Highlights
-
•
We developed a fast and accurate RT-LAMP detection method for Porcine Sapelovirus.
-
•
The amplification can be carried out at consistent 63 °C in a water bath.
-
•
The sensitivity of this method was 10 copies/μL.
-
•
The primers were specific to Porcine Sapelovirus with no cross-reaction with other related viruses.
-
•
The RT-LAMP for detection of Porcine Sapelovirus was better than SYBR RT-PCR and/or conventional RT-PCR.
Keywords: Porcine Sapelovirus, RT-LAMP, Detection, Picornaviridae
Abstract
The present study describes the development and validation of a one-step, single-tube, and real-time reverse transcription loop-mediated isothermal amplification (RT-LAMP) detecting Porcine Sapelovirus. RT-LAMP characterized by one strand displacement reaction with the specific stem-loop structure and Bst DNA polymerase could be finished in 60 min under isothermal condition at 63 °C. RT-LAMP assay showed higher sensitivity with 101 copies/μL than RT-PCR for the detection of Sapelovirus. The specificity of RT-LAMP assay was validated by the absence of any cross-reaction with other closely related virus in Picornaviridae group and other common virus causing porcine diarrhea. 7 positive Sapelovirus infection out of 63 fecal samples were identified using RT-LAMP, while 5 positive samples were determined by a conventional RT-PCR. A cost-effective method for Saplovirus detection with high sensitivity and specificity was developed and evaluated.
1. Introduction
Sapelovirus derived from Simian, Avian and Porcine viruses, is a new genus in the family Picornaviridae. This genus comprised three species, including Porcine Sapelovirus (PSV) (Fauquet et al., 2005), Simian Sapelovirus and Avian Sapelovirus. Porcine Sapelovirus 1 formally named Porcine Enterovirus 8 (PEV-8) and classified as a Porcine Enterovirus A (PEV A), Porcine Sapelovirus strain csh and Porcine Sapelovirus strain YC2011 are the three well acknowledged strains of PSV (Chen et al., 2012, Krumbholz et al., 2002, Lan et al., 2011). The genome of PSV is a single-stranded positive sense RNA with length of 7.5–8.3 kb nucleotides (Jauka, 2010). It contains a single open reading frame flanked by untranslated regions (UTRs) at both ends, and a long 5′UTR (600–1200 nts) (Tuthill et al., 2010). Similar to Porcine Enterovirus (PEV) and Porcine Teschovirus (PTV), PSV is the causative pathogen of neurological disorders, reproductive failure, pneumonia and diarrhea in porcine (Trefny, 1930). PSV is also found frequently to co-infection with other viral pathogen in pigs with diarrhea (Lan et al., 2011).
Routine laboratory diagnosis of PSV infection depends mainly on virus isolation and characterization, but this technique is less effective and time-consuming (Greig et al., 1961). Several molecular methods for detecting PSV have been developed, such as reverse transcription PCR (Palmquist et al., 2002), nest reverse transcription PCR (Zell et al., 2000), and real-time quantitative PCR (Krumbholz et al., 2003). However, these methods require expensive and sophisticated instruments, which restrict their wide use. Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification method which employs Bst DNA polymerase and four specially designed primers recognizing a total of six distinct sequences on the target DNA, and amplifies DNA under isothermal conditions within 1 h (Notomi et al., 2000). In amplification process, a large amount of pyrophosphate ions are produced and react with magnesium ions resulting in formation of magnesium pyrophosphate, a white precipitate that allows easy and rapid assessment of results (Mori et al., 2001). The present study aimed to develop the RT-LAMP assay for detection of Chinese Porcine Sapelovirus strain.
2. Materials and methods
2.1. Materials
Porcine Sapelovirus strain csh, Porcine Enterovirus 9 (PEV9), Porcine Epidemic Diarrhea Virus (PEDV), Porcine Transmissible Gastroenteritis Virus (TGEV), Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) were conserved in the laboratory. Porcine Teschovirus (PTV) and Encephalomyocarditis Virus (EMCV) were received from Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences (CAAS) and Lanzhou Veterinary Research Institute of CAAS respectively, and Foot-and-Mouth Disease Virus (FMDV) vaccine strain was purchased from China Animal Husbandry Industry Company Limited. Fecal samples were collected from healthy adult porcine in Shanghai piggeries for RT-LAMP evaluation. Viral RNA was extracted by using the TIANamp virus DNA/RNA kit (TianGen, Beijing, China) and stored at −80 °C until use.
2.2. Primer design
Primers used for RT-LAMP amplification of PSV were designed from the 5′UTR. The 5′UTR sequence of PSV (GenBank: HQ875059) was aligned with other related viruses by using CLUSTAL W to identify the conservative regions. The RT-LAMP primers were determined through the LAMP primer design software1 corresponding to the genome position from 41 bp to 235 bp. A set of four primers including two outer and two inner primers was selected (Table 1 ). Primers of SYBR RT-PCR were designed by Primer Premier 6.0 (Table 1). Primers of conventional RT-PCR were selected from the previous study (Palmquist et al., 2002).
Table 1.
The primers set for PSV RT-LAMP and SYBR RT-PCR.
| Primers | Genome positiona | Length (bps) | Sequence |
|---|---|---|---|
| F3 | 41–60 | 20 | CCATACCCTACCCTCCCTTC |
| B3 | 235–254 | 20 | GCCCATAGTTCACTGCCTAC |
| FIP(F1c-F2) | 121–142 + 61–80 | 42 | CACGCTACTGCCGTTAAGGTGT-CAAAGCGGATGGACACAAGG |
| BIP(B2-B1c) | 215–232 + 171–192 | 40 | ATAGCCATGTTAGTGACGCGCT-TCCTGTAACCAGTCGCCG |
| F | 67–85 | 18 | GGATGGACACAAGGACTT |
| B | 247–165 | 18 | GTTCATGCCTACTCTCC |
Porcine Sapelovirus strain csh (access no. HQ875059).
2.3. RT-LAMP development
The RT-LAMP reaction was carried out in 25 μL reaction total volume using the Loopamp RNA amplification kit (Eiken Chemical Company Limited, Japan) containing 40 pmol of each of the inner primers FIP and BIP, 5 pmol each of the outer primers F3 and B3, 12.5 μL reaction mix. The reaction liquid was mixed with 1 μL enzyme mixture and 1 μL fluorescent detection regent. The reaction mixture was incubated at 63 °C for 45 min in a Loopamp real-time turbidimeter LAC320 (Eiken Chemical Company Limited, Japan) or a 63 °C water bath, followed by heating at 80 °C for 5 min to terminate the reaction.
For real-time monitoring positive result, a sample having T p value of ≤45 min and turbidity above the threshold value of ≥0.1 was considered positive. Visualization results were completed by adding 1 μL fluorescent detection reagent Calcein (FDR, Eiken Chemical Company Limited, Japan). Orange changes to chartreuse fluorescence were regarded as positive reaction.
Sensitivity of the RT-LAMP assay was analyzed using 10-fold serial dilutions of viral RNA. The final concentrations of viral RNA were from 2.97 × 107 copies to 2.97 × 100 copies per reaction mix. Specificity of the assay was evaluated by cross reactivity tests with virus belonging to Picornaviridae and several other viral pathogens that may cause diarrhea and reproductive failure.
2.4. SYBR real-time RT-PCR (SYBR RT-PCR) and RT-PCR
SYBR RT-PCR was performed with the one step SYBR PrimeScript Plus RT-PCR Kit (TaKaRa Biotechnology (Dalian) Company Limited, Dalian, China) in the ABI7500 system in 20 μL reaction volume containing 2 μL total RNA, 0.8 μL forward and backward Primers (10 μM). The one step RT-PCR was carried out in a 50 μL total reaction volume by using Primerscript one step RT-PCR Kit Ver.2 (TaKaRa Biotechnology (Dalian) Company Limited, Dalian, China) with 8 μM forward and backward primers and 1 μL RNA, according to the manufacture's protocol.
3. Results
3.1. Optimization of RT-LAMP
Optimum specific amplification was achieved at 63 °C for 60 min. RT-LAMP was incubated at temperatures ranging from 61 °C to 65 °C and could be detected at 63 °C (data not shown). The symbol of a positive result was a T p value of ≤45 min and turbidity above the threshold value of ≥0.1. Sequencing of the product demonstrated the RT-LAMP result was consistent with the objective sequence.
3.2. Sensitivity and specificity
Sensitivity of the RT-LAMP assay detecting PSV was determined by testing serial 10-fold dilutions of virus RNA that had been quantified by infinite 200 pro (Tecan, Switzerland). The detection limit of the RT-LAMP assay was 10 copies/μL RNA, similar to SYBR RT-PCR, and higher than the RT-PCR (103 copies/μL) (Fig. 1, Fig. 2, Fig. 3 ). Specificity of the RT-LAMP assay was analyzed by cross-reactivity with viruses belonging to Picornaviridae, such as PTV, PEV9, FMDV, EMCV and other symptom related virus, like TGEV, PEDV and PRRSV. RT-LAMP demonstrated high specificity for PSV infection (Fig. 4 ).
Fig. 1.
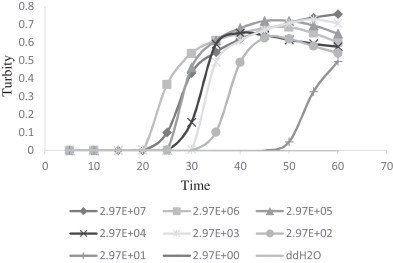
Sensitivity and dynamic range of PSV-specific RT-LAMP assay with 10-fold serial dilutions of RNA (from 2.97 × 107 copies/μL to 2.97 × 100 copies/μL). The detection limit is 101 copies/μL.
Fig. 2.

Visual detection of the sensitivity of RT-LMAP assay in 10-fold serials dilutions of PSV RNA corresponding to Fig. 1. The tubes from the left to the right represent 5 × 107 copies/μL, 5 × 106 copies/μL, 5 × 105 copies/μL, 5 × 104 copies/μL, 5 × 103 copies/μL, 5 × 102 copies/μL, 5 × 101 copies/μL, 5 × 100 copies/μL and Negative Control (RNase Free dH2O as a replacement of RNA).
Fig. 3.
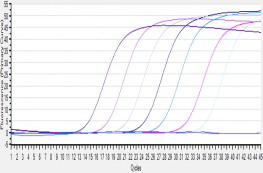
The sensitivity of SYBR rRT-PCR, the standard RNA was serially 10-fold diluted from 5 × 107 copies/μL to 5 × 100 copies/μL, and the amplification curves from the left to the right represent 5 × 107 copies/μL, 5 × 106 copies/μL, 5 × 105 copies/μL, 5 × 104 copies/μL, 5 × 103 copies/μL, 5 × 102 copies/μL, 5 × 101 copies/μL, 5 × 100 copies/μL respectively and Negative Control (RNase Free dH2O as a replacement of RNA).
Fig. 4.
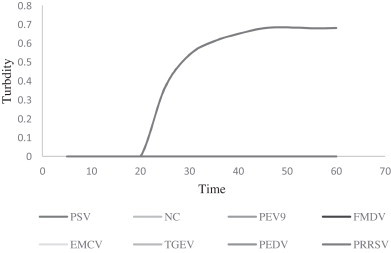
The amplification plot of the specificity of PSV-specific RT-LAMP assay. Here NC denoted Negative Control and the concentration of the PSV was 5 × 107 copies/μL. The picture revealed that the RT-LAMP assay had high specificity.
3.3. Evaluation of PSV RT-LAMP assay with clinical fecal sample
RT-LAMP for PSV was further assessed by detecting 63 fecal samples from healthy pigs. Detection was also performed by using SYBR RT-PCR and conventional RT-PCR. The RT-LAMP assay and SYBR RT-PCR both detected seven positive samples and the RT-PCR assay detected five positive samples. The RT-LAMP assay demonstrated higher sensitivity as compared to traditional RT-PCR by identifying two more positive cases.
4. Discussion
Since the LAMP technique was invented by Tsugunori Notomi, it has spread widely to many fields, such as Life Science, Veterinary Medicine, Food Science. LAMP or RT-LAMP have many advantages. Apart from rapidity, simplicity and precision, LAMP or RT-LAMP results can be assessed with calcein and an inexpensive turbidimeter. Furthermore, the reaction tube can be incubated in a water bath, which is economical (Mori et al., 2001). The RT-LAMP assay has developed as a powerful gene amplification tool to detect and diagnose infectious diseases, such as the Foot-and-Mouth Disease Virus (Dukes et al., 2006), West Nile Virus (Parida et al., 2004), Hepatitis E Virus (Lan et al., 2009), and SARS Coronavirus (Thai et al., 2004).
In this study, the utility of the RT-LAMP technique for rapid and accurate detection of PSV in clinical fecal specimens was developed. Detecting PSV in different animal origin clinical specimens could be achieved within 2 h including treatment of the samples. The sensitivity of the RT-LAMP was 10 copies/μL, and 100 times higher than RT-PCR. RT-LAMP detecting PSV infection is high specific with no cross-reaction with other viral pathogens. RT-LAMP also showed higher sensitivity by identification of two more positive samples than traditional RT-PCR (7 positive by RT-LAMP and 5 positive by RT-PCR). None of the RT-PCR detecting positive samples was missed by RT-LAMP, further showing the higher sensitivity of the RT-LAMP.
In conclusion, a simple, rapid, precise, sensitive and specific one-step PSV-RT-LAMP assay was developed and suitable for detection of clinical samples.
Conflict of interest statement
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Acknowledgments
Authors are thankful and pay their gratitude to Dr. Shangjin Cui of Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Zhongren Ma and Ruofei Feng of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
And they would like to thank Izhar Hyder Qazi, a Pakistani, for providing writing assistance.
Footnotes
References
- Chen J., Chen F., Zhou Q., Li W., Song Y., Pan Y., Zhang X., Xue Ch., Bi Y., Cao Y. Complete genome sequence of a novel Porcine Sapelovirus strain YC2011 isolated from piglets with diarrhea. J. Virol. 2012;86(19):10898. doi: 10.1128/JVI.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukes J., King D., Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch. Virol. 2006;151:1093–1106. doi: 10.1007/s00705-005-0708-5. [DOI] [PubMed] [Google Scholar]
- Fauquet C.M., Mayo M., Maniloff J., Desselberger U., Ball L.A. 2005. Virus taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses: Access Online via Elsevier. [Google Scholar]
- Greig A., Bannister G., Mitchell D., Corner A. Studies on pathogenic porcine enteroviruses: II. Isolation of virus in tissue culture from brain and feces of clinical cases. Can. J. Comp. Med. Vet. Sci. 1961;25(6):142. [PMC free article] [PubMed] [Google Scholar]
- Jauka T.I. Rhodes University; 2010. Generation of polyclonal antibodies against Theiler's Murine Encephalomyelitis virus protein 2C, and their use in investigating localisation of the protein in infected cells. [Google Scholar]
- Krumbholz A., Dauber M., Henke A., Birch-Hirschfeld E., Knowles N.J., Stelzner A., Zell R. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J. Virol. 2002;76(11):5813–5821. doi: 10.1128/JVI.76.11.5813-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbholz A., Wurm R., Scheck O., Birch-Hirschfeld E., Egerer R., Henke A., Wutzler P., Zell R. Detection of porcine teschoviruses and enteroviruses by LightCycler real-time PCR. J. Virol. Methods. 2003;113:51–63. doi: 10.1016/s0166-0934(03)00227-1. [DOI] [PubMed] [Google Scholar]
- Lan X., Yang B., Li B.Y., Yin X.P., Li X.R., Liu J.X. Reverse transcription-loop-mediated isothermal amplification assay for rapid detection of hepatitis E virus. J. Clin. Microbiol. 2009:2304–2306. doi: 10.1128/JCM.00498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan D., Ji W., Yang S., Cui L., Yang Z., Yuan C., Hua X. Isolation and characterization of the first Chinese Porcine Sapelovirus strain. Arch. Virol. 2011:1567–1574. doi: 10.1007/s00705-011-1035-7. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000:63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmquist J.M., Munir S., Taku A., Kapur V., Goyal S.M. Detection of porcine teschovirus and enterovirus type II by reverse transcription-polymerase chain reaction. J. Vet. Diagn. Invest. 2002;14:476–480. doi: 10.1177/104063870201400605. [DOI] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F., Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004;42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai H.T.C., Le M.Q., Vuong C.D., Parida M., Minekawa H., Notomi T., Hasebe F., Morta K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;28:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefny L. Massive illness of swine in Teschen area. Zveroleki Obzori. 1930;23:235–236. [Google Scholar]
- Tuthill T.J., Groppelli E., Hogle J.M., Rowlands D.J. Cell Entry by Non-Enveloped Viruses. Springer; 2010. Picornaviruses; pp. 43–89. [Google Scholar]
- Zell R., Krumbholz A., Henke A., Birch-Hirschfeld E., Stelzner A., Doherty M., Hoey E., Dauber M., Prager D., Wurm R. Detection of porcine enteroviruses by nRT–PCR: differentiation of CPE groups I–III with specific primer sets. J. Virol. Methods. 2000;88:205–218. doi: 10.1016/s0166-0934(00)00189-0. [DOI] [PubMed] [Google Scholar]


