Highlights
-
•
A real-time RT-LAMP assay was established for detection of a novel swine acute diarrhea syndrome coronavirus.
-
•
This assay in this study was demonstrated to be specific, sensitive and reproducible for detection of SADS-CoV.
-
•
This assay will provide a new method for the diagnosis, surveillance, and pathology studies of SADS-CoV.
Keywords: SADS-CoV, Real-time LAMP, Diagnosis
Abstract
A novel swine acute diarrhea syndrome Coronavirus (SADS-CoV) that causes severe diarrhea in suckling piglets was identified in southern China in 2017. A simple and rapid detection test was developed for this virus using real-time RT-LAMP based on the conserved N gene of the virus. The method had a detection limit of 1.0 × 101 copies/μL with no cross-reactions with classical swine fever virus, porcine and respiratory syndrome virus NA, porcine and respiratory syndrome virus EU, transmissible gastroenteritis coronavirus, foot and mouth disease virus, porcine epidemic diarrhea virus (S-INDEL and non-S-INDEL), swine influenza virus subtype H1N1, porcine circovirus type 2, seneca valley virus, porcine parvovirus, porcine deltacoronavirus and rotavirus. This method was also reproducible. Twenty of 24 clinical samples were identified as SADS-CoV RNA-positive by the real-time RT-LAMP and the results were consistent with that of the real time RT-PCR method. This new method for detecting SADS-CoV is specific and sensitive for the detection of SADS-CoV.
1. Introduction
Outbreaks of diarrhea in newborn piglets occurred on a pig farm in Guangdong province of southern China in February 2017 (Gong et al., 2017). At that time, several pathogens were tested for these cases of diarrhea, including enteric coronaviruses porcine deltacoronavirus (PDCoV), porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV). However, all samples were negative for the presence of these three pathogens (Pan et al., 2017). In September 2017, a novel swine enteric alphacoronavirus (SeACoV) also named as bat-HKU2–like porcine coronavirus (PEAV) or swine acute diarrhea syndrome coronavirus (SADS-CoV) was identified in southern China (Gong et al., 2017; Pan et al., 2017; Zhou et al., 2018a, Zhou et al., 2018b). The mortality rate was more than 35% in swine that were less than 10 days old and caused serious economic losses to the swine industry (Pan et al., 2017). All affected pigs showed acute vomiting and severe watery diarrhea, similar to clinical signs caused by PEDV, TGEV and PDCoV. A reliable and simple diagnostic method for the surveillance of SADS-CoV was needed to distinguish between these viral pathogens.
Loop-mediated isothermal amplification (LAMP) is a simple isothermal method for in vitro nucleic acid amplification. The only equipment needed is a constant temperature heating block or water bath. In addition, the results of the reaction can be directly observed with the naked eye when performed in the presence of a fluorescent dye (Notomi et al., 2000; Yuan et al., 2014). In addition, the dye enables the use of a fluorescent detector that can be used to follow the progress of the reaction (real-time LAMP) (Oscorbin et al., 2016).
LAMP assays have been successfully used for other pathogens (de Souza et al., 2012; Kurosaki et al., 2017; Li et al., 2012; Zhao et al., 2011). In this study, a real time RT-LAMP assay was developed for rapid diagnosis of novel SADS-CoV.
2. Materials and methods
2.1. Virus strains and clinical samples
Clinical samples were collected from suckling piglets with diarrhea on commercial pig farms in Guangdong, China. All specimens were suspended in phosphate-buffered saline (PBS) and stored at -80 °C until use. The SADS-CoV virus strain was isolated from diarrheal pigs in Guangdong Province of China (Zhou et al., 2018a). Other porcine viruses were used to assess the specificity of the real-time LAMP method. Classical swine fever virus (CSFV) C strain, porcine respiratory and reproductive syndrome virus (PRRSV) JXA1-R vaccine strain (NA type), foot-and-mouth disease virus (FMDV) O/HB/HK99 vaccine strain, porcine circovirus type 2 (PCV2) LG vaccine strain and swine influenza virus H1N1 TJ vaccine strain, were obtained from Guangdong Animal Epidemic Prevention and Material Reserve Center (Guangzhou, China). Porcine epidemic diarrhea virus (PEDV) CV777 vaccine strain (non-S-INDEL), rotavirus (RV) NX vaccine virus and transmissible gastroenteritis virus (TGEV) Huadu vaccine strain were purchased from Harbin Weike Biological Technology (Harbin, China). PRRSV ZD-1 strain (EU type) was acquired from Prof. Zhijun Tian of Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. PEDV GD1 strain (S-INDEL), seneca valley virus (SVV) CH-01–2015 strain and porcine deltacoronavirus (PDCoV) CHN-GD16-03 strain were stored in our lab. Porcine parvovirus (PPV) vaccine virus were obtained from Qilu Animal Health Products CO.,LTD. (Jinan, China).
2.2. RNA extraction
Frozen fecal samples were thawed and centrifuged for 10 min at 10,000 × g. Total RNA was extracted from the supernatants using a TGUide Virus DNA/RNA Kit and T-Guide instrument according to the manufacturer's instructions (Tiangen, Beijing, China). RNA was eluted into RNAse-free ddH2O and stored at −80 °C until use.
2.3. Design of primers for the real-time RT-LAMP
The genome sequences of SADS-CoV (Accession numbers: MF167434.1, MF370205.1, MF094681, MF094682, MF094683, MF094684, MG557844.1) were retrieved from GenBank. Based on the N gene region of SADS-CoV, six real-time LAMP primers were designed using the software http://primerexplorer.jp/e/ and synthesized by Shanghai Sangon Biotech (Shanghai, China) (GenBank: MG557844.1, Swine acute diarrhea syndrome coronavirus isolate SADS-CoV/CN/GDWT/2017). The primers included two inner primers (SADS-FIP and SADS-BIP) and two outer primers (SADS -F3 and SADS-B3) (Table 1 ).
Table 1.
Primers used to amplify the SADS-CoV N gene.
| Primers’ name | Sequence | Location in genome |
|---|---|---|
| SADS-F3 | 5′- CAGCCTTCTAACTGGCACTT -3′ | 25707bp–25726bp |
| SADS-B3 | 5′- ACAGTCAGGTCTGGTGGTAA -3′ | 25884bp–25903bp |
| SADS -FIP | 5′-CGTCAACAGCGACCCAATGCA-TCCTCACGCAGATGCTCC -3′ | (25786bp–25806bp)(25745bp–25762bp) |
| SADS -BIP | 5′-AACTAGCCCCACAGGTCTTGGT-AACCCAAACTGAGGTGTAGC -3′ | (25814bp–25835bp)(25860bp–25879bp) |
| SADS -LB | 5′- TCGCAATCGTAACAAAGAACCT -3′ | 25838bp–25859bp |
| SADS -LF | 5′- CACCCTGAATCCGTTTCCTG -3′ | 25766bp–25785bp |
Note: outer primers (SADS-F3 and SADS-B3); inner primers (SADS -FIP and SADS -BIP).
Loop primer (LF/LB).
2.4. Construction of plasmids containing the N gene of SADS-CoV
Reverse transcription of SADS-CoV RNA templates was performed with random primers following the manufacturer's recommendations using a One Step Prime Script RT-PCR kit (Takara, Shanghai, China). The target fragment was amplified using primers SADS-F3 and SADS-B3 (Table 1). PCR amplification from SADS-CoV N gene (1128bp) were amplified using standard PCR conditions and cloned into pGEM T easy vector (Promega, Madison, USA) between T7 RNA polymerase and SP6 polymerase promoter. The pGEM T easy plasmid was in vitro transcribed (IVT) by RiboMax Large Scale RNA production system T7 (Promega, Madison, USA) according to the manufacturer's instructions. To remove plasmid DNA, 40 units RNase-free DNase (Promega, Madison, USA) enzyme was used. Trizol LS reagent (Invitrogen, Carlsbad, CA) was used for RNA isolation according to manufacturer's instructions. The RNA concentrations were measured by UV spectrophotometry and converted into copy numbers as previously described (Parida et al., 2011). All RNA templates of SADS-CoV N gene were stored at - 80 °C until use.
2.5. Development of real-time RT-LAMP assay
The real-time RT-LAMP reaction was carried out in a 25 μL volume containing 2.5 μL 10 × Buffer (B0537S, New England Biolabs, Beverley, MA, USA), 0.2 mM outer primers (OF/OB), 1.6 mM inner primers (FIP/ BIP), 0.8 mM Loop primer (LF/LB), 1 μL Bst DNA polymerase (M5038 L, New England Biolabs), 3.5 μL dNTP Mix (D7373, Beyotime Biotechnology, Beijing, China), 4 μL betaine (BCBS087 V, Sigma, St. Louis, MO USA), 0.5 μL SYTO 9 green fluorescent nucleic acid stain (S34854, Invitrogen), 0.5 μL AMV Reverse Transcriptase (M510 A Promega) and 2 μL RNA template. Amplification reactions were carried out at 63 °C for 60 min and a Thermostatic Fluorescence Detector (DEAOU-308C, Diao, Guangzhou, China) was used to monitor reaction progress. Non-template control was used in each reaction.
Specificity of the method was determined using RNA templates of SADS-CoV and other swine viral RNA samples including CSFV, TGEV, FMDV, PEDV (S-INDEL and non-S-INDEL), PRRSV NA and EU, PCV2, SVV, PPV, PDCoV, RV, and SIV H1N1. Ten-fold serial dilutions of N gene RNA templates were used to calculate the analytical sensitivity and assess assay reproducibility. The method was further evaluated using 24 clinical specimens from pigs with diarrhea that had been previously evaluated by a real-time RT-PCR assay (Zhou et al., 2018a). Vero cell nucleic acid was used as a mock control. Nuclease free water was used as a negative control. All reactions were carried out in triplicate.
3. Results
3.1. Specificity, sensitivity and reproducibility
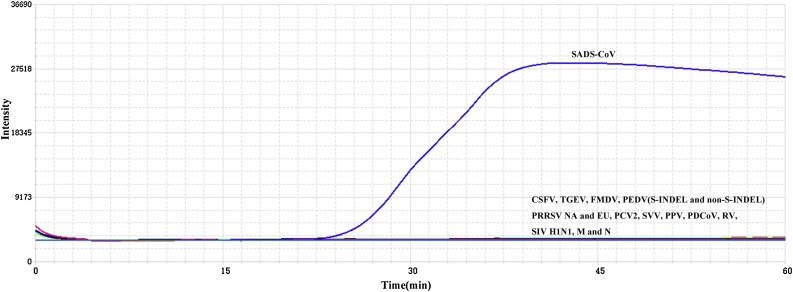
Only samples that contained SADS-CoV were positive while there was no amplification of other tested viruses (Fig. 1 ). This indicated that the assay was highly specific for the detection of SADS-CoV.
Fig. 1.
Specificity of the real time RT-LAMP assay for SADV-CoV. The SADV-CoV RNA was amplified by the assay, but not other swine viruses including classical swine fever virus (CSFV), porcine and respiratory syndrome virus (PRRSV) NA and EU, transmissible gastroenteritis corona virus (TGEV), foot and mouth disease virus (FMDV), porcine epidemic diarrhea virus (PEDV) (S-INDEL and non-S-INDEL), porcine circovirus type 2 (PCV2), seneca valley virus(SVV), porcine parvovirus (PPV), porcine deltacoronavirus (PDCoV), rotavirus (RV), and swine influenza virus (H1N1). Vero cell nucleic acid was used a mock control (M). Nuclease free water was used as a negative control (N). The assay was repeated thrice.
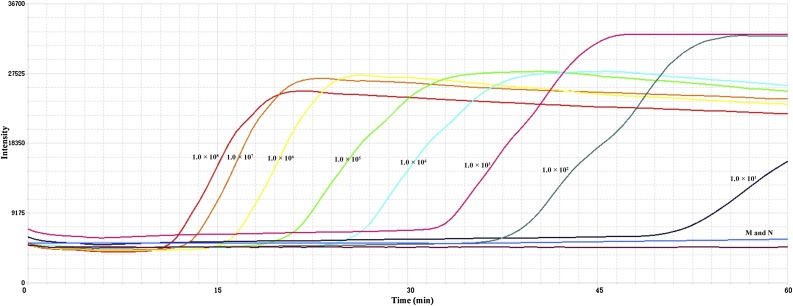
To evaluate RT-LAMP sensitivity, the SADS-CoV N gene was in vitro transcribed from a cloned fragment. The detection limit of this assay is 1.0 × 101 copies /μL (Fig. 2 ). This indicated that the real-time LAMP assay was a sensitive method for SADS-CoV detection. To investigate the reproducibility of the reaction, different concentrations of the SADS-CoV N gene RNA at 1.0 × 108, 1.0 × 107, 1.0 × 106, 1.0 × 105, 1.0 × 104, 1.0 × 103, 1.0 × 102 and 1.0 × 101 copies/μL. Our results obtained from three experiments showed intra coefficients of variation were less than 2.5% and inter coefficients of variation were less than 6.8% (Supplementary Table 1). This indicated that the method was repeatable.
Fig. 2.
Sensitivity of the real time RT-LAMP assay for SADS-CoV. Serial dilutions of in vitro transcribed copies of the cloned SADS-CoV N gene were tested in the assay. The detection limit of this assay was 1.0 × 101 copies. Vero cell nucleic acid was used a mock control (M). Nuclease free water was used as a negative control (N). The assay was repeated thrice.
3.2. Clinical sample testing
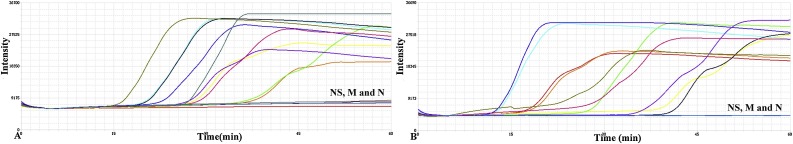
Our laboratory has previously tested 24 clinical samples from pigs with diarrheal symptoms. The real time RT-PCR tests indicated that twenty of these samples contained SADS-CoV (Zhou et al., 2018a). These 24 samples were again tested with the real-time RT-LAMP method. The real-time RT-LAMP assay could distinguish between the positive and negative samples (Fig. 3 ). Therefore, real-time LAMP was sufficiently accurate and reliable for the detection of SADS-CoV.
Fig. 3.
Detection of the SADS-CoV RNA from clinical samples by real time RT-LAMP assay. (A) and (B), Amplification of 20 SADS-CoV positive and 4 negative samples previously assayed using real-time RT-PCR were divided into two groups. NS, negative samples; M, mock control (vero cell nucleic acid), N, negative control (nuclease free water). Each group contained 10 positive samples, 2 negative samples, a mock control and a negative control. This assay was repeated thrice.
4. Discussion
SADS-CoV is a novel swine enteric alphacoronavirus identified in Guangdong province of China in 2017 (Gong et al., 2017; Pan et al., 2017). This disease has already caused huge economic losses in the Chinese swine industry. However, its detection was problematic since the disease symptoms are similar to other viruses (Jung et al., 2016; Pensaert and de Bouck, 1978; Stevenson et al., 2013). Although a TaqMan-based real-time RT-PCR assay had already been developed for SADS-CoV, this method required expensive equipment and supplies. Therefore, a simple and rapid method was needed for field studies of SADS-CoV infections. In this study, a real-time RT-LAMP assay was successfully developed using SADS-CoV N gene specific primers. The assay is suitable for SADS-CoV detection in less well-equipped laboratories.
LAMP technology is currently used for the nucleic acid detection of a number of animal pathogens (Niessen et al., 2013; Sahoo et al., 2016). The assay offers multiple advantages compared to conventional PCR including a shorter turnaround time (30–60 min) and a sensitivity 1–2 orders of magnitude higher (Huang et al., 2016; Yu et al., 2015). In the present study, the real-time RT-LAMP method was used to detect SADS-CoV and did not cross-react with other porcine pathogens. SADS-CoV N gene was specifically amplified with a detection limit of 1.0 × 101 copies/μL and had good reproducibility. The sensitivity of the LAMP assay is similar to real-time RT-PCR (Yu et al., 2015). In this study, the RT-LAMP correctly identified 20 out of 24 samples as positive, showing similar diagnostic sensitivity comparable to the real time RT-PCR. Together our data indicates that the real-time RT-LAMP assay was accurate and sensitive.
In conclusion, a novel real-time RT-LAMP method was developed for the detection of SADS-CoV in pigs. The assay was specific, sensitive and reproducible and will be helpful for the diagnosis, surveillance and pathogenesis studies of SADS-CoV infections.
Conflict of interest
Authors declare no competing interests.
Acknowledgments
The study was supported by the Science and Technology Research Program (2017A070702001, 2017B030314171, 2016A040403060 and 2016201604030059) of the Science and Technology Department of Guangdong Province and Guangzhou.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2018.06.010.
Contributor Information
Manlin Luo, Email: 710510116@qq.com.
Pengju Guo, Email: vetbio2016@hotmail.com.
Jingyun Ma, Email: majy2400@scau.edu.cn.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- de Souza D.K., Quaye C., Mosi L., Addo P., Boakye D.A. A quick and cost effective method for the diagnosis of mycobacterium ulcerans infection. BMC Infect. Dis. 2012;12:8. doi: 10.1186/1471-2334-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Li J., Zhou Q.F., Xu Z.C., Chen L., Zhang Y., Xue C.Y., Wen Z.F., Cao Y.C. A New Bat-HKU2-like coronavirus in swine, China, 2017. Emerg. Infect. Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wen F., Yue L., Chen R., Zhou W., Hu L., Chen M., Wang S. Exploration of fluorescence-based real-time loop-mediated isothermal amplification (LAMP) assay for detection of Isospora suis oocysts. Exp. Parasitol. 2016;165:1–6. doi: 10.1016/j.exppara.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Y., Martins D.B.G., Kimura M., Catena A.D.S., Borba M., Mattos S.D.S., Abe H., Yoshikawa R., de Lima Filho J.L., Yasuda J. Development and evaluation of a rapid molecular diagnostic test for Zika virus infection by reverse transcription loop-mediated isothermal amplification. Sci. Rep. 2017;7:13503. doi: 10.1038/s41598-017-13836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ma J.J., Xiao S.B., Zhang X.H., Wen L.B., Mao L., Ni Y.X., Guo R.L., Zhou J.M., Lv L.X., He K.W. Development of a loop-mediated isothermal amplification method for rapid detection of porcine boca-like virus. J. Virol. Methods. 2012;179:390–395. doi: 10.1016/j.jviromet.2011.11.027. [DOI] [PubMed] [Google Scholar]
- Niessen L., Luo J., Denschlag C., Vogel R.F. The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol. 2013;36:191–206. doi: 10.1016/j.fm.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscorbin I.P., Belousova E.A., Zakabunin A.I., Boyarskikh U.A., Filipenko M.L. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP) Biotechniques. 2016;61:20–25. doi: 10.2144/000114432. [DOI] [PubMed] [Google Scholar]
- Pan Y.F., Tian X.Y., Qin P., Wang B., Zhao P.W., Yang Y.L., Wang L.X., Wang D.D., Song Y.H., Zhang X.B., Huang Y.W. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Shukla J., Sharma S., Ranghia Santhosh S., Ravi V., Mani R., Thomas M., Khare S., Rai A., Kant Ratho R., Pujari S., Mishra B., Lakshmana Rao P.V., Vijayaraghavan R. Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for rapid and real-time detection of the swine-origin influenza a H1N1 virus. J. Mol. Diagnostics JMD. 2011;13:100–107. doi: 10.1016/j.jmoldx.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo P.R., Sethy K., Mohapatra S., Panda D. Loop mediated isothermal amplification: an innovative gene amplification technique for animal diseases. Vet. World. 2016;9:465–469. doi: 10.14202/vetworld.2016.465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Yu X.W., Shi L., Lv X.P., Yao W., Cao M.H., Yu H.X., Wang X.R., Zheng S.M. Development of a real-time reverse transcription loop-mediated isothermal amplification method for the rapid detection of porcine epidemic diarrhea virus. Virol. J. 2015;12 doi: 10.1186/s12985-015-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W.Z., Wang J.C., Zheng Y.S., Li L.M., Zhang X.Y., Sun J.G. Rapid detection of encephalomyocarditis virus by one-step reverse transcription loop-mediated isothermal amplification method. Virus Res. 2014;189:75–78. doi: 10.1016/j.virusres.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Zhao K., Shi W., Han F.T., Xu Y., Zhu L.L., Zou Y., Wu X.A., Zhu H., Tan F.R., Tao S.R., Tang X.M. Specific, simple and rapid detection of porcine circovirus type 2 using the loop-mediated isothermal amplification method. Virol. J. 2011;8 doi: 10.1186/1743-422X-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Sun Y., Wu J.L., Mai K.J., Chen G.H., Wu Z.X., Bai Y., Li D., Zhou Z.H., Cheng J., Wu R.T., Zhang X.B., Ma J.Y. Development of a TaqMan-based real-time RT-PCR assay for the detection of SADS-CoV associated with severe diarrhea disease in pigs. J. Virol. Methods. 2018;255:66–70. doi: 10.1016/j.jviromet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556 doi: 10.1038/s41586-018-0010-9. 255-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.