Highlights
-
•
This is the first report of a reverse transcription-nested real time PCR for CDV detection.
-
•
The technique was able to detect CDV RNA from vaccine and field strains.
-
•
This new technique (RT-nqPCR) was sensitive and specific.
-
•
Different clinical samples could be analysed by the method.
-
•
The method here described should be an important tool for diagnosis of canine distemper.
Keywords: Dog, Canine distemper virus, Diagnosis, Detection, RT-PCR
Abstract
Canine distemper virus (CDV) is the cause of a severe and highly contagious disease in dogs. Practical diagnosis of canine distemper based on clinical signs and laboratory tests are required to confirm CDV infection. The present study aimed to develop a molecular assay to detect and differentiate field and vaccine CDV strains. Reverse transcription followed by nested real time polymerase chain reaction (RT-nqPCR) was developed, which exhibited analytical specificity (all the samples from healthy dogs and other canine infectious agents were not incorrectly detected) and sensitivity (all replicates of a vaccine strain were positive up to the 3125-fold dilution – 100.7 TCID50). RT-nqPCR was validated for CDV detection on different clinical samples (blood, urine, rectal and conjunctival swabs) of 103 animals suspected to have distemper. A total of 53 animals were found to be positive based on RT-nqPCR in at least one clinical sample. Blood resulted in more positive samples (50 out of 53, 94.3%), followed by urine (44/53, 83.0%), rectal (38/53, 71%) and conjunctival (27/53, 50.9%) swabs. A commercial immunochromatography (IC) assay had detected CDV in only 30 conjunctival samples of these positive dogs. Nucleoprotein (NC) gene sequencing of 25 samples demonstrated that 23 of them were closer to other Brazilian field strains and the remaining two to vaccine strains. A single nucleotide sequences difference, which creates an Msp I restriction enzyme digestion, was used to differentiate between field and vaccine CDV strains by restriction fragment length polymorphism (RFLP) analysis. The complete assay was more sensitive than was IC for the detection of CDV. Blood was the more frequently positive specimen and the addition of a restriction enzyme step allowed the differentiation of vaccine and Brazilian field strains.
1. Introduction
Canine distemper (CD) is a severe and highly contagious disease that affects carnivores and is frequently fatal. The viral pathogen, canine distemper virus (CDV), belongs to genus Morbillivirus in the family Paramyxoviridae. Live attenuated vaccines were developed with classical strains (Onderstepoort, Snyder Hill and Rockborn) in the 1950s, which have been used widely to control the disease. Nevertheless, CDV continues to cause severe outbreaks worldwide, mainly in regions with poor vaccine coverage (Martella et al., 2008). Dogs with CD develop systemic clinical signs few days after infection. These clinical findings are variable, depending on the virulence of the virus strain, environmental conditions, age and immune status of the host. Acutely infected dogs shed the virus in several body secretions, thereby transmitting the virus to other animals (Greene and Appel, 2006).
Currently, practical diagnosis of distemper is still based on systemic clinical signs (respiratory, gastrointestinal, neurological or dermatological). A history of an unvaccinated puppy with depression and anorexia supports a suggestive diagnosis (Greene and Appel, 2006). However, many dogs lack the classical clinical presentation and/or are infected with other agents that are responsible for similar signs at the time of the first clinical visit to the veterinary, rendering CD diagnostic difficult (Amude et al., 2007). Some routine laboratorial tests may provide some useful clinical information because CD findings include partial and absolute lymphopenia and body inclusions in the buffy coat cells of conjunctival imprints. However, these tests are not conclusive and do not confirm infection, especially in atypical cases (Greene and Appel, 2006).
CDV can be specifically detected in clinical samples using different laboratory procedures, such as virus isolation, immunological techniques (immunofluorescence and immunochromatography) and molecular biology assays, the last of which mainly involves reverse transcription-polymerase chain reaction (RT-PCR) (Frisk et al., 1999, An et al., 2008). Virus isolation is fastidious and difficult to perform for routine diagnosis. Immunofluorescence has been the preferred technique up to a few years ago. In 2008, however, an immunochromatography rapid test was developed, which has been used as a routine test because it is fast and user friendly (An et al., 2008). In addition, different RT-PCR protocols have been developed to sensitive and specific CDV RNA detection in dogs (Frisk et al., 1999, Gebara et al., 2004, Shin et al., 2004, Castilho et al., 2007). These assays were targeted to the nucleoprotein (NC) gene, which encodes the most abundant structural viral protein and has a highly conserved nucleotide sequence (Yoshida et al., 1998). Furthermore, three different amplification schemes were described as practical diagnostic tools, as follows: RT-PCR with electrophoresis (Frisk et al., 1999, Calderon et al., 2007, Demeter et al., 2007), nested RT-PCR with electrophoresis (Shin et al., 2004, Józwik and Frymus, 2005, Kapil et al., 2008) and real time RT-PCR (Elia et al., 2006, Scagliarini et al., 2007).
The objective of the present study was to develop reverse transcription followed by a nested real time PCR (RT-nqPCR) for the sensitive and specific detection of CDV. The protocol was tested with different clinical samples (blood, urine, rectal and conjunctival swab) to define which sample was most effective in detecting CDV-infected dogs. Furthermore, an additional Msp I restriction enzyme digestion step was included to differentiate CDV field and vaccine strains using restriction fragment length polymorphism (RFLP) analysis.
2. Materials and methods
2.1. Vaccines
The following vaccines were kindly provided by the suppliers: Multi-Dog (Hertape Callier, Juatuba, Brazil), Vencomax 11 (Vencofarma, Londrina, Brazil), Duramune Max-5 (Zoetis, Campinas, Brazil) and Quantum Dog Da2PPvL (MSD Saude Animal, São Paulo, Brazil).
2.2. Animals and samples
Samples were collected from 127 dogs from the Veterinary Hospital of ULBRA University (Canoas, Rio Grande do Sul, Brazil) from March 2010 to June 2011. They were divided into the following two groups:
-
(1)
Twenty-four healthy dogs underwent neutering or spaying procedures.
-
(2)
One hundred and three dogs with at least one clinical sign suggestive of CDV or other viral infection (ocular and nasal discharge, coughing, dyspnoea, pneumonia, diarrhoea, vomiting, dermal pustules, enamel hypoplasia and hyperkeratosis of the foot pads and nose, neurologic disorder), with or without other generic disease signs (transient fever, loss of appetite, slight depression, tonsillitis) (Martella et al., 2008). Specific neurologic signs (circling, head tilt, nystagmus, partial or complete paralysis, convulsions, dementia, involuntary jerky twitching or contraction of muscles and convulsions preceded by chewing-gum movements of the mouth) and additional information about the animals (such as age, gender, breed and vaccination status) were also recorded.
After clinical examination, the following clinical samples were collected from all animals: 5 mL of whole blood (collected by jugular venepuncture using ethylenediaminetetraacetic acid – EDTA – as anticoagulant), 2 mL of urine, one rectal swab and two conjunctival swabs. One of the conjunctival swabs was freshly used for CDV antigen detection using a commercial immunochromatography (IC) assay. The remaining clinical samples were stored at −20 °C for further molecular analysis. Dogs with clinical signs of CD received supportive therapy in the Hospital or at home (when necessary) and were reassessed during treatment.
The project was approved by the Ethical Committee for Animal Use in Research from the Universidade Luterana do Brasil under study number CEP-ULBRA 2009-023A.
2.3. Immunochromatography (IC) assay
The qualitative detection of CDV antigen in one of the conjunctival samples was performed using IC assay Antigen Rapid CDV Ag Test Kit (Animal Genetics, Suwon, Korea) according to the manufacturer's instructions and previous description (An et al., 2008).
2.4. Primers and probe selection
Nucleotide sequence analysis of the CDV NC gene was performed using sequence data obtained from reference strains and Brazilian field samples published previously (Castilho et al., 2007, Headley et al., 2009). The following sequences were analysed: IP1407 (AY738624), IP1682 (DQ005126), IP2376 (DQ005131), IP2392 (DQ005127), IP2397 (AY738653), IP2705 (DQ005132), IP3045 (DQ005128), IP3258 (DQ005133), IP3288 (DQ005129), IP3683 (DQ005134), IP4712 (DQ005130), 2544/Hans (AJ009656), A75/17 (AF164967), 98-2654 (AY466011), 98-2645 (AY445077), Kaohsiung (DQ522030), Lederle (AY738625), Onderstepoort (AF305419), Snyder Hill (GU138403), Pingtung (DQ435615), TN-N (AY390348), AMA1-UEL/BR (EF193648), AMA4-UEL/BR (EF197736), st5804 (AY386315), and 007Lm (AB474397.1). All sequences were edited with EditSeq and aligned with MegAlign (DNAstar Lasergene package, Madison, WI, USA). Primers described in previous reports (Frisk et al., 1999, Shin et al., 2004, Elia et al., 2006, Castilho et al., 2007) were also compared to the complete alignment. Four primers and one probe were chosen after the comparative analysis between primers and the aligned sequences published previously. The primer sequences were almost identical to the original references, with few modifications and degenerations to recognise specific nucleotide sequences present in Brazilian field strains reported previously (Castilho et al., 2007). Probe sequence was the same as in the original article (Elia et al., 2006) (Table 1 ).
Table 1.
Oligonucleotide primers and probe used in the molecular assays.
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| CDV-1F (CDV-F)a | 5′-ACT GCT CCT GAT ACT GC-3′ | Castilho et al. (2007) |
| CDV-2R (CDV-R)a | 5′-TTC AAC ACC RAC YCC C-3′ | Castilho et al. (2007) |
| CDV-3F (p1)a | 5′-ACA GRA TTG CYG AGG ACY TRT-3′ | Frisk et al. (1999) |
| CDV-4R (p2)a | 5′-CAR RAT AAC CAT GTA YGG TGC-3′ | Frisk et al. (1999) |
| CDV-Pb | 5′-FAM-ACCCAAGAGCCGGATACATAGTTTCAATGC-TAMRA-3′ | Elia et al. (2006) |
Original denomination in the respective reference. Bold characters indicate modifications in the original sequences of the references.
2.5. RNA extraction
Total RNA of the clinical samples and viral vaccines was purified via a standard silica/GuSCN-based procedure (Boom et al., 1990) using the commercial kit Newgene (Simbios Biotecnologia, Cachoeirinha, Brazil). Briefly, 100 μL of each blood sample was mixed to 400 μL of lysis buffer (GuSCN 5 M, Tris–HCl 0.1 M [pH 8.0], EDTA 0.5 M and Triton X-100), incubated at 56 °C for 10 min before 20 μL of silica suspension was added and mixed. Tubes were centrifuged at 8609 × g for 30 s. The pellet was washed once with 500 μL washing buffer (GuSCN 5 M and Tris–HCl [pH 8.0] 0.1 M), once with 150 μL washing buffer, twice with 150 μL 75% ethanol and once with 150 μL of ethanol absolute. Silica suspension was dried at 56–60 °C for 15 min. RNA was eluted with 50 μL of TE buffer (Tris–HCl 0.1 M, EDTA 0.5 mM) after incubation at 60 °C for 5 min.
2.6. RT-nqPCR
Reverse transcription and the first round of PCR was performed in 30 μL with 75 mM of KCl, 50 mM of Tris–HCl (pH 8.3), 3 mM MgCl2, 2.5 mM DTT, 0.1 mM dNTPs, 0.2 μM of each primer, 24 U of MMLV-RT (Life Technologies, Carlsbad, USA), 4 U RnaseOut (Life Technologies, Carlsbad, CA, USA), 1 U Taq DNA polymerase (Ludwig Biotecnologia, Alvorada, Brazil) and 2 μL of extracted RNA. Amplification was carried out in Veriti 96 thermo cycler (Applied Biosystems, Norwalk, CT, USA) with the following conditions: 1 cycle at 37 °C for 30 min and 10 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 40 s and polymerisation at 72 °C for 1 min.
The second round of amplification (nested real time PCR) was carried out in 50 μL with 50 mM KCl, 10 mM Tris–HCl (pH 8.3), 1.5 mM MgCl2, 0.1 mM dNTPs, 0.2 μM of each primer and 0.15 μM of the probe, 1 U Taq DNA polymerase (Ludwig Biotecnologia, Alvorada, Brazil) and 2 μL of the first amplification reaction. Thermal cycling was performed using the StepOnePlus™ Real Time PCR System (Applied Biosystems, Norwalk, USA) with the following steps: 1 cycle at 94 °C for 3 min, 40 cycles of denaturation at 94 °C for 20 s, annealing at 60 °C for 40 s, polymerisation at 72 °C for 1 min and a final extension cycle at 72 °C for 5 min.
2.7. Sequencing
The amplification products of the four vaccines and 25 randomly selected positive RT-nqPCR samples were sequenced according the following procedures: forward and reverse sequencing reactions carried out using 15 to 30 ng of template DNA from PCR amplimers, 3.2 pmol of the inner primer and 4 μL of Big Dye Terminator v3.1 Cycle Sequencing (Applied Biosystems, Norwalk, USA) in a final volume of 20 μL. The sequencing rounds were performed in a Veriti 96 thermo cycler (Applied Biosystems, Norwalk, USA) with an initial step of 95 °C for 3 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 240 s. The samples were precipitated by ethanol/EDTA/sodium acetate protocol, diluted in 10 μL of formamide Hi-Di, denatured (95 °C for 2 min) and injected in the automated DNA sequencing ABI 3130 XL Genetic Analyzer (Applied Biosystems, Norwalk, USA). The sequence data were collected using the Data Collection program v1.0.1 with the parameters Dye Set “Z”. Quality analysis was performed using the Sequencing Analysis v.5.3.1 software by evaluating the main technical parameters, such as raw data, electropherogram and quality value of the sequenced bases (Applied Biosystems, Norwalk, USA). Sequences of the same amplicon (sense and antisense primers) were edited and assembled using SeqMan software (DNAStar, Madison, USA). All nucleotide sequences were edited, assembled and analysed using the Clustal W method available in the Bioedit software package. Brazilian and reference nucleotide sequences (previously described in Section 2.4) were compared using the neighbour-joining method with 1000 bootstrapping replicates for phylogenetic analysis (MEGA software version 5.0). Nucleotide and amino acid sequences were also aligned using the Clustal W method and compared for similarity analysis. NC gene fragments of all sequenced samples were deposited in Genbank under accession numbers JQ790531–JQ790555.
2.8. Restriction enzyme digestion
Digestion with Msp I restriction enzyme was performed after RT-nqPCR following the manufacturer's instructions (New England Biolabs, Ipswich, MA, USA). Restriction enzyme reactions were submitted to electrophoresis in 10% polyacrylamide gels, followed by silver nitrate staining.
2.9. Statistical analysis
Data analysis was conducted using Fisher's exact test or chi-square when indicated. In general, frequencies were compared between CDV positive and CDV negative groups. All p values presented were two-tailed and p < 0.05 was considered statistically significant.
3. Results
3.1. RT-nqPCR design and optimisation
Primers and probes were tested for the analysis of the four vaccines and five clinical samples (two positive and three negative based on the IC assay) via reverse transcription followed by a nested real time PCR (RT-nqPCR). The following combinations of primers/probe were used: CDV-1F and CDV-2R for RT and first PCR amplification; CDV-3F, CDV-4R and probe for second real time PCR amplification. In this system, CDV-positive samples (four vaccine and two positive IC assay field strains) presented a positive curve after Taqman amplification, with an expected amplified fragment of approximately 287 bp after gel electrophoresis (Frisk et al., 1999).
RNA samples extracted from one vial of Quantum Dog DA2PPvl + Cv live vaccine containing approximately 104.2 Tissue Cell Infectious Dose (TCID50) of CDV, according to the manufacturer's instructions, were serially diluted 5-fold (up to the 390,625-fold dilution) and submitted to the two following amplification procedures: RT-nqPCR proposed in the present study and the “one-step” RT-PCR method described previously (Frisk et al., 1999). CT values of the nested real time RT-PCR were measured in triplicate and were plotted against the dilutions of the vaccine RNA sample. RT-nqPCR presented a positive signal in all replicates up to the 3125-fold dilution (100.7 TCID50) and in one replicate of the 15,625-fold dilution (100.006 TCID50). Furthermore, this assay demonstrated to be 2 orders (25-fold) of magnitude more sensitive than was the “one-step” RT-PCR (data not shown).
Experiments were undertaken to assess diagnostic criteria such as specificity and reproducibility. Samples known to be positive for other infectious agents were run in this assay to identify any cross-reactivity with other targets. These samples were positive controls from other assays (canine coronavirus type I, canine coronavirus type II, canine parvovirus 2, reovirus and rotavirus). No cross-reactivity was observed with any other viruses. Similarly, no detectable fluorescence signal was obtained in the negative controls (negative samples and no template controls), confirming that the assay was specific for the detection of CDV RNA. Reproducibility was assessed in three experiments performed in triplicate on different days. Standard curves in all runs confirmed the previously obtained results, covering a range of four orders of magnitude (up to 15,625-fold dilution) and showing linearity over the entire quantitative range. In the three experiments, the mean of the coefficient of linear regression (R 2) was 0.94 (minimum of 0.92 and maximum of 0.96) and of the slope was −4.74 (minimum of −4.65 and maximum of −4.90).
3.2. Detection of CDV by IC assay and RT-nqPCR
IC assay and RT-nqPCR were used to detect CDV in different clinical samples (blood, urine, rectal and conjunctival swabs) of the 127 dogs. IC assay did not detect the CDV antigen in the conjunctival sample of the 24 healthy dogs, nor in 73 of the 103 dogs with clinical signs of viral disease. IC assay was positive in the conjunctival swab sample of the remaining 30 dogs suspected to have CD (29.1%). On the other hand, all 24 healthy dogs and 50 dogs with viral disease suggestive of clinical signs were negative according to the RT-nqPCR performed with four different samples (blood, urine, conjunctival and rectal swabs). The remaining 53 dogs suspected to have CD (51.5%) were positive based on this same assay in at least one clinical sample. A total of 23 samples (22.3%) were found to be positive in all four clinical samples, 14 (13.6%) in tree clinical samples, nine (8.7%) in two samples and seven (6.8%) in only one sample (blood or urine). A decreasing number of positive results were obtained using blood (50 samples), urine (44 samples), rectal (38 samples) and conjunctival swabs (27 samples) when the results for the different clinical samples were compared (Table 2 ). In the comparison of the four different samples tested from the same dog, significant difference (p < 0.05) was observed only between conjunctival swabs and the other samples, but not among blood, urine and rectal swabs.
Table 2.
RT-nqPCR results with the four different samples from the dogs with clinical signs suggestive of CDV infection.
| CDV | Blood | Urine | Rectal swab | Conjunctival swab | Samplesc, n (%)a | Samplesc, n (%)b |
|---|---|---|---|---|---|---|
| Positive | + | + | + | + | 23 (25.8) | 23 (22.3) |
| + | + | + | − | 10 (11.2) | 10 (9.7) | |
| + | + | − | + | 1 (1.1) | 2 (1.9) | |
| + | − | + | + | 1 (1.1) | 2 (1.9) | |
| + | + | − | − | 4 (4.5) | 6 (5.8) | |
| + | − | + | − | 2 (2.2) | 3 (2.9) | |
| + | − | − | − | 4 (4.5) | 4 (3.9) | |
| − | + | − | − | 1 (1.1) | 3 (2.9) | |
| Negative | − | − | − | − | 42 (47.2) | 50 (48.5) |
| Total | 88 | 103 | ||||
Column that shows only the animals from which it was possible to analyse all clinical specimens.
Column that shows all animals and clinical specimens analysed in the study (including animals with one or more missed clinical samples).
Seventeen clinical samples were missed and could not be evaluated. In 14 cases, only one clinical sample was missed (seven urine, four rectal swabs, two conjunctival swabs, one blood). In one case, three samples were missed (blood, rectal and conjunctival swabs).
IC assay and RT-nqPCR results were also compared using the same clinical samples (conjunctival swabs) with an overall concordance of 78%. The analysis of the discordant results showed a significant number (22) of conjunctival swab samples that were positive in one assay and negative in the other. The overall concordance was similar regarding the comparison between the IC assay using conjunctival swabs and nested-RT-PCR using other clinical samples (79.6% for anal swabs, 82.3% for urine and 77.2% for blood). However, the analysis of the discrepant results demonstrated that many positive samples in the RT-nqPCR were negative in the IC assay (21.8%, 16.7% and 16.3% for blood, urine and rectal swab samples, respectively), whereas only a few cases (one for blood and urine, four for rectal swab) had a positive result in the IC assay with a negative result in the RT-nqPCR (Table 3 ).
Table 3.
Comparative analysis of the IC and nested real time RT‘-PCR using different clinical samples.
| IC assay result | RT-nqPCR – different samples |
|||||||
|---|---|---|---|---|---|---|---|---|
| Conjunctival swab |
Anal swab |
Urine |
Blood |
|||||
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| Positive | 17 | 12 | 22 | 4 | 28 | 1 | 28 | 1 |
| Negative | 10 | 61 | 16 | 56 | 16 | 51 | 22 | 50 |
| Samples (n) | 100 | 98 | 96 | 101 | ||||
Considering blood as the reference clinical sample to be used in the RT-nqPCR, a more detailed analysis of the discrepant results was performed. Among the 22 dogs that were positive in the RT-nqPCR and negative in the IC assay, CDV was detected in three samples (urine, rectal and conjunctival swabs) of nine dogs, in two samples of five dogs (urine and rectal swab in four, rectal and conjunctival swab in one) and in one sample of four additional dogs. Eight of these 22 dogs developed the characteristic neurological signs (myoclonus, seizures, etc.) of the disease and were euthanised in extremis. In the only dog with a positive result for the IC assay and a negative result for the RT-nqPCR using blood, CDV was not detected in any other sample via molecular assay. This animal died soon after clinical evaluation without any other clinical signs of CDV infection.
3.3. Sequence analysis and restriction fragment length polymorphism (RFLP)
The amplified DNA of the four vaccines and 25 RT-nqPCR random positive samples were compared to those of 25 other reference and Brazilian strains described previously. The phylogenetic tree analysis revealed that the vaccine strain from Duramune Max-5 and Quantum Dog clustered with the classical vaccine strains (Lederle, Onderstepoort, and Snyder Hill), whereas Multi-dog and Vencomax 11 clustered with strains A75-17 and 98-2645. Two of the field samples also clustered with the first group (Lederle, Onderstepoort, and Snyder Hill), while the other 23 samples clustered with a group of Brazilian dog strains described previously (Castilho et al., 2007, Headley et al., 2009) (data not shown).
In the analysis of the aligned sequences, single nucleotide polymorphisms (SNPs) were observed at different nucleotide positions (Fig. 1 ). In two positions (nucleotides 809 and 977 of the reference strain Snyder Hill, GU138403, as highlighted in Fig. 1), all vaccine strains had nucleotide G and C, while all Brazilian dog strains (including all sequences of the Gene Bank) had A and T, respectively. Because the presence of C in position 977 provides a site for the Msp I restriction enzyme, it was selected and used to digest the amplified DNA to differentiate the vaccine and field samples via restriction fragment length polymorphism (RFLP) analysis. All four vaccines and two clinical samples that grouped with the classical strains presented with one common restriction fragment length pattern, while the amplified DNA of the other 51 positive dogs (23 described previously and 28 from this work) presented with a different restriction fragment length pattern (Fig. 2 ).
Fig. 1.
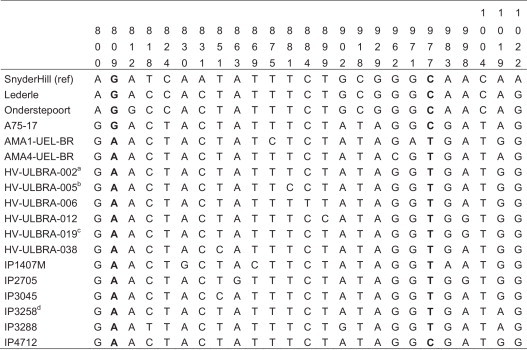
Polymorphic sites in the sequenced fragments of the N gene. The position of each polymorphic site is shown above the figure numbered according to the reference sequence of strain Snyder Hill (NCBI accession number: GU138403). aThere were 18 additional sequences identical to HV-ULBRA-002: HV-ULBRA-003, HV-ULBRA-004, HV-ULBRA-011, HV-ULBRA-013, HV-ULBRA-018, HV-ULBRA-024, HV-ULBRA-026, HV-ULBRA-029, HV-ULBRA-040, HV-ULBRA-041, HV-ULBRA-042, HV-ULBRA-046, HV-ULBRA-048, HV-ULBRA-049, IP1682, IP2376, IP2392, IP2397M. bThere was one more sequence identical to HV-ULBRA-005: HV-ULBRA-009. cThere were two additional sequences identical to HV-ULBRA-019: HV-ULBRA-020, HV-ULBRA-023. dThere was one more sequence identical to IP3258: IP3683.
Fig. 2.
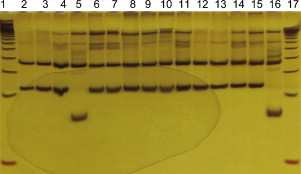
Polyacrylamide gel stained with silver nitrate showing the different banding patterns after RT-PCR followed by restriction endonuclease digestion with. Numbers 1 and 17 – 50 bp molecular marker; numbers 2–4, 6–15 – pattern of the Brazilian field strains; numbers 5 and 16 – vaccine samples.
4. Discussion
The diagnosis of CD is based on clinical signs and should be confirmed with available laboratory tests. Haematological and cytological analyses are still recommended and can help to confirm the disease in some clinical situations. A more specific laboratory diagnosis is based on viral isolation and detection of CDV antigens or nucleic acids in scrapings and body fluids (Greene and Appel, 2006). For many years, antigen detection has been performed via direct immunofluorescence assay (IFA), although false negative results were common in some clinical conditions (Józwik and Frymus, 2005). Recently, a rapid immunochromatography (IC) test with good performance was developed (An et al., 2008), which has been used mostly in veterinary laboratories and clinics worldwide because of its simple and fast analytical characteristics.
On the other hand, different RT-PCR protocols were developed in the last 15 years, which have been used to obtain fast, sensitive and specific CDV detection in different clinical samples of infected dogs (Frisk et al., 1999, Elia et al., 2006, Scagliarini et al., 2007). RT-nested-PCR has proven to be more sensitive than “one-step” RT-PCR in detecting CDV in different clinical samples (Shin et al., 2004, An et al., 2008). In these previous studies, the NC gene was the main target for the RT-PCR because it is one of the most conserved regions of the CDV genome, presenting with high homology among different strains around the world (Yoshida et al., 1998, Castilho et al., 2007).
In the present study, a reverse transcription followed by a nested real time PCR (RT-nqPCR), based on the amplification of the NC gene, was developed and tested for CDV detection in different clinical samples. The whole procedure used primers and probes described previously and involved testing via inclusion assays with field and vaccine strains from different CDV lineages (Frisk et al., 1999, Elia et al., 2006, Castilho et al., 2007). RT-nqPCR detected all vaccine strains and Brazilian CDV field samples, displaying greater sensitivity than did the “one-step” RT-PCR (without a nested amplification). It was also faster than other RT-nested-PCR protocols because of the addition of the probe in the second amplification and “real time” analysis. Although the method is based on two amplification rounds, the strategy of using a first round of RT-PCR amplification, with ten cycles, followed by a second round of real-time PCR, with 40 cycles, presented very good linear and reproducible results, similar to another real-time technique described previously (Elia et al., 2006). A comparison with this developed real time RT-PCR was attempted, although it was not successful because the majority of the positive Brazilian field samples presented with negative results with this procedure (data not shown). In the analysis of the aligned sequences, the comparison between the primers and probe of the previous developed technique and Brazilian NC gene field strains demonstrated four single nucleotide modifications in the sequence of the primers; two in CD-F, at nucleotide positions 918 and 929; and two in CD-R, at nucleotide positions 978 and 983 (Fig. 1). This situation could prevent the primers from annealing when using the protocol described by Elia et al. (2006).
This new molecular procedure detected viral RNA in three recommended body fluids (blood, urine and conjunctival swabs) and one additional sample (rectal swab) of CDV-infected dogs. In comparison among the different samples, blood and urine appeared to be the most appropriate samples to be tested with the nested real time RT-PCR. These body fluids also displayed a higher number of PCR positive results than did other samples (saliva, nasal and conjunctival swabs) in previous studies (Shin et al., 2004, An et al., 2008). A new finding was that the great majority of the positive blood and urine samples were also positive in the rectal swab samples. The rectal (or even faecal) swab is easier to collect than the other samples and has already been used to detect other gastroenteritis viral agents, such as canine parvovirus 2, rotavirus and coronavirus (Decaro et al., 2005, Calderon et al., 2011, Pinto et al., 2012).
Conjunctival swab was also a suitable specimen for CDV detection using the RT-nqPCR in the present study, although the number of positive results was lower than with the other samples. Conjunctival swab was previously recommended for CDV detection, mainly for the early diagnosis of CD (Kim et al., 2006). Another study demonstrated a concordance of 100% between RT-nested-PCR and IC assays using conjunctival swabs. The authors emphasised that this sample was the most suitable specimen for early antemortem diagnosis of CD, most likely because of the persistent shedding of CDV in the eye, unlike in other compartments (An et al., 2008). Based on these results, a conjunctival swab is the main specimen recommended for testing with the commercial IC assay. In the present study, the comparison between RT-nqPCR and IC assays (when both methods used conjunctival swabs as samples) displayed a high percentage of discordant results (22%) and false-negative results (13.6%). One possible reason is that the sampled infected dogs were in different stages of the disease. The majority of the positive dogs (64.1%) developed neurologic signs (according to the clinical exam records), which usually start 20 days after infection, when the virus reaches the epithelial tissues and the central nervous system (CNS) (Martella et al., 2008). Another possibility is that insufficient samples were collected to perform these tests, although the two conjunctival swabs (the first for the IC assay and the second for the RT-nqPCR) were collected carefully from different eyes at the time of clinical examination.
The comparison between IC assay and RT-nqPCR with other samples showed the higher analytical sensitivity of the latter procedure. Twenty of the RT-nqPCR-positive dogs developed the severe form of the disease with neurologic disorders two to three weeks after the clinical examination and were euthanised. In contrast, only twelve of these dogs were positive in the IC assay (data not shown), which means that 40% were false negative results based on the IC assay. The superior performance of nested-RT-PCR over the IC assay has been reported previously (An et al., 2008). These authors suggested that the IC assay requires large amounts of viral antigens to produce a clear positive result, which could limit the sensitivity of the assay. Because sensitive and rapid detection of the virus can reduce the mortality of the disease and to start appropriate treatment before full signs become evident, the use of a more sensitive test should be chosen when available.
Although the NC gene is highly conserved and a small coding portion was sequenced (only 245 base pairs, excluding the primers), nucleotide polymorphisms were observed among the field and vaccine samples. Interestingly, the nucleotide mutations did not result in any amino acid substitution in all 25 field samples, vaccine strains or other Brazilian strains. However, the few mutations were used to compare all sequences. The great majority of the field CDV nucleotide sequences (23 out of 25) were similar to other Brazilian strains sequenced previously. All were identified in central-nervous-system (CNS) samples collected between 2000 and 2006 from dogs with CD (Castilho et al., 2007, Headley et al., 2009). Interestingly, two dog samples showed nucleotide sequences similar to the classical strain Lederle used as a vaccine. These dogs were positive according to RT-nqPCR only in the blood samples and did not progress to CD, suggesting they could be circulating live vaccines. Furthermore, the nucleotide sequence differences could be used in RFLP analysis to differentiate vaccine strains from field samples. A similar procedure, using RFLP analysis of the NC gene, was proposed previously to differentiate between vaccine and field strains in China (Wang et al., 2011). Other studies have proposed RFLP analysis for CDV differentiation, although all of them involved the more variable haemagglutinin (H) gene as the target (Mochizuki et al., 1999, Calderon et al., 2007, Di Francesco et al., 2012).
5. Conclusions
This is the first report of reverse transcription followed by a nested real time PCR (RT-nqPCR) protocol for the sensitive detection of CDV in different clinical samples. The combination of two primer pairs in a nested format using a universal probe was able to detect both vaccine and field strains of CDV. The RT-nqPCR demonstrated sensitivity and specificity similar to other DNA amplification methods and was more sensitive than a commercial IC assay. The samples that allowed the detection of a greater number of positive samples were blood, followed by urine, rectal and conjunctival swabs. Furthermore, the nucleotide sequence of the NC gene from Brazilian field strains allowed the development of an additional RFLP analysis to differentiate them from vaccine strains. The method described herein should be an important diagnostic tool for the diagnosis of canine distemper.
Acknowledgements
The authors would like to thank Hertape Calier Saude Animal, Laboratórios Vencofarma do Brasil, Zoetis do Brasil and MSD Saúde Animal for providing the vaccine strains; the veterinarians of the Veterinary Hospital of the Brazilian Lutheran University (ULBRA) who treated the animals, collected and analysed the clinical samples; and the technicians of Simbios Biotecnologia and Laboratory of Molecular Diagnostic (ULBRA) who performed technical support. This work was supported by Simbios Biotecnologia and ULBRA.
References
- Amude A.M., Alfieri A.A., Alfieri A.F. Clinicopathological findings in dogs with distemper encephalomyelitis presented without characteristic signs of the disease. Research in Veterinary Science. 2007;82:416–422. doi: 10.1016/j.rvsc.2006.08.008. [DOI] [PubMed] [Google Scholar]
- An D.J., Kimb T.Y., Songc D.S., Kangc B.K., Park B.K. An immunochromatography assay for rapid antemortem diagnosis of dogs suspected to have canine distemper. Journal of Virological Methods. 2008;147:244–249. doi: 10.1016/j.jviromet.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R., Sol C.J., Salimans M.M., Jansen C.L., Wertheim-van Dillen P.M., van der Noordaa J. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon M.G., Romanutti C., D’Antuono A., Keller L., Mattion N., La Torre J. Evolution of canine parvovirus in Argentina between years 2003 and 2010: CPV2c has become the predominant variant affecting the domestic dog population. Virus Research. 2011;157:106–110. doi: 10.1016/j.virusres.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon M.G., Remorini P., Periolo O., Iglesias M., Mattion N., La Torre J. Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinate and non-vaccinate dogs in Argentina. Veterinary Microbiology. 2007;125:341–349. doi: 10.1016/j.vetmic.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Castilho J.G., Brandão P.E., Carnieli J.R.P., Oliveira R.N., Macedo C.I., Peixoto Z.M.P., Carrieri M.L., Kotait I. Molecular analysis of the N gene of canine distemper virus in dogs in Brazil. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2007;59:654–659. [Google Scholar]
- Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Di Trani L., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. Journal of Virological Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter Z., Lakatos B., Palade E.A., Kozma T., Forgách P., Rusvai M. Genetic diversity of Hungarian canine distemper virus strains. Veterinary Microbiology. 2007;122:258–269. doi: 10.1016/j.vetmic.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Francesco C.E., Di Francesco D., Di Martino B., Speranza R., Santori D., Boar i.A., Marsilio F. Detection by hemi-nested reverse transcription polymerase chain reaction and genetic characterization of wild type strains of Canine distemper virus in suspected infected dogs. Journal of Veterinary Diagnostic Investigation. 2012;24:107–115. doi: 10.1177/1040638711425700. [DOI] [PubMed] [Google Scholar]
- Elia G., Decaro N., Martella V., Cirone F., Lucente M.S., Lorusso E., Di Trani L., Buonavoglia C. Detection of canine distemper virus in dogs by real-time RT-PCR. Journal of Virological Methods. 2006;136:171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Frisk A.L., König M., Moritz A., Baumgärtner W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. Journal of Clinical Microbiology. 1999;37:3634–3643. doi: 10.1128/jcm.37.11.3634-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebara C.M.S., Wosiacki S.R., Negrão F.J., Oliveira D.B.de, Beloni S.N.E., Alfieri A.A., Alfieri A.F. Detecção do gene da nucleoproteína do vírus da cinomose canina por RT-PCR em urina de cães com sinais clínicos de cinomose. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2004;56:480–487. [Google Scholar]
- Greene C.E., Appel M.J. Canine distemper. In: Greene C.E., editor. Infectious Diseases of the Dog and Cat. Philadelphia: Saunders Company; Philadelphia: 2006. pp. 25–41. [Google Scholar]
- Headley S.A., Amude A.M., Alfieri A.F., Bracarense A.P., Alfieri A.A., Summers B.A. Molecular detection of canine distemper virus and the immunohistochemical characterization of the neurologic lesions in naturally occurring old dog encephalitis. Journal of Veterinary Diagnostic Investigation. 2009;21:588–597. doi: 10.1177/104063870902100502. [DOI] [PubMed] [Google Scholar]
- Józwik A., Frymus T. Comparison of the immunofluorescence assay with RT-PCR and nested PCR in the diagnosis of canine distemper. Veterinary Research Communications. 2005;29:347–359. doi: 10.1023/b:verc.0000048528.76429.8b. [DOI] [PubMed] [Google Scholar]
- Kapil S., Allison R.W., Johnston L., III, Murray S.H., Meinkoth J., Johnson B. Canine distemper virus strains circulating among North American dogs. Clinical and Vaccine Immunology. 2008;15:707–712. doi: 10.1128/CVI.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Jeoung S.Y., Ahn S.J., Lee J.H., Pak S.I., Kwon H.M. Comparison of tissue and fluid samples for the early detection of canine distemper virus in experimentally infected dogs. Journal of Veterinary Medical Science. 2006;68:877–879. doi: 10.1292/jvms.68.877. [DOI] [PubMed] [Google Scholar]
- Martella V., Elia G., Buonavoglia C. Canine distemper virus. Veterinary Clinics of North America. Small Animal Practice. 2008;38:787–797. doi: 10.1016/j.cvsm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mochizuki M., Hashimoto M., Hagiwara S., Yoshida Y., Ishiguro S. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. Journal of Clinical Microbiology. 1999;37:2936–2942. doi: 10.1128/jcm.37.9.2936-2942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L.D., Streck A.F., Gonçalves K.R., Souza C.K., Corbellini A.O., Corbellini L.G., Canal C.W. Typing of canine parvovirus strains circulating in Brazil between 2008 and 2010. Virus Research. 2012;165:29–33. doi: 10.1016/j.virusres.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliarini A., Dal Pozzo F., Gallina L., Vaccari F., Morganti L. TaqMan based real time PCR for the quantification of canine distemper virus. Veterinary Research Communications. 2007;31(Suppl. 1):261–263. doi: 10.1007/s11259-007-0020-9. [DOI] [PubMed] [Google Scholar]
- Shin Y.J., Cho K.O., Cho H.S., Kang S.K., Kim H.J., Kim Y.H., Park H.S., Park N.Y. Comparison of one-step RT-PCR and a nested PCR for the detection of canine distemper virus in clinical samples. Australian Veterinary Journal. 2004;82:83–86. doi: 10.1111/j.1751-0813.2004.tb14651.x. [DOI] [PubMed] [Google Scholar]
- Wang F., Yan X., Chai X., Zhang H., Zhao J., Wen Y., Wu W. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in China. Virology Journal. 2011;8:85. doi: 10.1186/1743-422X-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E., Iwatsuki K., Miyashita N., Gemma T., Kay C., Mikami T. Molecular analysis of the nucleocapsid protein of recent isolates of canine distemper virus in Japan. Veterinary Microbiology. 1998;59:237–244. doi: 10.1016/s0378-1135(97)00194-6. [DOI] [PubMed] [Google Scholar]


