Highlights
-
•
A duplex real-time RT-PCR test for both detection and differentiation of variant and virulent PEDV was developed.
-
•
The detection limit of the assay was 1 copy for both the virulent and variant strains.
-
•
The assay may be useful for control and prevention of PED in the United States.
Keywords: Porcine epidemic diarrhea virus (PEDV), Virulent PEDV, Variant PEDV, Duplex real-time RT-PCR, Differentiation
Abstract
Porcine epidemic diarrhea virus (PEDV) has caused significant economic losses in the US swine industry since May 2013. A new variant strain of PEDV emerged in the US in the late December, 2013. This variant strain of PEDV differs from the virulent strain of PEDV currently circulating in the US in 1170 nt of the 5’end of the S1 domain in the spike gene. Importantly, the variant PEDV caused significantly less mortality in piglets than the virulent PEDV, based on clinical observations. This suggests it may be a potential vaccine candidate for PED. Variant PEDV has been detected in samples from multiple states by our laboratory as well as other laboratories in the US. It is critical to detect and differentiate variant PEDV from the virulent PEDV during outbreaks to enhance control and to prevent PED associated disease. In this study, the development and validation of a duplex real-time RT-PCR assay for detection and differentiation of the variant and the virulent strains of PEDV currently circulating in the US was reported.
1. Introduction
Porcine epidemic diarrhea (PED) virus is a member of the order Nidovirales, family Coronaviridae, subfamily Coronavirinae, genus Alphacoronavirus. PED is a highly contagious diarrheal disease, characterized by severe watery diarrhea and high mortality in piglets. PED was originally identified in England in 1971 (Oldham, 1972). Since then, it has been reported in several European and Asian countries including China and Korea. Since 2010, a highly virulent strain of PEDV emerged in China and caused significant loss in the pig industry (Sun et al., 2012). In May 2013, this virulent strain of PEDV was recognized in the United States (US). By March 8 of 2014, PED had been detected in 27 US states and a total 4458 cases were confirmed (http://www.aasv.org/pedv/PEDv_weekly_report_140312.pdf). The disease has caused severe economic losses to the swine industry in the US. Recently the virus was found in Canada.
Studies have shown that pigs either naturally or experimentally infected with virulent strain of PEDV developed characteristic gross (thin and dilated intestinal walls) and histologic lesions (severe atrophy of villi) (Stevenson et al., 2013, Jung et al., 2014). Complete genomic analysis of the virulent PEDV strains from the US showed that they cluster in a single clade and are closely related with the AH2012 strain reported in China (Stevenson et al., 2013, Huang et al., 2013). Recently, we reported a new variant PEDV detected in Ohio (Wang et al., 2014). Clinical observation indicated that this new variant virus (OH 851) caused mild clinical disease with low mortality in newborn piglets (unpublished data), making it a potential vaccine candidate. Strain OH 851 is distinct from the virulent strains of PEDV in the US and is most closely related to CH/HBQX/10 reported in central China (Zheng et al., 2013), based on the phylogenetic analysis of the full-length spike gene (Wang et al., 2014). Further analysis showed that the strain OH851 differs from the virulent strains of PEDV in the first 1170 nucleotides (nt) of spike gene, indicating that at least two genotypes of PEDV are circulating in the US pigs (Stevenson et al., 2013, Huang et al., 2013, Wang et al., 2014).
Since effective vaccines are not currently available in North America, accurate diagnosis combined with biosecurity is the only reliable method for control and prevention of PED. Electron microscopy was widely used in the diagnosis of the initial outbreaks of PEDV (Pospischil et al., 1981). Since then, several methods have been developed for laboratory diagnosis of PEDV, including the direct immunofluorescence test for detection of PEDV antigen (Guscetti et al., 1998), enzyme-linked immunosorbant assays (ELISA) for detection of either PEDV antigen or antibodies (Carvajal et al., 1995), and reverse transcription-polymerase chain reaction (RT-PCR) (Kwon et al., 1997, Ishikawa et al., 1997). In recent years, real-time RT-PCR has increasingly been used to detect viral pathogens because of its advantages including high specificity and sensitivity, fast turnaround, and quantification of pathogen loads. Currently, the Animal Disease Diagnostic Laboratory in the Ohio Department of Agriculture uses a real-time RT-PCR method which targets the membrane (M) gene for detection of PEDV. However, since the M gene is highly conserved between the virulent and the variant strains of PEDV in the US, this real-time RT-PCR assay cannot differentiate between the two strains of PEDV. Therefore, the present study sought to develop and evaluate a duplex real-time RT-PCR method to distinct between the virulent and the variant strains of PEDVs.
2. Methods
2.1. Primer and probe design
Multiple-sequence alignments with the virulent and the variant strains of PEDV sequences detected in the US were carried out with the Mega 6.05 program. Primers were designed by targeting the conserved regions between virulent and variant PEDV, while the probes were designed by targeting the location where variant PEDVs have two deletions. For multiplexing, the probe for the virulent PEDV was labeled with the 5′-reported dye Cy5 and the 3′-quencher BHQ2, and the probe for the variant PEDV was labeled with the 5′-reported dye 6-carboxyfluorescein (FAM) and double quencher of the internal ZEN and 3‘Iowa Black® FQ (3IABkFQ). The sequences and amplicon sizes of the primers and probes are listed in Table 1 and Fig. 1 . Sequences of primers and probe for the real-time RT-PCR targeting M gene were listed in Table 1.
Table 1.
Sequences of primers and probes used in this study.
| Name | Primer/probe sequence | Amplicon size (bp) |
|---|---|---|
| PEDV S1 forward | 5′-AGGCGGTTCTTTTCAAAATTTAATG-3′ | |
| PEDV S1 reverse | 5′-GAAATGCCAATCTCAAAGCC-3′ | 191 for virulent PEDV |
| Virulent PEDV S1 probe | 5′-/5Cy5/TATTGGTGAAAACCAGGGTGTCAAT/3BHQ_2/-3′ | 179 for variant PEDV |
| Variant PEDV S1 probe | 5′-/56-FAM/TGGTTATCTACCTAGTATGAACTCCTCTAGC/3IABkFQ/-3′ | |
| PEDV-M-forward | 5′-CATGGGCTAGCTTTCAGGTC-3′ | |
| PEDV-M-reverse | 5′-CGGCCCATCACAGAAGTAGT-3′ | 181 for both virulent |
| PEDV-M-probe | 5′/56-FAM/CATTCTTGGTGGTCT TTCAATCCTGA/ZEN 3IABkFQ/3′ | and variant PEDVs |
| P160 forward | 5‘-ATCCATTAGTGATGTTGTGTTAG-3′ | 1075 for virulent PEDV |
| P161 reverse | 5‘-TAATATTAAACCTCAGAGCCTCTG-3′ | 1066 for variant PEDV |
PEDV: porcine epidemic diarrhea virus.
Fig. 1.
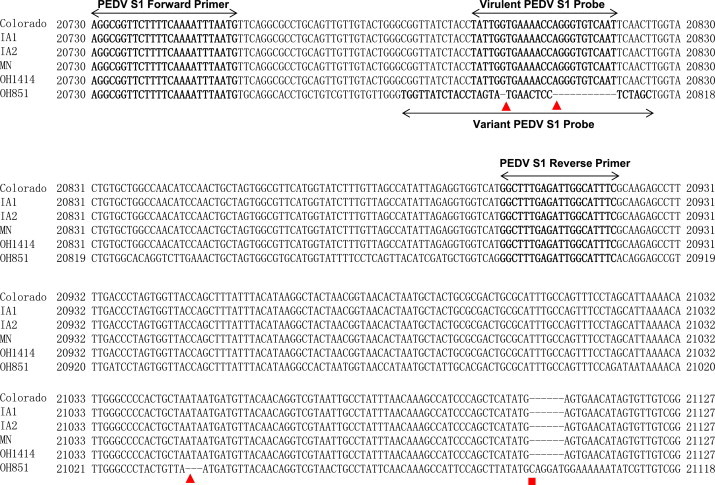
Consensus sequence alignment of virulent PEDVs of Colorado (accession no. KF272920), IA1 (accession no. KF468754), IA2 (accession no. KF468A753), MN (accession no. KF468752), OH1414 (accession no. KJ408801) isolates and variant (v) PEDV OH851 (accession no. KJ399978) isolates in the amplified region, including the primer and probe target sequences (bold). Red triangles indicate the three deletion sites, and red square indicates the insertion site. Names of primers and probes are indicated in the corresponding regions. Nucleotide numbering of virulent PEDV and variant PEDV is indicated based on their complete genome sequences.
2.2. RNA extraction and amplification conditions
RNA was extracted with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Amplification was performed with the QIAgen One Step RT-PCR kit (Valencia, CA, USA) in a SmartCycler II instrument. The amplification conditions were 50 °C for 30 min; 95 °C for 15 min; and 45 cycles of 94 °C, 10 s, 54 °C, 30 s, and 72 °C, 12 s. Primers (Integrated DNA Technologies, Coralville, Iowa, USA) at 240 nM and each probe (Integrated DNA Technologies, Coralville, Iowa, USA) at 240 nM were used for one reaction. The primer set and individual probe were tested first in a single real-time RT-PCR assay and then in a duplex real-time RT-PCR assay.
2.3. The specificity of duplex real-time RT-PCR
Intra-specificity of the duplex RT-PCR assay was determined using single probe of either virulent PEDV probe for variant PEDV strain or variant PEDV probe for virulent PEDV strain and two probes for both types of PEDVs for positive control. Inter-specificity of duplex RT-PCR assay was examined using various swine virus strains available in our lab. These viruses include porcine reproductive and respiratory syndrome virus, swine influenza virus (H3N2), transmissible gastroenteritis virus, encephalomyocarditis virus, porcine coronavirus HKU15, porcine parvovirus, and pseudorabies virus. For DNA virus porcine parvovirus and pseudorabies virus, DNA samples were extracted using DNeasy Blood & Tissue Kit (QIAgen, Valencia, CA, USA). In the assay, 2.5 μl RNA or DNA samples were used, 2.5 μl each of virulent PEDV OH1715 strain and variant PEDV OH851 were used as positive control in the duplex RT-PCR and 2.5 μl distilled water was used as negative control.
2.4. Validation of sensitivity
The PCR products amplified by using RNAs from OH851 (variant PEDV) and OH1715 (virulent PEDV) and the primer set P160–P161 covering the region where contains the majority of sequence variations between the virulent and variant PEDVs were cloned into the pCR 2.1 vector (Invitrogen, Carlsbad, CA, USA). The plasmids with the OH851 (pCR 2.1-OH851) or OH1715 (pCR 2.1-OH1715) genes were confirmed by sequencing. The detection limit of the real-time RT-PCR assay was determined through serial dilutions of each plasmid. Duplicates for each dilution were examined for separate and duplex reactions.
2.5. Field samples
Clinical fecal and intestinal samples submitted to the Animal Disease Diagnostic Laboratory in Ohio Department of Agriculture were processed for RNA extraction. RNA samples were first tested for PEDV by a real-time RT-PCR targeting the M gene. If positive, the duplex real-time RT-PCR was then used to differentiate the variant and the virulent strains of PEDV.
3. Results
3.1. Primer-probe set selection
Based on the sequence alignment and analysis of both virulent and variant PEDV partial S1 region, in addition to several sequence variations, there were 3 deletions and one insertion present in the variant PEDV as compared with virulent PEDV (Fig. 1). The primers were designed by targeting the conserved regions between the two viruses whereas the probes targeting the region where the first two-deletion regions were located in the variant strain of PEDV.
3.2. Specificity of the duplex real-time RT-PCR assay.
The duplex RT-PCR assay can detect specifically the virulent strain of PEDV by the Cy5 probe or the variant strain of PEDV by the FAM probe. In contract, the duplex RT-PCR did not cross-react with any other pig viruses used in the study, the Cy5 probe did not cross-react with variant PEDV strain, and the FAM probe did not cross-react with virulent PEDV strain (Table 2 ).
Table 2.
Specificity of the duplex real-time RT-PCR assay#.
| Virus name | Result | Note | |
|---|---|---|---|
| Intra-specificity | Virulent PEDV OH1715 (variant PEDV probe) | Negative | Single probe used |
| Variant PEDV OH851 (Virulent PEDV probe) | Negative | ||
| Inter-specificity | Transmissible gastroenteritis virus* | Negative | Two probes used |
| Porcine coronavirus HKU15 KY4813 | Negative | ||
| Porcine reproductive and respiratory syndrome virus OH28372 | Negative | ||
| Swine influenza virus OH27361(H3N2) | Negative | ||
| Encephalomyocarditis virus* | Negative | ||
| Porcine parvovirus* | Negative | ||
| Pseudorabies virus* | Negative |
Positive controls were run in the duplex real-time using RNA samples of both OH1715 and OH851; Negative control were run using distilled water.
Virus strains purchased from National Veterinary Service Laboratory.
3.3. Detection limit
The sensitivity of the duplex real-time RT-PCR assay was validated through serial dilutions of pCR 2.1-OH851 and pCR 2.1-OH1715 constructs. The detection limit was 1 copy for both variant and virulent strains of PEDVs. Standard curves were plotted using 10-fold serial dilutions of plasmid DNA of virulent and variant PEDV for the duplex real-time RT-PCR. As shown in Fig. 2 , there is a strong linear correlation (r 2 > 0.99) between C t values and the corresponding amount of plasmid copy numbers for both virulent and variant PEDV. The standard curves of virulent and variant PEDV were plotted with slopes of −3.40 and −3.31, respectively (Fig. 2A and B). The duplex real-time RT-PCR detected 1 genomic copy for both virulent and variant strain of PEDVs.
Fig. 2.
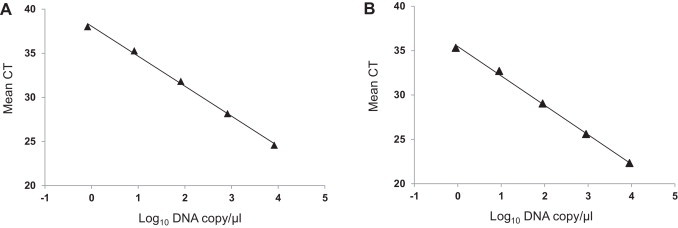
Standard curves for the duplex real-time RT-PCR assay. (A) Plasmid DNA standard curve for virulent PEDV strain, y = −3.40x + 38.07, r2 = 0.997 and (B) plasmid DNA standard curve for variant PEDV strain, y = −3.31x + 35.45, r2 = 0.998.
3.4. Application of the duplex real-time RT-PCR to clinical samples
A total of 295 positive samples tested by the real-time RT-PCR targeting on M gene were run again by the duplex real-time RT-PCR. Forty five samples tested positive for the variant PEDV and the remaining 250 samples were positive for the virulent PEDV. The results were confirmed by sequencing using P160–P161 primer set.
4. Discussion
PEDV causes diarrhea in pigs and high mortality in piglets. Since May of 2013, PEDV has been identified in the US resulting in severe economic losses to the US swine industry. Data on PED outbreaks have been collected and complied by the US National Animal Health Laboratory Network each week since June 17 of 2013. Since there is no PEDV vaccine available in North America, it is important to take biosecurity strategies to control PED. The findings of a recent study demonstrated that PEDV was found in 5.2% of trailers used to transport pigs, highlighting the importance of strict biosecurity (Lowe et al., 2014).
In the late December 2013, a variant strain of PEDV was detected in Ohio by our laboratory (Wang et al., 2014). Genetic analysis showed that this virus differed from the virulent strains of PEDV currently circulating in the US in the 5′ end of the S1 domain (mainly located in the first 1170 nt of spike gene), thus the real-time RT-PCR targeting on the M gene does not provide differentiation of the variant PEDV from virulent strain of PEDV. Therefore, the aim of this study was to develop a duplex real-time RT-PCR which would detect and differentiate the virulent strain from the variant strain of PEDV.
To optimize the ability of the assay to detect both variant and virulent strains of PEDV, the two primers were located in the conserved region of S1 region. To differentiate specifically the variant PEDV from the virulent strains of PEDV, probes were designed by targeting the highly variable region containing the first two deletions present in the variant PEDV (Fig. 1). It is possible that the third deletion and insertion site may also be used as a target for designing primers and probes (Fig. 1). The assay is specific for PEDVs, since cross-reaction with non-PED viral genomes used in this study was not detected. In addition, the assay is highly sensitive, being able to detect 1 copy in 25 μl reaction of either variant or virulent strains of PEDV.
The efficiency of the duplex real-time RT-PCR assay was determined by testing clinical samples which were positive by the real-time RT-PCR assay targeting the M gene. Of the 45 clinical samples that tested positive for the variant PEDV, all of them were confirmed by sequencing of the spike gene.
In conclusion, we have developed a duplex real-time RT-PCR assay that reliably detects and differentiates the virulent strain and variant strain of PEDV. This assay may be used by veterinary diagnostic laboratories to detect the new variant strain and the virulent strains of PEDV currently circulating in the US.
References
- Carvajal A., Diego R., Lanza I., Diego R., Rubio P., Carmenes P. Evaluation of a blocking ELISA using monoclonal antibodies for the detection of porcine epidemic diarrhea virus and its antibodies. J. Vet. Diagn. Invest. 1995;7:60–64. doi: 10.1177/104063879500700109. [DOI] [PubMed] [Google Scholar]
- Guscetti F., Bernasconi C., Tobler K., Van Reeth K., Pospischil A., Ackermann M. Immuno-histochemical detection of porcine epidemic diarrhea virus compared to other methods. Clin. Diagn. Lab. Immunol. 1998;5:412–414. doi: 10.1128/cdli.5.3.412-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737-13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K., Sekiguchi H., Ogino T., Suzuki S. Direct and rapid detection of porcine epidemic diarrhea virus by RT-PCR. J. Virol. Methods. 1997;69:191–195. doi: 10.1016/s0166-0934(97)00157-2. [DOI] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:668–671. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.H., Lee J.G., Han M.G., Kang Y.B. Rapid diagnosis of porcine epidemic diarrhea virus infection by polymerase chain reaction. J. Vet. Med. Sci. 1997;59:231–232. doi: 10.1292/jvms.59.231. [DOI] [PubMed] [Google Scholar]
- Lowe J., Gauger P., Harmon K., Zhang J., Connor J., Yeske P., Loula T., Levis I., Dufresne L., Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg. Infect. Dis. 2014;20:872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham J. Letter to the editor. Pig Farming. 1972:72–73. [Google Scholar]
- Pospischil A., Hess R.G., Bachmann P.A. Light microscopy and ultrahistology of intestinal changes in pigs infected with epizootic diarrhoea virus (EDV): comparison with transmissible gastroenteritis (TGE) virus and porcine rotavirus infections. J. Vet. Med. B. 1981;28:564–577. doi: 10.1111/j.1439-0450.1981.tb01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.H., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F.M., Huo J.Y., Zhao J., Chang H.T., Wang X.M., Chen L. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus field strains in central China during 2010–2012 outbreaks [in Chinese] Bing Du Xue Bao. 2013;29:197–205. [PubMed] [Google Scholar]


