Highlights
-
•
Sample quality is a critical consideration in nucleic acid-based diagnostic assay performance.
-
•
Educational efforts are important to ensure that specimen collection occurs in the correct disease timeframe.
-
•
FTA paper stabilizes nucleic acids for veterinary diagnostic testing.
Keywords: Diagnostic specimen, FTA Card, Bovine Respiratory Disease Complex
Abstract
In order to improve the analytic quality of respiratory specimens collected from cattle for nucleic acid-based diagnosis, a study was undertaken to verify realtime PCR efficiency of specimens collected and stabilized on FTA Cards™, filter paper which is treated chemically. Nucleic acids collected using FTA Cards without the need for a cold-chain or special liquid media handling provided realtime PCR results consistent (96.8% agreement, kappa 0.923 [95% CI = 0.89–0.96]) with the same specimens collected using traditional viral transport media and shipped on ice using the U.S. Department of Transportation mandated liquid handling requirements. Nucleic acid stabilization on FTA Cards was evaluated over a temperature range (−27 °C to +46 °C) for up to 14 days to mimic environmental conditions for diagnostic sample handling between collection and processing in a routine veterinary laboratory. No significant difference (P ≥ 0.05) was observed in realtime PCR cycle threshold values over the temperature range and time storage conditions for Bovine Viral Diarrhea virus, Bovine Respiratory Syncytial virus, Bovine Coronavirus, and Bovine Herpesvirus I. The four viruses evaluated in the study are associated with Bovine Respiratory Disease Complex where improvements in ease and reliability of specimen collection and shipping would enhance the diagnostic quality of specimens collected in the field, and ultimately improve diagnostic efficiency.
Testing of specimens collected from live animals for pathogen detection has become increasingly important for the control and prevention of production-limiting diseases. Although the detection of a specific virus does not correlate perfectly with presence of clinical disease, the identification of specific viruses which animals are exposed to can help determine production management decisions. Nucleic acid detection assays, predominately realtime PCR, have become a routine and highly valued component of laboratory diagnosis, providing increased timeliness and reliability for detection of a wide range of animal pathogens. The accuracy and diagnostic sensitivity of nucleic acid-based methodologies are however impacted critically by the quality and integrity of the diagnostic specimens. Factors contributing to low-quality, non-representative diagnostic specimens often include microbial overgrowth and enzyme-induced degradation of nucleic acids (Blacksell et al., 2004, Wang et al., 2011, Malentacchi et al., 2013, Pazzagli et al., 2013) which occur between the time of sample collection in the field and processing in the laboratory. This is of particular concern for veterinary diagnostics where animals, particularly wildlife and range animals such as sheep and cattle, are not located near a veterinary clinic, laboratory, or appropriate courier system. A variety of stabilizing agents and fixatives are available for the preservation of DNA and RNA, but for various reasons, including cost and complexity of use, they have not found wide application for collection and transport of veterinary diagnostic specimens. A commercial technology used initially to preserve DNA for long-term storage, has since been applied to stabilization of both RNA and DNA for use in forensic and diagnostic applications (Salvador and De Ungria, 2004, Picard-Meyer et al., 2007, Tack et al., 2007). The commercial product, FTA Cards™,1 is comprised of filter paper treated chemically, that when the biological specimen is applied to the paper, serves to lyse cells, denature proteins, and stabilize the nucleic acids. The nucleic acids in the specimen captured by and dried onto the FTA Card are protected from nucleases (Liang et al., 2006), oxidation, UV damage, and microbial overgrowth. Importantly, the stabilized specimen can be transported between the collection site and the laboratory without the need for a cold chain, liquid media handling concerns, time-sensitive shipping arrangements, or associated risk of disseminating an infectious agent. The FTA Card technology has been applied successfully to a range of tissues to capture and stabilize nucleic acids for subsequent testing, including human DNA (Dobbs et al., 2002), wildlife DNA (Smith and Burgoyne, 2004), pathogen nucleic acids from plant specimens (Ndunguru et al., 2005), and a range of human and animal pathogen nucleic acids, including from parasites, bacteria, and viruses. (Moscoso et al., 2004, Purvis et al., 2006, Picard-Meyer et al., 2007, Kraus et al., 2011).
The purpose of the current work was to verify the use of the FTA technology for collection and transport of diagnostic specimens used in the detection and identification of key viral pathogens associated with, or predisposing cattle to, Bovine Respiratory Disease (BRD) Complex (Fulton, 2009) including bovine viral diarrhea virus (family flaviviridae, genus pestivirus, BVDV), bovine respiratory syncytial virus (family paramyxoviridae, subfamily penumovirinae, genus pneumovirus, BRSV), bovine coronavirus (family coronaviridae, genus coronavirus, BCoV), and bovine herpesvirus 1 (family herpesviridae, subfamily alphaherpesvirinae, BHV-1). Bovine Respiratory Disease was targeted specifically for this study as a disease syndrome impacting the beef cattle industry (Fulton, 2009) and where early and reliable diagnosis is considered to be compromised due to the difficulty of collecting and submitting samples to the laboratory in a sufficiently suitable manner to fully insure the integrity and quality of the specimens. The FTA Card was considered an easy and reliable field-sampling and shipping option not requiring liquid handling, shelf-life sensitive laboratory media that must be pre-ordered or stored on-site by veterinary practitioners and producers, and having no requirements for cold storage. These characteristics are lacking for liquid media traditionally used, and thus the FTA Card approach offers an improvement in the practicality and feasibility of sampling from at-risk animals in the field, and in turn providing enhanced nucleic acid-based diagnostic and disease surveillance support for producers and practitioners in rural or remote environments.
The objective of the study was to verify that nucleic acids from specimens collected onto FTA Cards would be of equal or improved quality for PCR-based diagnosis (Horwood and Mahony, 2011, Thonur et al., 2012) as compared to specimens collected and transported using traditional liquid media. The study design included bench validation to demonstrate the successful recovery of target viral nucleic acids, both DNA and RNA, from the selected pathogens following stabilization on FTA Cards. The efficiency of nucleic acids recovered from FTA Cards and used in realtime PCR testing for the 4 representative BRD-associated viruses was compared to nucleic acids recovered from the traditional method of specimen collection and transport using viral transport media. The study assessed the stability of nucleic acids stored on FTA cards at a temperature range representing the extremes of environmental heat (13° to 46 °C) and cold specimen handling conditions (−7° to −27 °C), and a timeframe from specimen collection to laboratory processing consistent with the expected extremes of diagnostic sample shipping (7–14 days). For field validation, a combination of archival virus positive diagnostic specimens (n = 120) and specimens collected by cooperating large animal veterinarians who were provided with the FTA Cards and minimal sampling instruction were utilized (n = 81). The samples included respiratory tract specimens obtained from animals in beef herds presenting with clinical respiratory disease and consistent with current BRD herd surveillance and veterinary diagnostic practices. Archival samples included diagnostic swab fluids previously tested as realtime PCR positive for BRSV (n = 30), BHV-1 (n = 30), BCoV (n = 30), and BVDV (n = 30). Thirty virus negative archival samples were additionally tested for each of the four viruses. Samples were removed from a −70 °C freezer archive, thawed, and aliquots were spotted onto FTA cards and allowed to air-dry at room temperature. Additionally, eighty-one deep nasal swabs were collected in duplicate in the field using the traditional method of a Dacron or cotton-tipped swab, transferred into tubes of viral transport media, packaged with wet ice packs and absorbent packing materials, then shipped via overnight road delivery. The second swab collected at the same time from the same animal was transferred onto FTA Cards, allowed to air dry, and shipped in a sealed envelope (Fig. 1 ).
Fig. 1.
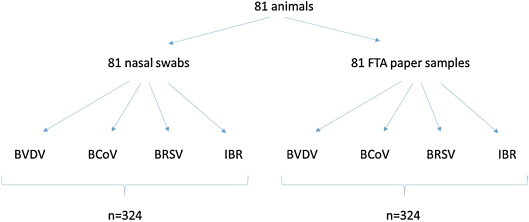
Nasal swabs were obtained from 81 calves. The sampling method was nasal swabs placed in viral transport media or onto FTA Cards. Each sample was tested for the presence of 4 viruses using independent real-time PCR assays, resulting in 324 PCR tests per sample collection method.
Reference strains of the 4 BRD-associated viruses used in the study included Bovine Viral Diarrhea virus TGAC 11-14-92 kindly provided by Dr. J. Ridpath, National Animal Disease Center (NADC) in Ames, Iowa; Bovine Respiratory Syncytial Virus strain NVSL 110BVD1011; Bovine Coronavirus strain NVSL 020BDV1001; and Bovine Herpesvirus-1 stain VR-188 (Lot number 214729) obtained from the American Tissue Culture Collection. Stock virus was serially diluted from 1:10 to 1:10−8 in viral transport media, and stored at −70 °C prior to use. Four drops of each virus dilution were spotted onto individual Indicator FTA Cards, each drop containing approximately 20 μl of fluid. The FTA Cards were air dried at room temperature for approximately 30 min. Each specimen collected onto FTA Cards and into viral transport media was extracted and tested in triplicate by the routine realtime PCR methodology used by the California Animal Health and Food Safety Laboratory System for diagnostic sample testing. An internal control, Xeno™,2 was added prior to processing in order to evaluate the extraction efficiency from the FTA cards. The FTA Cards were processed by punching 5 mm disks from the inoculated area of 0, placing the punches in individual wells of a 96 well microtiter plate, and adding 100 μl of Rapid Extraction Solution3 to release the nucleic acids from the FTA paper. After 20 min of incubation at room temperature, the plate was shaken for 5 min at 500 rpm. Fifty microliters of fluid was transferred into a new well and the plate with the disks was discarded. From this step on, the extraction procedure was identical for the FTA Card and Viral Transport Media specimens. The MagMAX-96™ viral Isolation4 Kit was used to prepare samples for realtime PCR testing following the recommendations of the manufacturer. Real-time PCR was performed using previously published primer sets for BVDV (Mahlum et al., 2002), BHV-1 (Brower et al., 2008) and BRSV (Boxus et al., 2005); the primer and probe information for BCoV was kindly provided by Dr K. Kurth (WVDL, Madison, WI). All PCR reactions utilized the Path-ID™ Multiplex One-Step RT-PCR Kit5 and were performed under the same conditions on an Applied Biosystems 7500 Fast Real-Time PCR System.6 The PCR thermocycler setting for the Path-ID™ Multiplex One-Step RT-PCR Kit was as follows: stage 1 at 50 °C for 10 min; stage 2 at 95 °C for 10 min; and stage 3 at 95 °C for 15 s; followed by stage 4 at 60 °C for 1 min; stages 3–4 were repeated for 40 cycles, whereas the commercial BVDV detection kit7 was performed per recommendation of the manufacturer. To evaluate operator variability, side-by-side testing was performed and compared between two laboratory technicians. No difference in nucleic acid recovery was detected between technicians.
The realtime PCR Ct values for the Xeno™ Internal Control provided a standard deviation of less than 0.3 cycle threshold (Ct) values, indicating consistent recovery of the viral nucleic acids from the FTA Cards. There was 100% agreement of PCR results for the FTA Card specimens and the specimens in viral transport media across all dilutions for the 4 individual viruses in the bench validation phase of the study. All virus dilution and nucleic acid recovery series showed linearity for the respective assays over at least 4 dilutions with R 2 values above 0.99. A t-test assessing variations among the different storage conditions revealed no statistically significant difference (P ≥ 0.05) in Ct values for the range of temperature and time storage conditions for any of the 4 viruses tested. Realtime PCR efficiencies calculated within the temperature studies were above 89% for each of the 4 viruses, and linearity measurements were consistently above R 2 values of 0.92. Inter-assay variability was highest in the BVDV assay with a standard deviation of 1.5 Cts among all dilutions evaluated and lowest in the BRSV assay with a calculated standard deviation of 0.5 Cts. Agreement for the archival samples when comparing FTA-stabilized to the original swab fluid specimens was 99.58%, kappa 0.992 (95% CI = 0.975–1.0), related to a single BHV-1 discrepant specimen that was detected as positive from viral transport media and negative from the FTA Card. (Table 1, Table 2 ). A noteworthy observation was higher Ct values indicating less viral nucleic acid detected for the archived samples on FTA cards for BRSV (average difference 6.8 Ct), BHV-1 (average difference 8.2 Ct), and BCoV (average difference 5.9 Ct). The lower detection sensitivity for FTA card samples generated from archived case material is presumed to be due to the dilution of the original swab material in viral transport media prior to archiving, as opposed to collecting directly onto FTA Cards at the time of sampling. In a recent field study (Foster et al., unpublished) 100% agreement was shown for FTA Card samples collected from nasal cavities of known BVDV PI and direct contact animals (n = 69) when compared to EDTA blood from the same animals. For the field validation component of the current study where samples were collected directly onto FTA Cards prior to laboratory submission (Fig. 1), agreement was 94.75% (307/324) based on dichotomous results (positive versus negative) comparing the specimens on FTA Cards to those in viral transport media; 4.01% (13/324) of specimens were PCR positive only using the FTA Card, and 1.23% (4/324) were positive only using the traditional viral transport media method of collection and shipping. Kappa comparison (Bland and Altman, 1986, von Kummer et al., 1996) yielded a 0.779 (95% CI = 0.678–0.880) agreement (good) between the FTA Cards and the viral transport media method of sample stabilization and transport for the field sample portion of the study (Table 2). Bovine Coronavirus was the most prevalent virus detected by realtime PCR during this phase of the study, with 60% animals testing positive (49/81). In 35 of the 49 positive animals, BCoV was detected using both sample handling approaches, in 13/49 cases only the FTA Card specimen yielded a positive result, and in 1/49 only the specimen in viral transport media tested PCR positive for BCoV. The overall agreement between the FTA Card specimens and VTM specimens in viral transport media for BCoV was 82.7% (67/81). For the remaining viruses, there was an insufficient incidence of the virus among the field specimens collected directly onto FTA Cards to statistically compare the FTA Card to the viral transport media approach to sample handling. A single specimen collected onto a FTA Card yielded a low concentration of BVDV (near the assay's defined limit of detection), leading to a predicted agreement of 98.76% (80/81 animals) for BVDV. Bovine Herpesvirus-1 was detected in a low concentration in two specimens in viral transport media, but not in the paired FTA specimens yielding a predicted agreement for BHV-1 of 97.53% (79/81). Bovine Respiratory Syncytial virus was detected using both the specimens in viral transport media and those on FTA Cards for 100% agreement; however the realtime PCR positive test result was for only one animal (Table 1, Table 2).
Table 1.
Comparison of real-time PCR results stratified into field (n = 81/virus) and archived diagnostic case samples (n = 30 positive plus 30 negative/virus).
| FTA Card positive |
FTA Card negative |
|||||||
|---|---|---|---|---|---|---|---|---|
| Swab fluid positive | BRSV | Corona | BHV-1 | BVDV | BRSV | Corona | BHV-1 | BVDV |
| Field samples | 1 | 35 | 0 | 0 | 0 | 1 | 2 | 1 |
| Archive samples | 30 | 30 | 29 | 30* | 0 | 0 | 1 | 0 |
| Swab fluid negative | BRSV | Corona | BHV-1 | BVDV | BRSV | Corona | BHV-1 | BVDV |
|---|---|---|---|---|---|---|---|---|
| Field samples | 0 | 13 | 0 | 0 | 80 | 32 | 79 | 80 |
| Archive samples | 0 | 0 | 0 | 0 | 30 | 30 | 30 | 30 |
Foster, personal communication.
Table 2.
Kappa values (95% CI) and percentage agreement for specimens collected into viral transport media compared to specimens collected onto FTA Cards. Table rows demonstrate overall agreement for all viruses by test sample origin; columns show overall agreement for test sample origin by individual virus.
| BRSV | Corona | BHV-1 | BVDV | Overall agreement | |
|---|---|---|---|---|---|
| Field samples (n = 81/virus) | 1.0 | 0.66 (0.51–0.82) | N/Aa | N/Aa | 0.78 (0.68–0.88) |
| 100% | 82.72% | 97.53% | 98.76% | 94.75% | |
| Archive samples (n = 60/virus) | 1.0 | 1.0 | 0.97 (0.90–1.0) | 1.0 | 0.99 (0.98–1.0) |
| 100% | 100% | 98.33% | 100% | 99.58% | |
| Overall agreement | 1.0 | 0.81 (0.71–0.90) | 0.94 (0.87–1.0) | 0.98 (0.94–1.0) | 0.92 (0.89–0.96) |
| 100% | 91.49% | 97.87% | 99.29% | 96.99% |
Not applicable; kappa calculation not performed due to a numerator or denominator value equal to zero.
The study provides bench data and initial field data to support the use of FTA Card technology as a simple and efficient alternative method for diagnostic sample collection and shipment. The overall agreement between FTA Cards and viral transport media for sample handling was 0.923 (95% CI = 0.888–0.958) (Table 1, Table 2). The primary drawback for the use of FTA Card technology is that the sample collected cannot be used for virus isolation attempts, and is suitable only for nucleic acid-based diagnostic testing. The FTA Card approach demonstrates equivalent and potentially superior performance in delivering nucleic acids for nucleic acid-based testing, while overcoming critical sample quality drawbacks frequently associated with inconsistent cold chains, producer/practitioner on-site storage or rapid accessibility to quality laboratory transport media and supplies, and shipping concerns related to liquid biological materials. In the course of the field study, an unexpected observation was that practitioners and producers selected animals for sampling that were showing relatively advanced clinical signs of respiratory disease, apparently assuming that clinical signs equated with better diagnostic recovery of the infectious agent. In the case of BRD, as with many other disease syndromes, the inciting agent is often not detectable by the time the clinical signs are severe or even observable. This sampling observation clearly indicates that in addition to ease and convenience of the sampling tool, an equally important criterion for quality specimens must include appropriate educational efforts to ensure that specimen collection occurs in the optimum timeframe and from the animals most likely to be shedding the specific infectious agent(s) of interest.
Conflict of interests
The authors declared no potential conflict of interest with respect to the research, authorship, or publication of this article.
Funding
Financial support for the execution of this study was provided by the Rustici Rangeland Foundation, 2012.
Footnotes
Whatman FTA Cards (WB120206), GE Healthcare, Township, NJ.
TaqMan® BVDV and Xeno RNA controls, Life Technologies, Carlsbad, CA.
Rapid Extraction Solution (AM 9775), Life Technologies, Carlsbad, CA.
MagMax96Viral RNA Isolation well kit (AM 1836-5), Life Technologies, Carlsbad, CA.
Path-ID™ Multiplex One-Step RT-PCR Kit, Life Technologies, Carlsbad, CA.
ABI 7500fast PCR system, Life Technologies, Carlsbad, CA.
VetMAX-Gold BVDV Detection Kit, Life Technologies, Carlsbad, CA.
References
- Blacksell S.D., Khounsy S., Westbury H.A. The effect of sample degradation and RNA stabilization on classical swine fever virus RT-PCR and ELISA methods. J. Virol. Methods. 2004;118:33–37. doi: 10.1016/j.jviromet.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- Boxus M., Letellier C., Kerkhofs P. Real Time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus. J. Virol. Methods. 2005;125:125–130. doi: 10.1016/j.jviromet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Brower A., Homb K.M., Bochsler P., Porter R., Woods K., Ubl S., Krueger D., Cigel F., Toohey-Kurth K. Encephalitis in aborted bovine fetuses associated with Bovine herpesvirus 1 infection. J. Vet. Diagn. Invest. 2008;20:297–303. doi: 10.1177/104063870802000306. [DOI] [PubMed] [Google Scholar]
- Dobbs L.J., Madigan M.N., Carter A.B., Earls L. Use of FTA gene guard filter paper for the storage and transportation of tumor cells for molecular testing. Arch. Pathol. Lab. Med. 2002;126:56–63. doi: 10.5858/2002-126-0056-UOFGGF. [DOI] [PubMed] [Google Scholar]
- Fulton R.W. Bovine respiratory disease research (1983–2009) Anim. Health Res. Rev. 2009;10:131–139. doi: 10.1017/S146625230999017X. [DOI] [PubMed] [Google Scholar]
- Horwood P.F., Mahony T.J. Multiplex real-time RT-PCR detection of three viruses associated with the bovine respiratory disease complex. J. Virol. Methods. 2011;171:360–363. doi: 10.1016/j.jviromet.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Kraus R.H., van Hooft P., Waldenstrom J., Latorre-Margalef N., Ydenberg R.C., Prins H.H. Avian influenza surveillance with FTA cards: field methods, biosafety, and transportation issues solved. JoVE. 2011 doi: 10.3791/2832. http://www.jove.com/details.php?id=2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.L., Quirk D., Zhou A. RNase L: its biological roles and regulation. IUBMB Life. 2006;58:508–514. doi: 10.1080/15216540600838232. [DOI] [PubMed] [Google Scholar]
- Mahlum C.E., Haugerud S., Shivers J.L., Rossow K.D., Goyal S.M., Collins J.E., Faaberg K.S. Detection of bovine viral diarrhea virus by TaqMan reverse transcription polymerase chain reaction. J. Vet. Diagn. Invest. 2002;14:120–125. doi: 10.1177/104063870201400205. [DOI] [PubMed] [Google Scholar]
- Malentacchi F., Pazzagli M., Simi L., Orlando C., Wyrich R., Hartmann C.C., Verderio P., Pizzamiglio S., Ciniselli C.M., Tichopad A., Kubista M., Gelmini S. SPIDIA-DNA: an external quality assessment for the pre-analytical phase of blood samples used for DNA-based analyses. Clin. Chim. Acta. 2013;424:274–286. doi: 10.1016/j.cca.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Moscoso H., Thayer S.G., Hofacre C.L., Kleven S.H. Inactivation, storage, and PCR detection of Mycoplasma on FTA filter paper. Avian Dis. 2004;48:841–850. doi: 10.1637/7215-060104. [DOI] [PubMed] [Google Scholar]
- Ndunguru J., Taylor N.J., Yadav J., Aly H., Legg J.P., Aveling T., Thompson G., Fauquet C.M. Application of FTA technology for sampling, recovery and molecular characterization of viral pathogens and virus-derived transgenes from plant tissues. Virol. J. 2005;2:45. doi: 10.1186/1743-422X-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazzagli M., Malentacchi F., Simi L., Orlando C., Wyrich R., Gunther K., Hartmann C.C., Verderio P., Pizzamiglio S., Ciniselli C.M., Tichopad A., Kubista M., Gelmini S. SPIDIA-RNA: first external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. Methods. 2013;59:20–31. doi: 10.1016/j.ymeth.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Picard-Meyer E., Barrat J., Cliquet F. Use of filter paper (FTA) technology for sampling, recovery and molecular characterisation of rabies viruses. J. Virol. Methods. 2007;140:174–182. doi: 10.1016/j.jviromet.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Purvis L.B., Villegas P., Perozo F. Evaluation of FTA paper and phenol for storage, extraction and molecular characterization of infectious bursal disease virus. J. Virol. Methods. 2006;138:66–69. doi: 10.1016/j.jviromet.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Salvador J.M., De Ungria M.C. DNA stability of forensic STR markers in FTA-extracted buccal DNA of betel-quid chewers. Oral Oncol. 2004;40:231–232. doi: 10.1016/j.oraloncology.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Smith L.M., Burgoyne L.A. Collecting, archiving and processing DNA from wildlife samples using FTA databasing paper. BMC Ecol. 2004;4:4. doi: 10.1186/1472-6785-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack L.C., Thomas M., Reich K. Automated forensic DNA purification optimized for FTA card punches and identifiler STR-based PCR analysis. Clin. Lab. Med. 2007;27:183–191. doi: 10.1016/j.cll.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Thonur L., Maley M., Gilray J., Crook T., Laming E., Turnbull D., Nath M., Willoughby K. One-step multiplex real time RT-PCR for the detection of bovine respiratory syncytial virus, bovine herpesvirus 1 and bovine parainfluenza virus 3. BMC Vet. Res. 2012;8:37. doi: 10.1186/1746-6148-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kummer R., Holle R., Gizyska U., Hofmann E., Jansen O., Petersen D., Schumacher M., Sartor K. Interobserver agreement in assessing early CT signs of middle cerebral artery infarction. Am. J. Neuroradiol. 1996;17:1743–1748. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zoueva O., Zhao J., Ye Z., Hewlett I. Stability and infectivity of novel pandemic influenza A (H1N1) virus in blood-derived matrices under different storage conditions. BMC Infect. Dis. 2011;11:354. doi: 10.1186/1471-2334-11-354. [DOI] [PMC free article] [PubMed] [Google Scholar]


