying a large number of test samples. Calculation of fold change relative to housekeeping gene(s), with kinetic PCR efficiency adjustment, incorporates corrections for reverse transcription efficiency and PCR efficiency, as well as individual sample addition, by using normalizer housekeeping genes. In our study, although initial assays using DNA plasmids demonstrated that the efficiencies of all primer/probe sets in the TaqMan real time PCR assays were consistent, data from cultures of ferret lymph nodes and our preliminary data with ferret respiratory samples (not shown) indicate that the efficiency of reactions clusters for each set of cDNAs and appropriate housekeeping genes need to be determined for each sample type. The importance of careful assessment of suitable housekeeping genes for real time PCR has also been demonstrated for human, canine and ferret tissues (Peters et al., 2007, Mane et al., 2008, Bruder et al., 2010). As real time PCR measures the Ct in the exponential phase of the PCR, it has been argued that the effect of differences in PCR efficiency is minor (Giulietti et al., 2001). However, we anticipate this TaqMan assay will be useful for various ferret samples, particularly nasal washes or bronchoalveolar lavages, and respiratory tissues in virus-infected ferrets, which may have low amounts of RNA, and variable levels of expression of cytokine and chemokine genes. Given the variability in the cellular composition of samples from different sites (e.g. lung tissue compared to lymph node), incorporation of corrections for reverse transcription and PCR efficiency and housekeeping gene variability are necessary in these samples.
The cytokine profiles induced by stimulation of lymph node cells from naïve ferrets with mitogens were consistent with those reported in studies using human and other animal leukocytes. Stimulation of ferret lymph node cells with ConA induced similar profiles to cultures of mouse splenocytes (Candolfi et al., 1995), human PBMCs (Al Wabel et al., 1993, Yaqoob and Calder, 1998, Radke et al., 2012), woodchuck PMBCs (Menne et al., 2002) and calf CD4+ and CD8+ PBMCs (Tanaka et al., 2007) stimulated with ConA. Of interest, ConA has been shown to induce expression of IL1β and low levels of IL1α in human PBMCs (Yaqoob and Calder, 1998), but we did not detect increases in either cytokine in ferret lymph node cells. We also used the other published primers for detecting IL1β (Fang et al., 2010, Rowe et al., 2010) with our samples but could not detect expression (data not shown). Increased levels of TNFα, IL2 and IFNγ were reported following culture of human PBMCs with PHA (Godoy-Ramirez et al., 2004, Anderson and Teuber, 2010), and we obtained similar results using ferrets cells. Ionomycin and PMA have been shown to increase production of IL2, IL4 and IFNγ in human PBMCs (Jung et al., 1993) and IL4, IL8, IFNγ and TNFα in ferret BAL, splenocytes or PBLs (Rutigliano et al., 2008, Martel and Aasted, 2009); all of these genes were also upregulated in this current study following stimulation of ferret leukocytes with either Ionomycin or PMA. LPS is a potent stimulator of IL1α and β, IL6, and TNFα from human PBMCs (Al Wabel et al., 1993, Yaqoob and Calder, 1998, Matera et al., 2009, Coch et al., 2013) and IL6 and TNFα from mouse PBMCs and macrophages (Kawai et al., 2004). Similarly, IL1α and β, IL6, and IL8, but not TNFα, were increased in LPS-stimulated ferret lymph node cultures. A similar study in which ferret PBMCs were cultured with LPS detected expression of IL6, IFNγ, IL10 and TNFα by TaqMan real time RT-PCR at earlier timepoints, suggesting the kinetics of the response are important (Nakata et al., 2009). We also assessed TNFα expression using previously published TaqMan primers and probe (Nakata et al., 2009) and SYBR Green primers (Fang et al., 2010, Rowe et al., 2010) with our samples and the profiles were consistent with those obtained with the TaqMan assay designed in this study (data not shown).
Stimulation of ferret lymph node cultures with live influenza virus resulted in upregulated expression of IFNγ and Granzyme A, and these markers of T lymphocyte (and NK cell) activation, are also upregulated following stimulation of human PBMCs with influenza virus (Forbes et al., 2012, Vanders et al., 2013). IL2 was not detected in ferret cells, although it has been detected by flow cytometry in human cells following in vitro influenza virus stimulation (Scheible et al., 2011, Guérin-El Khourouj et al., 2012). Increased expression of type I interferons (also seen upon virus stimulation in human PBMCs (Forbes et al., 2012)) and a corresponding decrease in MCP1 expression was induced in ferret cells following exposure to influenza virus. This inverse relationship has also been reported in mice co-infected with bacteria and influenza virus and is suggested to be due to type I interferon-mediated suppression of macrophages (Nakamura et al., 2011). This may also explain the reductions in IL1 and IL8 in ferret lymph node cells cultured with virus as both these mediators would be typically produced by macrophages in the lymph node. The significant down-regulation of IFNα and variable expression of MCP-1 and IL12p40 in cultures stimulated with mitogens may also be due to altered activation of cells of the innate immune system. The magnitude and differential patterns of gene expression detected here indicate that these assays can be used for more detailed investigations of the cell types and pathways (Ghosh et al., 2006) that contribute to immune responses in the ferret.
In summary, we have described a series of TaqMan real time RT-PCR assays to quantify a large number of cytokines and chemokines in the ferret model. Our study highlights key technical aspects of these assays to maximize the analysis of immune mediators induced from ferret cells. The particular cytokine and chemokine profiles induced in ferret cells following stimulation with mitogens or influenza virus were consistent with those reported for other species. Overall these data demonstrate the usefulness of these assays to enhance our understanding of influenza virus infection and other diseases that use the ferret model.
Acknowledgements
The authors are grateful for advice provided by Dr Wa-Chin Boon, Dr John Roiniotis, Professor Paul Hertzog and Mr Steve Vander Hoorn. The authors are also grateful for technical assistance provided by bioCSL Limited Animal House staff. The authors also acknowledge the contribution of gifts of reagents from Dr Liyen Loh and Ms Sarina Camuglia. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
References
- Al Wabel A., Al-Janadi M., Raziuddin S. Cytokine profile of viral and autoimmune chronic active hepatitis. J. Allergy Clin. Immunol. 1993;92(6):902–908. doi: 10.1016/0091-6749(93)90068-q. [DOI] [PubMed] [Google Scholar]
- Anderson K., Teuber S. Ellagic acid and polyphenolics present in walnut kernels inhibit in vitro human peripheral blood mononuclear cell proliferation and alter cytokine production. Ann. N.Y. Acad. Sci. 2010;1190:86–96. doi: 10.1111/j.1749-6632.2009.05259.x. [DOI] [PubMed] [Google Scholar]
- Andersson L., Nordling S., Häyry P. Proliferation of B and T cells in mixed lymphocyte cultures. J. Exp. Med. 1973;138(1):324–329. doi: 10.1084/jem.138.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric, R., Engelhardt, J., Gibbs, R., Kawaoka, Y., Sur, M., Weinstock, G., Jensen, K., Paeper, B., Palermo, R., Katze, M. Genomic and EST sequencing of the ferret (Mustela putorius furo). Retrieved 13 December 2013 from http://www.genome.gov/pages/research/sequencing/seqproposals/ferretseq.pdf.
- Belser J.A., Katz J.M., Tumpey T.M. The ferret as a model organism to study influenza A virus infection. Disease Models & Mechanisms. 2011;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J., Maines T., Gustin K., Katz J., Tumpey T. Kinetics of viral replication and induction of host responses in ferrets differs between ocular and intranasal routes of inoculation. Virology. 2013;438:56–60. doi: 10.1016/j.virol.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Bossart K., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J., McEachern J., Green D., Hancock T., Chan Y., Hickey A., Dimitrov D., Wang L., Broder C. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5:e10000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder C.E., Yao S., Larson F., Camp J., Tapp R., McBrayer A., Powers N., Granda W., Jonsson C. Transcriptome sequencing and development of an expression microarray platform for the domestic ferret. BMC Genomics. 2010;11:251. doi: 10.1186/1471-2164-11-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron C., Cameron M., Bermejo-Martin J., Ran L., Xu L., Turner P., Ran R., Danesh A., Fang Y., Chan P., Mytle N., Sullivan T., Collins T., Johnson M., Medina J., Rowe T., Kelvin D. Gene expression analysis of host innate immune responses during lethal H5N1 infection in ferrets. J. Virol. 2008;82(22):11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candolfi E., Hunter C., Remington J. Roles of gamma Interferon and other cytokines in suppression of the spleen cell proliferative response to Concanavalin A and toxoplasma antigen during acute toxoplasmosis. Infect. Immun. 1995;63(3):751–756. doi: 10.1128/iai.63.3.751-756.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilson O., Kelly-Chilson A. Mitogenic lectins bind to the antigen receptor on human lymphocytes. Eur. J. Immunol. 1989;19:389–396. doi: 10.1002/eji.1830190225. [DOI] [PubMed] [Google Scholar]
- Coch C., Lück C., Schwickart A., Putschli B., Renn M., Höller T., Barchet W., Hartmann G., Schlee M. A human in vitro whole blood assay to predict the systemic cytokine response to therapeutic oligonucleotides including siRNA. PLoS One. 2013;8(8):e71057. doi: 10.1371/journal.pone.0071057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh A., Seneviratne C., Cameron C., Banner D., Devries M., Kelvin A., Xu L., Ran L., Bosinger S., Rowe T., Czub M., Jonsson C., Cameron M., Kelvin D. Cloning, expression and characterization of ferret CXCL10. Mol. Immunol. 2008;45(5):1288–1297. doi: 10.1016/j.molimm.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh A., Cameron C., León A., Ran L., Xu L., Fang Y., Kelvin A., Rowe T., Chen H., Guan Y., Jonsson C., Cameron M., Kelvin D. Early gene expression events in ferrets in response to SARS coronavirus infection versus direct interferon-alpha2b stimulation. Virology. 2011;409(1):102–112. doi: 10.1016/j.virol.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Palma F., Alfoldi J., Johnson J., Jaffe D., Berlin A., Gnerre S., Grabherr M., Hall G., Lara M., MacCallum I., Mauceli E., Przyblyski D., Ribeiro F., Russell P., Sharpe T., Turner-Maier J., Walker B.J., Young S., Birren B., Lindblad-Toh K. 2013. Ferret Genome Project. Retrieved 8 January 2013 from http://www.broadinstitute.org/scientific-community/science/projects/mammals-models/ferret-genome-project. [Google Scholar]
- Easlick J., Szubin R., Lantz S., Baumgarth N., Abel K. The early interferon alpha subtype response in infant macaques infected orally with SIV. J. Acquir. Immune Defic. Syndr. 2010;55(1):14–28. doi: 10.1097/QAI.0b013e3181e696ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Rowe T., Leon A., Banner D., Danesh A., Xu L., Ran L., Bosinger S., Guan Y., Chen H., Cameron C., Cameron M., Kelvin D. Molecular characterization of in vivo adjuvant activity in ferrets vaccinated against influenza virus. J. Virol. 2010;84(17):8369–8388. doi: 10.1128/JVI.02305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes R., Wark P.B., Murphy V., Gibson P. Pregnant women have attenuated innate interferon responses to 2009 pandemic influenza A virus subtype H1N1. J. Infect. Dis. 2012;206(5):646–653. doi: 10.1093/infdis/jis377. [DOI] [PubMed] [Google Scholar]
- Geisbert T., Feldmann H., Broder C. Animal challenge models of henipavirus infection and pathogenesis. Curr. Top. Microbiol. Immunol. 2012;359:153–177. doi: 10.1007/82_2012_208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T.K., Mickelson D.J., Fink F., Solberg J.C., IngleWeld J.R., Hook D., Gupta S.K., Gibson S., Alkan S.S. Toll-like receptor (TLR) 2-9 agonists-induced cytokines and chemokines: I. Comparison with T cell receptor-induced responses. Cell. Immunol. 2006;243:48–57. doi: 10.1016/j.cellimm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Giulietti A., Overbergh L., Valckx D., Decallonne B., Bouillon R., Mathieu C. An overview of real-time quantitiative PCR: applicationd to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- Godoy-Ramirez K., Franck K., Mahdavifar S., Andersson L.G.H. Optimum culture conditions for specific and nonsepcific acitvation of whole blood and PBMC for intracellular cytokine assessment by flow cytometry. J. Immunol. Methods. 2004;292:1–15. doi: 10.1016/j.jim.2004.04.028. [DOI] [PubMed] [Google Scholar]
- Guérin-El Khourouj V., Duchamp M., Krivine A., Pédron B., Ouachée-Chardin M., Yakouben K., Frémond M., Baruchel A., Dalle J., Sterkers G. Cellular and humoral immunity elicited by influenza vaccines in pediatric hematopoietic-stem cell transplantation. Hum. Immunol. 2012;73(9):884–890. doi: 10.1016/j.humimm.2012.07.039. [DOI] [PubMed] [Google Scholar]
- Hamelin M., Baz M., Abed Y., Couture C., Joubert P., Beaulieu E., Bellerose N., Plante M., Mallet C., Schumer G., Kobinger G., Boivin G. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 2010;6(7):e1001015. doi: 10.1371/journal.ppat.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer P., Mane V., Schramm L., Puig M., Verthelyi D., Chen A., Zhao Z., Navarro M., Kirschman K., Bykadi S., Jubin R., Rabin R. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-depenedent. Immunol. Cell Biol. 2012;90:774–783. doi: 10.1038/icb.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.S.H., Banner D., Degousee N., Leon A.J., Xu L., Paquette S.G., Kanagasabai T., Fang Y., Rubino S., Rubin B., Kelvin D.J., Kelvin A.A. Differential pathological and immune responses in newly weaned ferrets are associated with a mild clinical outcome of pandemic 2009 H1N1 infection. J. Virol. 2012;86(24):13187–13201. doi: 10.1128/JVI.01456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A.C., Lowther S., Middleton D., Barr I.G. Assessing the development of oseltamivir and zanamivir resistance in A(H5N1) influenza viruses using a ferret model. Antiviral Research. 2010;87:361–366. doi: 10.1016/j.antiviral.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Jung T., Schauer U., Heusser C., Neumann C., Rieger C. Detection of intracellular cytokines by flow cytometry. J. Immunol. Methods. 1993;159:197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- Kang Y., Song B., Lee J., Kim H., Seo S. Pandemic H1N1 influenza virus causes a stronger inflammatory response than seasonal H1N1 influenza virus in ferrets. Arch. Virol. 2011;156(5):759–767. doi: 10.1007/s00705-010-0914-7. [DOI] [PubMed] [Google Scholar]
- Kawai K., Kuwahara K., Oizumi N., Kitagaki H., Fujisawa S. Effects of carteolol hydrochloride on the in vitro production of LPS-induced proinflammatory cytokines by murine macrophage. J. Ocul. Pharmacol. Ther. 2004;20(3):237–245. doi: 10.1089/1080768041223620. [DOI] [PubMed] [Google Scholar]
- Keiser N., Engelhardt J. New animal models of cystic fibrosis: what are they teaching us? Curr. Opin. Pulm. Med. 2011;17(6):478–783. doi: 10.1097/MCP.0b013e32834b14c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kim H., Cho S., Seo S. Influenza B virus causes milder pathogenesis and weaker inflammatory responses in ferrets than influenza A virus. Viral Immunol. 2009;22(6):423–430. doi: 10.1089/vim.2009.0045. [DOI] [PubMed] [Google Scholar]
- Kobinger G., Meunier I., Patel A., Pillet S., Gren J., Stebner S., Leung A., Neufeld J., Kobasa D., von Messling V. Assessment of the efficacy of commercially available and candidate vaccines against a pandemic H1N1 2009 virus. J. Infect. Dis. 2010;201(7):1000–1006. doi: 10.1086/651171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie K.L., Carolan L.A., Middleton D., Lowther S., Kelso A. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. Journal of Infectious Diseases. 2010;202:1011–1020. doi: 10.1086/656188. [DOI] [PubMed] [Google Scholar]
- Maines T., Belser J., Gustin K., van Hoeven N., Zeng H., Svitek N., von Messling V., Katz J., Tumpey T. Local innate immune responses and influenza virus transmission and virulence in ferrets. J. Infect. Dis. 2012;205(3):474–485. doi: 10.1093/infdis/jir768. [DOI] [PubMed] [Google Scholar]
- Mane V., Heuer M., Hillyer P., Navarro M., Rabin R. Systematic method for determining an ideal housekeeping gene for real-time PCR analysis. J. Biomol. Tech. 2008;19:342–347. [PMC free article] [PubMed] [Google Scholar]
- Martel C., Aasted B. Characterization of antibodies against ferret immunoglobulins, cytokines and CD markers. Vet. Immunol. Immunopathol. 2009;132(2–4):109–115. doi: 10.1016/j.vetimm.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Matera G., Muto V., Vinci M., Zicca E., Abdollahi-Roodsaz S., van de Veerdonk F.L., Kullberg B.-J., Liberto M.C., van der Meer J.W.M., Focà A., Netea M.G., Joosten L.A.B. Receptor recognition of and immune intracellular pathways for Veillonella parvula lipopolysaccharide. Clin. Vaccine Immunol. 2009;16(12):1804–1809. doi: 10.1128/CVI.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne S., Wang Y., Butler S., Gerin J., Cote P., Tenant B. Real-time polymerase chain reaction assays for leukocyte CD and cytokine mRNAs of the Eastern woodchuck (Marmota monax) Vet. Immunol. Immunopathol. 2002;87:97–105. doi: 10.1016/s0165-2427(02)00121-6. [DOI] [PubMed] [Google Scholar]
- Meunier I., von Messling V. NS1-mediated delay of type I interferon induction contributes to influenza A virulence in ferrets. J. Gen. Virol. 2011;92(Pt 7):1635–1644. doi: 10.1099/vir.0.032193-0. [DOI] [PubMed] [Google Scholar]
- Meunier I., von Messling V. PB1-F2 modulates early host responses but does not affect the pathogenesis of H1N1 seasonal influenza virus. J. Virol. 2012;86(8):4271–4278. doi: 10.1128/JVI.07243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier I., Embury-Hyatt C., Stebner S., Gray M., Bastien N., Li Y., Plummer F., Kobinger G., von Messling V. Virulence differences of closely related pandemic 2009 H1N1 isolates correlate with increased inflammatory responses in ferrets. Virology. 2012;422(1):125–131. doi: 10.1016/j.virol.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Davis K.M., Weiser J.N. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 2011;121(9):3657–3665. doi: 10.1172/JCI57762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M., Itou T., Sakai T. Molecular cloning and phylogenetic analysis of inflammatory cytokines of the ferret (Mustela putorius furo) J. Vet. Med. Sci. 2008;70(6):543–550. doi: 10.1292/jvms.70.543. [DOI] [PubMed] [Google Scholar]
- Nakata M., Itou T., Sakai T. Quantitative analysis of inflammatory cytokines expression in peripheral blood mononuclear cells of the ferret (Mustela putorius furo) using real-time PCR. Vet. Immunol. Immunopathol. 2009;130(1–2):88–91. doi: 10.1016/j.vetimm.2008.12.009. [DOI] [PubMed] [Google Scholar]
- NHMRC . 2013. Australian Code for the Care and Use of Animals for Scientific Purposes. [Google Scholar]
- Nishezuka Y. The role of protein kinase C in cell surface signal transduction and tumor promotion. Nature. 1984;308:693–696. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- Ochi A., Danesh A., Seneviratne C., Banner D., Devries M., Rowe T., Xu L., Ran L., Czub M., Bosinger S., Cameron M., Cameron C., Kelvin D. Cloning, expression and immunoassay detection of ferret IFN-gamma. Dev. Comp. Immunol. 2008;32(8):890–897. doi: 10.1016/j.dci.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister J., Middleton D., Crameri G., Yamada M., Klein R., Hancock T., Foord A., Shiell B., Michalski W., Broder C., Wang L. Chloroquine administration does not prevent Nipah virus infection and disease in ferrets. J. Virol. 2009;83:11979–11982. doi: 10.1128/JVI.01847-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister J., Middleton D., Wang L., Klein R., Haining J., Robinson R., Yamada M., White J., Payne J., Feng Y., Chan Y., Broder C. A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine. 2011;29:5623–5630. doi: 10.1016/j.vaccine.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters I., Peeters D., Helps C., Day M. Development and application of multiple internal reference (housekeeper) gene assays for accurate normalisation of canine gene expression studies. Vet. Immunol. Immunopathol. 2007;117:55–66. doi: 10.1016/j.vetimm.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A–Z of Quantitative PCR. International University Line (IUL); La Jolla, CA, USA: 2004. Quantification strategies in real-time PCR. (Chapter 3) [Google Scholar]
- Pillet S., Kobasa D., Meunier I., Gray M., Laddy D., Weiner D., von Messling V., Kobinger G. Cellular immune response in the presence of protective antibody levels correlates with protection against 1918 influenza in ferrets. Vaccine. 2011;29(39):6793–6801. doi: 10.1016/j.vaccine.2010.12.059. [DOI] [PubMed] [Google Scholar]
- Qin S., Klamar C., Fallert Junecko B., Craigo J., Fuller D., Reinhart T. Functional characterization of ferret CCL20 and CCR6 and identification of chemotactic inhibitors. Cytokine. 2013;61:924–932. doi: 10.1016/j.cyto.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke L., Lopez Hemmerling D., Lubitz A., Giese C., Frohme M. Induced cytokine response of human PMBC-cultures: correlation of gene expression and secretion profiling and the effect of cryopreservation. Cell. Immunol. 2012;272:144–153. doi: 10.1016/j.cellimm.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Roberts A., Lamirande E., Vogel L., Jackson J., Paddock C., Guarner J., Zaki S., Sheahan T., Baric R., Subbarao K. Animal models and vaccines for SARS-CoV infection. Virus Res. 2008;133(1):20–32. doi: 10.1016/j.virusres.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman S., Middleton D.J., Pearse M.J., Barr I.G., Lowther S. Control of pandemic (H1N1) 2009 influenza virus infection of ferret lungs by non-adjuvant-containing pandemic and seasonal vaccines. Vaccine. 2012;30:3618–3623. doi: 10.1016/j.vaccine.2012.03.043. [DOI] [PubMed] [Google Scholar]
- Rowe T., León A., Crevar C., Carter D., Xu L., Ran L., Fang Y., Cameron C., Cameron M., Banner D., Ng D., Ran R., Weirback H., Wiley C., Kelvin D., Ross T. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology. 2010;401(2):257–265. doi: 10.1016/j.virol.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutigliano J., Doherty P., Franks J., Morris M., Reynolds C., Thomas P. Screening monocloncal antibodies for cross-reactivity in the ferret model of influenza infection. J. Immunol. Methods. 2008;336:71–77. doi: 10.1016/j.jim.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible K., Zhang G., Baer J., Azadniv M., Lambert K., Pryhuber G., Treanor J., Topham D. CD8+ T cell immunity to 2009 pandemic and seasonal H1N1 influenza viruses. Vaccine. 2011;29(11):2159–2168. doi: 10.1016/j.vaccine.2010.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.I.E., Hammarstrom L., Bird A.G., Kunori T., Gustafsson B., Holme T. Lipopolysaccharide and lipid A-induced human B cell activation. Eur. J. Immunol. 1979;9:619–625. doi: 10.1002/eji.1830090809. [DOI] [PubMed] [Google Scholar]
- Suguitan A.J., Matsuoka Y., Lau Y., Santos C., Vogel L., Cheng L., Orandle M., Subbarao K. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J. Virol. 2012;86(5):2706–2714. doi: 10.1128/JVI.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Sui H., Fisher J., Yan Z., Liu X., Cho H., Joo N., Zhang Y., Zhou W., Yi Y., Kinyon J., Lei-Butters D., Griffin M., Naumann P., Luo M., Ascher J., Wang K., Frana T., Wine J., Meyerholz D., Engelhardt J. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J. Clin. Invest. 2010;120(9):3149–3160. doi: 10.1172/JCI43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitek N., von Messling V. Early cytokine mRNA expression profiles predict Morbillivirus disease outcome in ferrets. Virology. 2007;362(2):404–410. doi: 10.1016/j.virol.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitek N., Rudd P., Obojes K., Pillet S., von Messling V. Severe seasonal influenza in ferrets correlates with reduced interferon and increased IL-6 induction. Virology. 2008;376(1):53–59. doi: 10.1016/j.virol.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Aso H., Miyazawa K., Nagai Y., Watanabe K., Ohwada S., Kobayashi J., Yamaguchi T. Differential cytokine gene expression in CD4+ and CD8+ T cell subsets of calves. Vet. Immunol. Immunopathol. 2007;118:84–91. doi: 10.1016/j.vetimm.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Team R.C. R Foundation for Statistical Computing; 2013. R: A Language and Environment for Statistical Computing.http://www.R-project.org [Google Scholar]
- Vanders R.L., Gibson P.G., Wark P.A.B., Murphy V.E. Alterations in inflammatory, antiviral and regulatory cytokine responses in peripheral blood mononuclear cells from pregnant women with asthma. Respirology. 2013;18(5):827–833. doi: 10.1111/resp.12068. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., de Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. research:0034.1-0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Messling V., Svitek N., Cattaneo R. Receptor (SLAM [CD150]) recognition and the V protein sustain swift lymphocyte-based invasion of mucosal tissue and lymphatic organs by a morbillivirus. J. Virol. 2006;80(12):6084–6092. doi: 10.1128/JVI.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqoob P., Calder P.C. Cytokine production by human peripheral blood mononuclear cells: differential sensitivity to glutamine availability. Cytokine. 1998;10(10):790–794. doi: 10.1006/cyto.1998.0358. [DOI] [PubMed] [Google Scholar]
- Zeng H., Goldsmith C.S., Maines T.R., Belser J.A., Gustin K.M., Pekosz A., Zaki S.R., Katz J.M., Tumpey T.M. Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. J. Virol. 2013;87(5):2597–2607. doi: 10.1128/JVI.02885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


