Highlights
-
•
We established a TaqMan-based real-time RT-PCR assay for the detection of a novel swine diarrhea virus SADS-CoV.
-
•
The assay in this study was of high sensitivity, specificity and reproducibility.
-
•
We compared the accuracy between conventional PCR and TaqMan-based RT-PCR assay with adequate clinical samples.
-
•
This approach will help to provide knowledge on epidemiology and pathogenesis studies of SADS-CoV.
Keywords: SADS-CoV, TaqMan-based real-time RT-PCR, Diagnosis, Quantification
Abstract
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a novel coronavirus which was first reported in southern China in 2017. It can cause severe diarrhea disease in pigs. In order to detect this new emerging virus rapidly and reliably, a TaqMan-based real-time RT-PCR assay was established in this study. Specific primers and probe were designed and synthesized based on the conserved region within the N gene of the viral genome. Results showed that the lowest limit of detection was 3.0 × 101 copies/μL. This approach was specific for SADS-CoV, and there were no cross-reaction observed against other 15 swine viruses. It was 10 times more sensitive than the conventional PCR and gave higher SADS-CoV positive detection rate (70.69%, 123/174) than the conventional PCR (51.15%, 89/174) from clinical samples. These data indicated that the TaqMan-based real-time RT-PCR assay established here was an effective method with high sensitivity, specificity and reproducibility for faster and more accurate detection and quantification of SADS-CoV.
1. Introduction
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a newly discovered virus, which is an enveloped, positive and single-stranded sense RNA virus and belongs to the order Nidovirales, family Coronaviridae and genus Alphacoronavirus (Zhou et al., 2018; Pan et al., 2017; Woo et al., 2012). SADS-CoV can cause severe and acute diarrhea and rapid weight loss in piglets less than 6-day-old. From January 2017 to May, the outbreak of SADS-CoV has led to the death of almost 25,000 piglets in southern China and resulted in significant economic losses (data unpublished). Considering the clinical symptoms of this new emerging virus that are similar to those caused by other known swine enteric coronaviruese in China, there is an urgent need to establish a faster and reliable diagnostic technique for identification and quantification of SADS-CoV (Dong et al., 2015; Sun et al., 2016).
The real-time PCR using the fluorogenic probe system has been widely used to quantify virus nucleic acids in recent years (Li et al., 2011; Gava et al., 2015; Niu et al., 2016; Dall Agnol et al., 2017). Compared to the conventional PCR, it allows the accurate and reproducible quantitation of viral gene copies, and has advantages in rapidity, accuracy, reproducibility, and high sensitivity (Li et al., 2016; Fowler et al., 2016). So far, there is no diagnostic method have been established for the detection of SADS-CoV. In this study, a TaqMan-based real-time RT-PCR was designed for SADS-CoV diagnosis and quantification in clinical swine tissue samples.
2. Material and methods
2.1. Virus and samples collection
SADS-CoV virus used in this study was isolated from clinical samples with diarrheal symptoms from pig herds in Guangdong Province of China. Specimens from piglets with signs of severe diarrhea and high mortality were collected from February to August 2017 in four SADS-CoV positive farms.
2.2. Nucleic acid extraction and reverse transcription
Specimens were homogenized in phosphate-buffered saline (PBS) (20% w/v), and frozen and thawed three times, and then centrifuged for 10 min at 10,000 × g. Viral nucleic acid was extracted following the manufacturer’s recommendations of AxyPrepTM Body Fluid Viral DNA/RNA Miniprep Kit (Axygen Scientific, Inc). Reverse transcription was carried out according to the manufacturer's protocol of Prime ScriptTM RT Reagent Kit with gDNA Eraser (Takara, Dalian, China), the first step was performed in a total volume of 10 μL of a mixture containing 2 μL 5 × gDNA Eraser Buffer, 1μL gDNA Eraser, 1 μL total RNA and 6 μL RNase Free dH2O at the condition of 42 ℃ for 2 min to remove residual genomic DNA, and then, ten microliters of mixture was used in a reaction volume of 20 μL containing 1 μL PrimeScript RT Enzyme Mix I, 1 μL RT Primer Mix, 4 μL 5 × PrimeScript Buffer 2, 4 μL RNase Free dH2O, reactions were conducted with a program that included 37 °C for 15 min, 87 °C for 5 s. All DNA and cDNA templates were stored at −80 °C until use.
2.3. Design of primers and probe for the real-time RT-PCR
The genome sequences of SADS-CoV (Accession number: MF094681-MF094684) were retrieved from GenBank. Conserved regions of the N gene were identified by nucleotide sequence alignments with MegAlign7.1.0 (DNAStar, USA). Primers and probe were selected and designed using Primer premers 5.0 to generate a 449 bp amplicon for plasmid and 155 bp real-time RT-PCR product. The probe was labeled with 5-carboxyflourescein(FAM) at the 5′-end and N, N, N' N'-tetramethyyl-6-carboxyrhodamine(TAMRA) at the 3′-end. Nucleotide information about primers and probe are summarized in Table 1 .
Table 1.
Primers and probe for SADS-CoV detection.
| primers | Sequence(5’–3’) | amplicon |
|---|---|---|
| SADS-N-F | CAGGTCTTGGTGTTCGCAATCG | 449 bp |
| SADS-N-R | ACCGTGCTGAACGAGGTCACT | |
| qSADS-N-F | CTGACTGTTGTTGAGGTTAC | 155 bp |
| qSADS-N-R | TCTGCCAAAGCTTGTTTAAC | |
| probe | 5, -FAM -TCACAGTCTCGTTCTCGCAATCA- TAMRA-3, |
2.4. Construction of standard plasmids DNA for the real-time RT-PCR
The target fragment was obtained with SADS-CoV-N-F/R by PCR and then cloned into pMD19-T vector (TaKaRa, Dalian, China). PCR product was transformed into DH5α Escherichia coli cells following the manufacture's recommendations (TaKaRa, Dalian, China) and purified by Plasmid Mini Kit 2 (Omega Bio-tek, USA). The positive recombinant plasmid was confirmed by sequencing (BGI, Shenzhen, China). Subsequently, the SADS-CoV plasmid (pSADS-CoV-N) was quantified by spectrophotometric analysis (NanoDrop ND-1000) and converted into copy number. The plasmid DNA was stored at −80 ℃ in aliquots until use.
2.5. TaqMan-based real-time RT-PCR assay
The optimization of the real-time RT-PCR reaction components and cycling conditions was performed using DNA standards. Concentrations of primers, probe and cDNA were selected based on the greatest fluorescence obtained and the lowest threshold cycle. The optimized real-time RT-PCR assays were carried out in 20 μL reaction mixture containing 10 μL of Premix Ex Taq (Probe qPCR) (2×), 0.8 μL of qSADS-CoV-N-F (10 μM), 0.8 μL of qSADS-CoV-N-R (10 μM), 0.8 μL (10 μM) of probe, 1 μL of cDNA template, and 6.6 μL of ddH2O. The PCR amplification was performed with a CFX96 Real-Time PCR Detection System (BIO-RAD) under the following conditions: 95 ℃ for 30 s for initial denaturation followed by 45 cycles of 95 ℃ for 5 s and 62 ℃ for 30 s. Ten-fold serial dilutions of pSADS-CoV-N, ranging from 109 to 102 copies/μL, were tested in five replicates with real-time RT-PCR to generate the standard curve.
2.6. Sensitivity and specificity of the real-time RT-PCR
The concentration of the plasmid DNA was 196.455 ng/μL, equivalent to 3.0 × 1010 copies/μL before dilution. To examine the detection limit of the real-time RT-PCR, plasmid DNA was diluted serially 10-fold with RNase Free ddH2O (109–102 copies/μL) as templates. Briefly, conventional RT-PCR was performed by using 1 μL of diluted cDNA template and 10 μmol of primers in a 20 μL reaction volume with the following thermal profile: 50 °C for 30 min, 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and a final step of 72 °C for 5 min. And then the detection limit between the real-time RT-PCR and conventional PCR were compared.
The specificity of the TaqMan-based real-time RT-PCR assay was demonstrated with other porcine viruses, including classical swine fever virus (CSFV), foot-and-mouth disease virus (FMDV), porcine circovirus 3 (PCV3), porcine rotavirus (RV), porcine deltacoronavirus (PDCoV), porcine epidemic diarrhea virus (PEDV), porcine kobuvirus (PKV), porcine reproductive and respiratory syndrome virus (PRRSV), porcine sapelovirus (PSV), porcine bocavirus (PBoV), swine vesicular disease virus (SVDV), seneca valley virus (SVA), swine influenza virus (SIV), transmissible gastroenteritis virus (TGEV) and staphylococcus aureus, which were stored at our laboratory. Negative control (RNase Free ddH2O) were also contained in the run.
2.7. Reproducibility of the real-time RT-PCR
Ten-fold serial plasmid dilutions ranging from 3.0 × 102 to 3.0 × 109 copies/μL were used to assess the coefficients of variation of the real-time RT-PCR. Intra-and inter-assay for Cq values were both included in three different times and quintuplicate.
2.8. Detection of the clinical tissue samples
In order to evaluate the real-time RT-PCR assay, a total of 174 clinical tissue samples with diarrheal symptoms, including heart, liver, spleen, lung, kidney, jejunum, ileum, duodenum, tonsil, lymphoglandulae mesentericae and lymphonodi abdominals were collected and quantified by the developed method. The samples amplified before 40 cycles and showed the typical amplification curves were considered to be SADS-CoV positive. Conventional PCR was preformed simultaneously.
3. Results
3.1. Standard curve and dynamic range
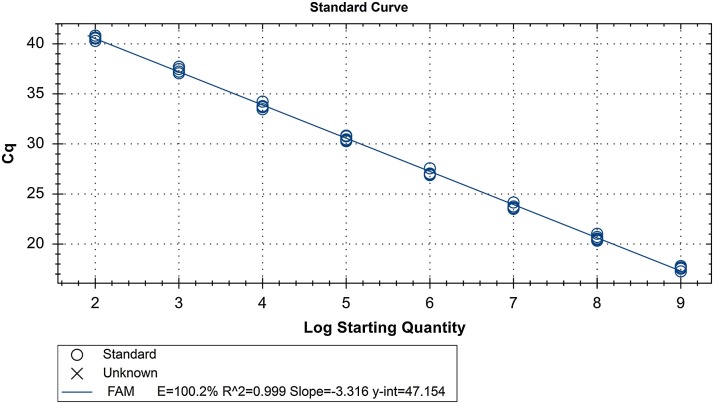
Ten-fold serial dilutions of DNA plasmids were used to establish a standard curve by plotting the logarithm of the plasmid copy number against the measured Cq values. The standard curve generated had a wide dynamic range of 109–102 copies/μL with a linear correlation (R 2) of 0.999 between the Cq value and the logarithm of the plasmid copy number (Fig. 1 ).
Fig. 1.
Standard curve of TaqMan-based real-time RT-PCR assay. The assay was conducted using duplicated ten-fold serial dilutions (3.0 × 109–3.0 × 102 copies/μL) of the plasmid DNA; y = −3.316x + 47.154.
3.2. Specificity of the real-time RT-PCR
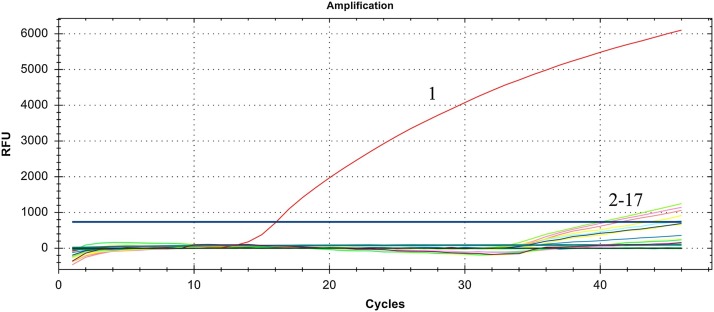
The real-time RT-PCR gave positive results for the standard plasmid of SADS-CoV strain (3.0 × 107 copies/μL) and negative results (Cq > 40) for other porcine viruses, including PEDV, PDCoV, RV, PRRSV, PKV, PSV, PBoV, FMDV, SVDV, SVA, SIV, CSFV, PCV3, TGEV, SA and negative control (RNase Free ddH2O) (Fig. 2 , Table 2 ).
Fig. 2.
Specificity of the real-time RT-PCR assay. 1:SADS-CoV; 2-17: PEDV, PDCoV, RV, PRRSV, PKV, PBoV, PSV, FMDV, SVDV, SVA, SIV, CSFV, PCV3, TGEV, Staphylococcus aureus and negative control.
Table 2.
The samples' information of specificity assay for SADS-CoV Taqman-baesd real-time RT-PCR. N/A: negative.
| Number | Samples | Type | Cq Mean |
|---|---|---|---|
| 1 | SADS-CoV | isolate strain | 16.07 |
| 2 | PEDV | isolate strain | 41.49 |
| 3 | TGEV | vaccine strain | N/A |
| 4 | RV | vaccine strain | N/A |
| 5 | PDCoV | isolate strain | N/A |
| 6 | FMDV | isolate strain | 45.37 |
| 7 | PRRSV | vaccine strain | N/A |
| 8 | CSFV | vaccine strain | N/A |
| 9 | SVA | isolate strain | N/A |
| 10 | PCV3 | wild strain | 40.06 |
| 11 | SVDV | isolate strain | 40.57 |
| 12 | SIV | vaccine strain | N/A |
| 13 | PKV | wild strain | N/A |
| 14 | PBoV | wild strain | N/A |
| 15 | PSV | wild strain | N/A |
| 16 | SA | isolate strain | N/A |
| 17 | ddH2O | negative control | N/A |
3.3. Sensitivity of the real-time RT-PCR
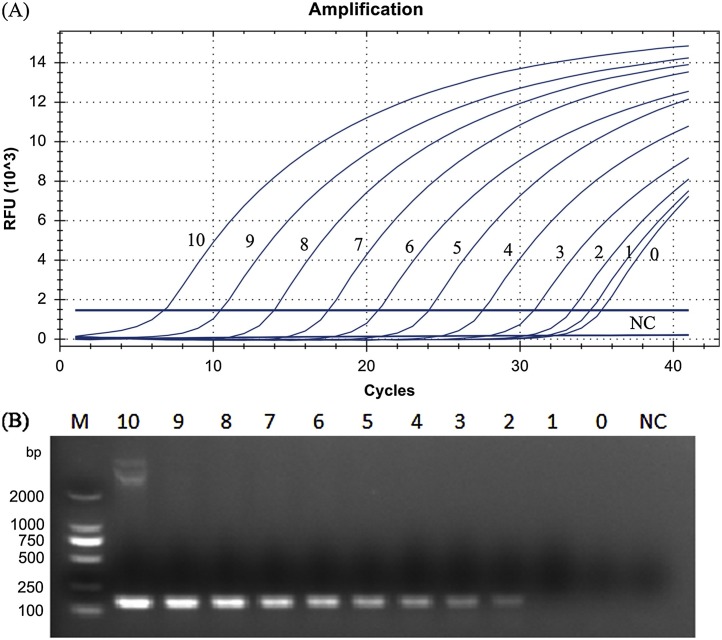
To compare the sensitivity of the TaqMan-based real-time RT-PCR and the conventional PCR, ten-fold serial dilutions of the plasmid pSADS-N were analyzed. The detection limits for the TaqMan-based real-time RT-PCR and the conventional PCR assays were 3.0 × 101 and 3.0 × 102 DNA copies/μL, respectively (Fig. 3 ).
Fig. 3.
(A) Sensitivity of the TaqMan-based real-time RT-PCR assay for SADS-CoV detection. (B) Sensitivity of the conventional PCR for SADS-CoV detection. 10−0: a serial of ten-fold dilutions plasmid DNA (3.0 × 1010–3.0 × 10° copies/μL); NC: negative control; M: DL 2000 marker.
3.4. Reproducibility of the TaqMan-based real-time RT-PCR assay
The coefficient of variation (C.V) in the intra-assay and inter-assay was evaluated. The intra-assay C.Vs of real-time PCR ranged from 0.31% to 1.93%, while the inter-assay C.Vs ranged from 0.14% to 1.86% (Table 3 ).
Table 3.
Reproducibility of intra-assay and inter-assay TaqMan-based real-time RT-PCR for SADS-CoV detection. SD = Standard deviation; C.V (%) = Percentage of coefficient variation.
| SADS-CoV clone | Intra-assay |
Inter-assay |
||||
|---|---|---|---|---|---|---|
| (copies/μL) | Cq Mean | SD | C.V(%) | Cq Mean | SD | C.V(%) |
| 3.0 × 109 | 8.77 | 0.12 | 1.33% | 8.68 | 0.16 | 1.86% |
| 3.0 × 108 | 12.99 | 0.25 | 1.93% | 13.00 | 0.02 | 0.16% |
| 3.0 × 107 | 16.68 | 0.09 | 0.56% | 16.78 | 0.09 | 0.53% |
| 3.0 × 106 | 20.09 | 0.07 | 0.33% | 20.08 | 0.05 | 0.23% |
| 3.0 × 105 | 23.25 | 0.12 | 0.54% | 23.28 | 0.04 | 0.19% |
| 3.0 × 104 | 26.52 | 0.12 | 0.45% | 26.51 | 0.04 | 0.14% |
| 3.0 × 103 | 30.17 | 0.09 | 0.31% | 30.09 | 0.16 | 0.52% |
| 3.0 × 102 | 32.64 | 0.21 | 0.63% | 32.84 | 0.16 | 0.48% |
3.5. Detection of the clinical tissue samples
The developed TaqMan-based real-time RT-PCR assay was evaluated with 174 clinical samples. 123 out of 174 samples were positive for SADS-CoV by the TaqMan-based real-time RT-PCR. When these samples were detected by the conventional PCR, only 89 out of 174 (51.15%) were positive. The established technique increased the positive detection positive rated by 20%. Samples that were tested positively by the conventional PCR were also positive by the real-time RT-PCR. 34 samples were detected negatively by the conventional PCR, while using the TaqMan-based real-time RT-PCR, they were actually positive for SADS-CoV (Table 4 ).
Table 4.
Comparative results of clinical samples tested by conventional PCR and TaqMan-based real-time RT-PCR for SADS-CoV detection. LA: Lymphonodi abdominals; LM: Lymphoglandulaemesentericae.
| samples | Number of samples | Number of SADS-CoV positive samples |
|
|---|---|---|---|
| Conventional PCR | Real-time RT-PCR | ||
| Heart | 18 | 8 | 11 |
| Liver | 18 | 8 | 12 |
| Spleen | 18 | 9 | 13 |
| Lung | 18 | 9 | 13 |
| Kidney | 18 | 8 | 14 |
| Jejunum | 17 | 13 | 15 |
| Ileum | 18 | 17 | 17 |
| Duodenum | 18 | 11 | 13 |
| Tonsil | 13 | 1 | 5 |
| LA | 9 | 1 | 2 |
| LM | 9 | 4 | 8 |
| Total | 174 | 89 | 123 |
4. Discussion
SADS-CoV is a newly identified virus in south China in 2017. Previous methods for the detection of this novel virus have relied on qualitative rather than quantitative PCR (Zhou et al., 2018; Pan et al., 2017; Gong et al., 2017). The conventional PCR is time-consuming, and it couldn't estimate the amount of viral gene copies, which could be crucial for virus diagnosis and correlations with clinical diseases (Mackay et al., 2002).
In this study, we described the development and validation of a TaqMan-based real-time RT-PCR assay for SADS-CoV diagnosis and quantification in 174 clinical samples from piglets affected by severe diarrhea in southern China. The hydrolysis probe employed in this assay was located in the conserved region within the N gene of the viral genome. Results showed that the correlation coefficient of the standard curve in this study was 0.999, much closing to the R2 of 1 in theory. No cross-reaction signals were observed for fifteen other porcine diseases. The lowest limit of detection was 3.0 × 101 copies/μL, 10 times more sensitive than the conventional PCR. The intra-assay C.Vs were equal or less than 1.93% and the inter-assay C.Vs were equal or less than 1.86%, indicating the good reproducibility of this assay. The positive rate determined by the new technique was 70.69%, whereas that of the conventional PCR was merely 51.15%, further demonstrating the higher sensitivity of the former method. All these data showed that the TaqMan-based real-time RT-PCR approach was reliable and sensitive.
In conclusion, the TaqMan-based real-time RT-PCR assay was established in present study for detection and quantification of SADS-CoV in pigs. Because of it remarkable sensitivity, reproducibility and rapidity compared to the conventional PCR, this assay may not only help to resolve diagnosis investigation, but also help to provide knowledge on epidemiology and pathogenesis studies of SADS-CoV.
Conflict of interest
The authors declare no conflict of interests with any organization.
Acknowledgments
This work was supported by the National Key Research and Development Program, Grant/Award Number: 2016YFD0501304. We would acknowledge Guangdong Wen’s Foodstuffs Group Co., Ltd. China, for providing us with piglets’ tissue samples.
Contributor Information
Xiang-bin Zhang, Email: Zhangxb@scau.edu.cn.
Jing-yun Ma, Email: majy2400@scau.edu.cn.
References
- Dall Agnol A.M., Otonel R.A., Leme R.A., Alfieri A.A., Alfieri A.F. A TaqMan-based qRT-PCR assay for Senecavirus A detection in tissue samples of neonatal piglets. Mol. Cell. Probes. 2017;33:28–31. doi: 10.1016/j.mcp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V.L., Ransburgh R.H., Poulsen E.G., Wads worth J., King D.P., Mioulet V., Knowles N.J., Williamson S., Liu X., Anderson G.A. Development of a novel real-time RT-PCR assay to detect Seneca Valley virus associated with emerging cases of vesicular disease in pigs. J. Virol. Methods. 2016;239:34. doi: 10.1016/j.jviromet.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Gava D., Souza C.K., Schaefer R., Vincent A.L., Cantao M.E., Coldebella A., Ciacci-Zanella J.R. A TaqMan-based real-time PCR for detection and quantification of porcine parvovirus 4. J. Virol. Methods. 2015;219:14–17. doi: 10.1016/j.jviromet.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new Bat-HKU2–like Coronavirus in Swine, China. Emerg. Infect. Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Xiao S., Ma J., Liu Y., Mao L., Wen L., Mao A., Zhang X., Ni Y., Guo R., Zhou J., Yu Z., Lv L., Wang X., Fang L., Chen H., He K. Development of a novel TaqMan-based real-time PCR assay for the detection of porcine boca-like virus (Pbo-likeV) Virol. J. 2011;8:357. doi: 10.1186/1743-422X-8-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li W., Mao L., Hao F., Yang L., Zhang W., Jiang J. Rapid detection of novel caprine parainfluenza virus type 3 (CPIV3) using a TaqMan-based RT-qPCR. J. Virol. Methods. 2016;236:126–131. doi: 10.1016/j.jviromet.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Mackay I.M., Arden K.E., Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Chen Yang, Yu X., Ti J., Wang A., Diao Y. Development of a TaqMan-based real-time PCR assay for the detection of Novel GPV. J. Virol. Methods. 2016;237:32–37. doi: 10.1016/j.jviromet.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y., Wang L., Wang D., Song Y., Zhang X., Huang Y. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang X., Wei Chen, Feng J. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J. Vet. Med. Sci. 2016;78:355–363. doi: 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai Teng, J.L Tsang, C.C Wang, Zheng B., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and Avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and Avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Fan, H., Lan, T., Yang, X.L., Zhang, W., Zhu, Y., et al., 2018. Fatal swine acute diarrhea syndrome caused by an HKU2-related coronavirus of bat origin. Nature (Accepted for publication). [DOI] [PMC free article] [PubMed]