Abstract
Replication of eukaryotic positive-stranded RNA viruses is usually linked to the presence of membrane-associated replicative organelles. The purpose of this review is to discuss the function of proteins responsible for formation of the coronavirus replicative organelle. This will be done by identifying domains that are conserved across the order Nidovirales, and by summarizing what is known about function and structure at the level of protein domains.
Highlights
-
•
Bioinformatics reveals a new domain-level map of coronavirus nsp3-nsp6.
-
•
Domain-level protein variability is a tool for functional annotation.
-
•
Ten nsp3 domains are conserved in all known coronaviruses.
-
•
Review of the role of the nsp5 main protease in RNA synthesis.
1. Introduction
The order Nidovirales includes several families of large RNA viruses, arranged from longest to shortest genome as the Coronaviridae, Roniviridae, Mesoniviridae and Arteriviridae. The Coronaviridae currently contains the Torovirinae and Coronavirinae lineages, though analysis of recently reported divergent toro-like viruses suggests that the Torovirinae may be better represented as an independent family in the Nidovirales (Stenglein et al., 2014). The Coronavirinae currently contains four genera – Alpha- and Betacoronavirus that infect mammals, and Gamma- and Deltacoronavirus that infect birds and mammals.
Members of the Nidovirales infect metazoan hosts, have several replicative genes in common (Lauber et al., 2013), express their structural genes via subgenomic mRNAs which usually join sequences from both genomic termini (Sawicki et al., 2007), express their replicases as polyproteins via a ribosomal frameshift, and replicate in association with viral transmembrane proteins on intracellular paired membranes (reviewed in (V'Kovski et al., 2015)). Coronavirus growth is accompanied by a variety of intracellular membrane rearrangements, as illustrated for the coronavirus Mouse hepatitis virus (MHV) in Fig. 1 . Regions of these paired-membrane structures have been given a variety of names in the literature, including double-membrane vesicles (DMVs), convoluted membranes, spherules, zippered endoplasmic reticulum, but it is not clear whether the different parts of the organelle have different functions. Furthermore, recent studies (Al-Mulla et al., 2014, Maier et al., 2016) suggest that there may be considerable plasticity and overlap among coronavirus paired membrane replicative structures. For this reason, it seems preferable to break with past practice of focussing on double-membrane vesicles in particular, to consider the double-membrane organelle (DMO) as a whole. The term DMO encompasses DMVs as well as all of the other associated paired-membrane structures like convoluted membranes and spherules that make up the viral replicative organelle. The term DMO will be used throughout this review except when specifically referring to the DMV component of the replicative organelle. This review summarizes what we know about coronavirus DMOs, and what bioinformatics can tell us about DMO-making proteins.
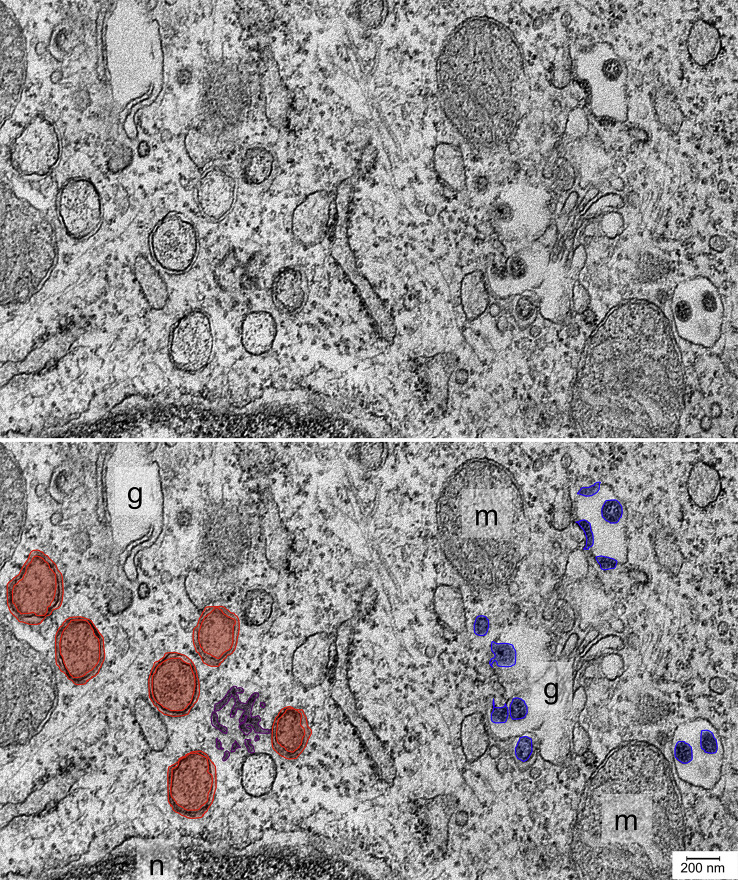
Fig. 1.
Intracellular changes following coronavirus infection. Electron micrograph of an ultrathin section of a murine DBT cell infected with Mouse hepatitis virus. Double-membrane vesicles (red) and convoluted membranes (purple) associated with viral RNA synthesis can be seen at left and new virions (blue) can be seen associated with Golgi membranes at right.
2. Origin of DMO membranes
Membrane-bound replicative organelles are a widespread but not universal intracellular feature associated with positive-strand RNA virus replication (Neuman et al., 2014a). Coronaviruses form a double-membrane organelle (DMO) derived from the endoplasmic reticulum (ER) that contains viral RNA and replicase proteins (Deming et al., 2007, Hagemeijer et al., 2010, Oostra et al., 2007, Reggiori et al., 2010, Shi et al., 1999, van der Meer et al., 1999). In the DMO, two lipid bilayers are held at a constant distance of about 20 nm (Angelini et al., 2013). Electron microscopy and tomography studies have revealed that the replicative organelles of coronaviruses and the related arteriviruses are drawn from a repertoire of paired-membranes, including open-ended spherules, closed double-membrane vesicles, and both planar and convoluted paired membranes (Knoops et al., 2008, Knoops et al., 2012, Maier et al., 2013). Nonstructural proteins nsp3, nsp4 and nsp6 are required to form structures similar to the DMOs observed in SARS coronavirus (SARS-CoV) infected cells (Angelini et al., 2013). And since protease domains of nsp5 are required to release nsp4 and nsp6 from the polyprotein precursor, the region from nsp3 to nsp6 can collectively be thought of as the DMO-forming apparatus of coronaviruses. Phylogenetic analysis and comparison of domain architecture can be taken as evidence of homology of the coronavirus DMO-making proteins across the Nidovirales (Fig. 2 ).
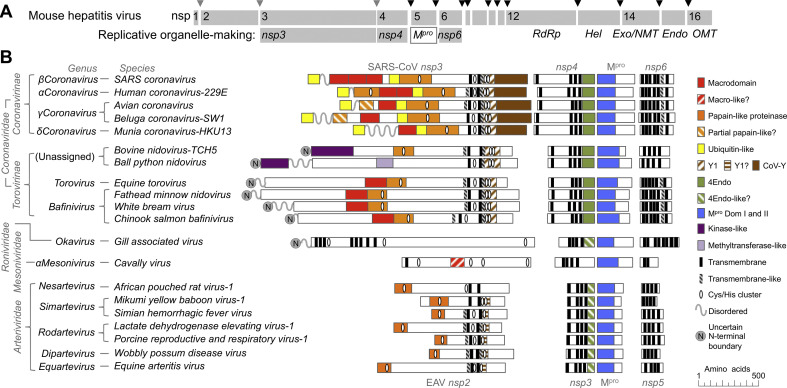
Fig. 2.
Organization and conservation of genes involved in DMO formation throughout the Nidovirales. (A) A schematic of the SARS coronavirus polyprotein showing the organization of nonstructural proteins (nsp) and the location of components important for RNA synthesis and replicative organelle formation. Sites where the polyprotein is cleaved by the main protease (Mpro) are marked with black triangles, and cleavage sites of nsp3 proteases are marked by grey triangles. The RNA-dependent RNA polymerase (RdRp), superfamily 1 helicase (Hel), exonuclease (Exo), endonuclease (Endo), and the two viral cap methyltransferases (NMT, OMT) are shown. (B) Organization of the proteins most likely to be involved in formation of replicative organelles across the order Nidovirales. Conserved domains were identified by amino acid alignment, or by comparison of predicted protein secondary structure. Domain function was predicted by hhpred (Soding et al., 2005). Potential cleavage sites were annotated by analogy to known cleavage sites and proximity to conserved domains. Clusters with 4 histidine or cysteine residues within 25 amino acid residues that may bind metal ions are indicated. Conserved transmembrane regions (black) and regions predicted as transmembrane helices by TMHMM 2.0 (Sonnhammer et al., 1998) but shown experimentally not to be transmembrane are zebra striped.
The formation of paired membranes probably involves interactions on both sides of the membrane, and perhaps within the membrane, and a few of these interactions have been confirmed biochemically. SARS-CoV nsp3-nsp3 interactions have been detected in cells by yeast two-hybridization (Pan et al., 2008) and GST pulldown (Imbert et al., 2008), and in purified protein by perfluorooctanoic acid polyacrylamide gel electrophoresis (Neuman et al., 2008). While SARS-CoV nsp4-nsp4 interactions were not found in yeast-two hybrid or mammalian two-hybrid screens (Pan et al., 2008, von Brunn et al., 2007) studies with MHV did detect nsp4-nsp4 interactions by Venus reporter fluorescence (Hagemeijer et al., 2011). To date, homotypic interactions have not been demonstrated for nsp6 despite several attempts (Imbert et al., 2008, Pan et al., 2008, von Brunn et al., 2007). Heterotypic interactions between coronavirus nsp3 and nsp4 have been observed by mammalian two-hybridization (Pan et al., 2008) and Venus reporter fluorescence (Hagemeijer et al., 2011), although because of differences in the parts of nsp3 that were used, these may actually represent two distinct modes of interaction between nsp3 and nsp4. Nsp4-nsp6 interaction has been demonstrated by Venus reporter fluorescence (Hagemeijer et al., 2011) and indirectly because co-expression of nsp4 abrogates vesicle accumulation due to expression of nsp6 (Angelini et al., 2013). Each of these proteins also potentially interacts with host proteins, which may have a downstream effect on pathogenesis (Pfefferle et al., 2011).
Coexpression of nsp3 and nsp4 produces large areas of paired membranes, apparently arranged in parallel tubes (Angelini et al., 2013, Hagemeijer et al., 2014). In terms of topology, nsp3-4 membrane pairing involves linking opposite sides of the ER across the ER lumen, so it is likely that the luminal domains of both proteins are involved in this interaction. Membrane pairing would not necessarily need to involve high-affinity interactions, as demonstrated by a study where paired membranes were induced by low-affinity interactions between membrane-linked green fluorescent protein (Snapp et al., 2003). The minimal requirements for DMO-like membrane pairing appear to be the C-terminal region of nsp3 that contains both transmembrane regions and the luminal ectodomain (Hagemeijer et al., 2014), and at least the N-terminal region of nsp4 including the first three transmembrane regions of nsp4 (Sparks et al., 2007). Mutations in the glycosylated luminal domain of nsp4 result in either non-recoverable virus or less consistent membrane pairing (Gadlage et al., 2010). While the final transmembrane C-terminal cytosolic domain of nsp4 is dispensible for coronavirus replication (Sparks et al., 2007), it is not clear whether partial deletion of nsp4 affects DMO structure. These observations together suggest that coronavirus membranes most likely pair through heterotypic interactions involving the luminal domains of nsp3 and nsp4, though interactions between cytosolic domains that lead to nsp3 and nsp4 clustering may also be important for membrane pairing.
The ER is the likely source of coronavirus DMO membranes, which may be obtained by co-opting the ER-associated degradation (ERAD) tuning pathway, a cellular degradation pathway that is responsible for the turnover of misfolded proteins in the ER (Reggiori et al., 2010). The ERAD tuning pathway is modulated by stress-inducible positive regulators of protein disposal such as EDEM1 (ER degradation-enhancing alpha mannosidase-like 1) and OS-9 (osteosarcoma amplified 9), which assist in transporting misfolded proteins into the cytosol for proteasomal degradation. Under physiological conditions, low concentrations of EDEM1 and OS-9 are maintained in the ER lumen in order to avoid premature degradation of proteins that are in the process of folding (Calì et al., 2008). In this case, EDEM1 and OS-9 are selectively confined by interacting with the transmembrane-anchored cargo receptor SEL1L (suppressor of lin-12-like) and later released from the ER lumen in small short-lived vesicles, called EDEMosomes, which rapidly fuse with the endolysosomal compartments (Bernasconi et al., 2012). In infected cells, viral double-stranded RNA colocalizes with EDEM1, OS-9, SEL1L and LC3-I, which is recruited to autophagosomes. Moreover, replication of MHV, which does not require an intact autophagy pathway, is impaired upon knockdown of LC3 or SEL1L (Bernasconi et al., 2012). Taken together, this is evidence that MHV exploits the ERAD-tuning machinery to establish DMOs for replication. A summary of nsp3-6 interactions and induced membrane rearrangements is shown in Fig. 3 .
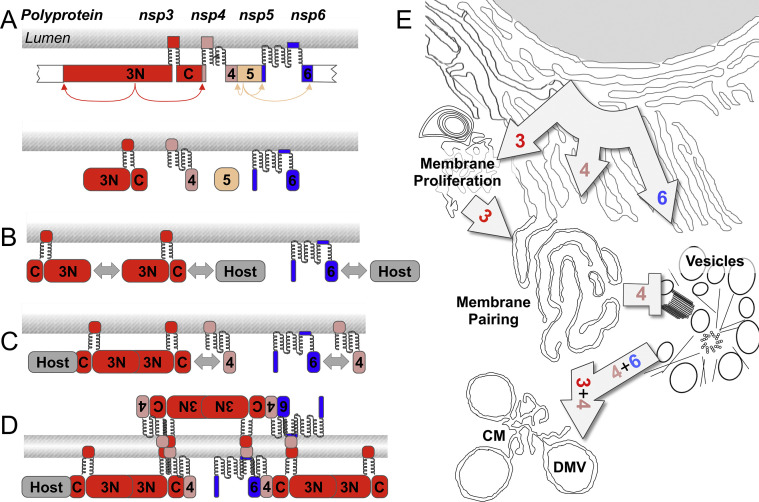
Fig. 3.
Interactions and intracellular changes associated with expression of SARS-CoV nonstructural protein 3, 4 and 6. Panel A is a schematic view of the topology and processing of nsp3-6 from polyprotein precursors. The amino-terminal (3N) and carboxyl-terminal (C) regions of nsp3 are labelled. Panels B–D show putative nsp-nsp and nsp-host interactions that may contribute to membrane phenotypes and membrane pairing. Panel E summarizes the effects of SARS-CoV nsp3, 4 and 6 expression on intracellular membrane appearance.
3. Nsp3
Coronavirus nsp3 is a large multidomain protein ranging from around 1450 amino acid residues in Deltacoronavirus (Woo et al., 2012) to nearly 2100 amino acid residues in the unpublished Hipposideros pratti betacoronavirus-Zhejiang2013 (GenBank accession NC_025217). Most nsp3s are predicted to be ∼200 kDa, and are cleaved from the polyprotein 1a or 1ab papain-like proteases (PLpro) that are encoded within nsp3.
To make it easier to discuss specific parts of such a large protein in terms of both structure and function, we previously published a domain-level annotation of nsp3 (Neuman et al., 2008), which we have updated for this review (Fig. 4 ).
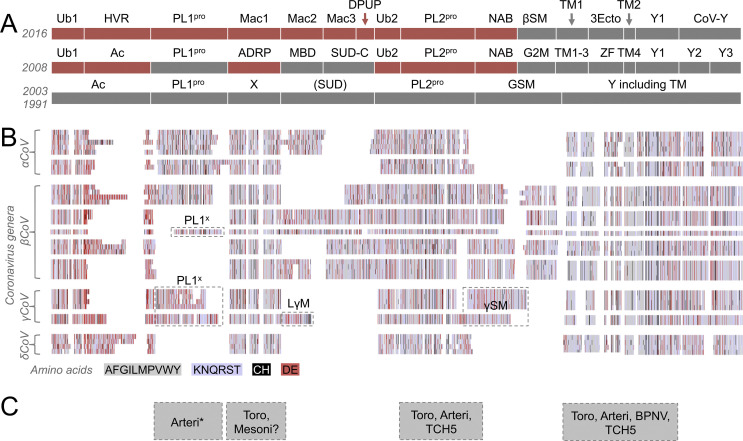
Fig. 4.
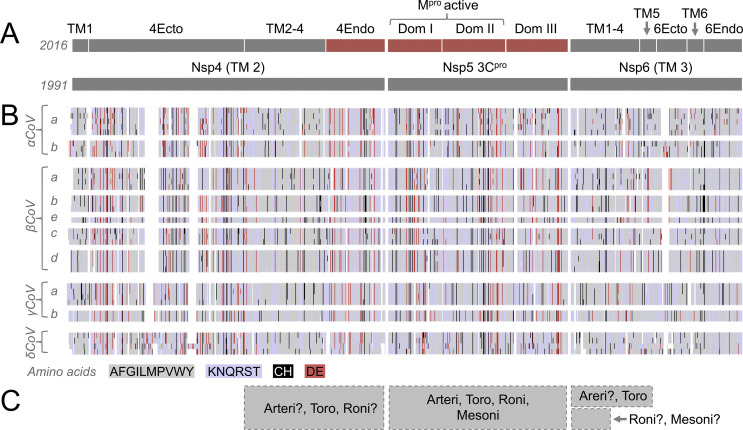
Revised domain-level organization of nsp3 in the Coronavirinae. In panel A, domain annotations based on the ongoing work of Gorbalenya and collaborators (1991 and 2003), our previous annotation (2008) and our new revised annotation (2016) are shown for comparison. Structures that were solved at the time of domain assignment are shown in red, and domain assignments based on amino acid alignments are shown in grey. Panel B is based on a multiple sequence alignment of nsp3 amino acid sequences from the genera Alphacoronavirus (αCoV), Betacoronavirus (βCoV), Gammacoronavirus (γCoV) and Deltacoronavirus (δCoV). Four to fifteen amino nsp3 protein sequences are shown per genus, organized into phylogenetic clusters. Amino acid residues in the alignment have been converted to colored blocks to show domain-level conservation and highlight areas with clusters of well-conserved hydrophobic (grey), polar (light blue), acidic (red) and cysteine/histidine (black) residues. Detailed alignments can be made available by writing to the corresponding author. Domain designations include ubiquitin-like (Ub1, Ub2), hypervariable region (HVR) previously described as acidic (Ac), full (PL1pro, PL2pro) or partial (PL1x) papain-like protease that is missing at least one domain and one catalytically important amino acid, macro domains Mac1 (previously X or ADP-ribose 1″ phosphatase), Mac2 (previously SARS-CoV unique domain N-terminal or metal-binding domain MBD), Mac3 (previously SARS-CoV unique domain middle), domain preceding Ub2 and PL2pro (DPUP), nucleic acid binding (NAB), group specific markers (GSM), Betacoronavirus specific marker (βSM, previously group 2 specific marker G2M), Gammacoronavirus specific marker (γSM), large Gammacoronavirus marker (LγM), transmembrane region (TM1, TM2), nsp3 ectodomain (3Ecto, formerly zinc finger ZF), Y domain regions Y1 and coronavirus-specific Y domain (CoV—Y). In panel C, grey boxes show regions of apparent homology between coronavirus nsp3 and the equivalent proteins of Arteriviridae (Arteri), Mesoniviridae (Mesoni), Torovirinae including Bafinivirus (Toro), Ball python nidovirus (BPNV) and Bovine nidovirus-TCH5, and present in Arteriviridae but processed into separate polypeptides (Arteri*).
Based on phylogenetic analysis of nidovirus nsp3 homologues, results from previously published studies (Gorbalenya et al., 2006, Ratia et al., 2006, Saikatendu et al., 2005, Serrano et al., 2007b, Thiel et al., 2003, Ziebuhr et al., 2001) and de novo domain prediction software (Jaroszewski et al., 2005), we estimate that the full repertoire of sequenced coronavirus nsp3 genes encodes 18 domains, with individual viruses having 10–16 domains each. Several of these domains are duplicated, including two ubiquitin-like domains Ub1 and Ub2, two PLpro, and three macrodomains (Mac1, Mac2 and Mac3). Ten of these domains form the core of coronavirus nsp3, and are found in every currently known coronavirus, including both ubiquitin-like domains, the second PLpro, the first macrodomain Mac1, a hypervariable region consisting of mostly acidic residues, and a region including the transmembrane regions TM1 and TM2, nsp3 ectodomain (3Ecto), a nidovirus-conserved domain of unknown function (Y) and a region predicted to contain two structural domains which are only found in coronaviruses (CoV—Y). Six of the ten domains conserved in all coronaviruses are also found in other members of the Nidovirales, with evidence that the region from TM1 to Y is present in all nidoviruses except the two families that infect arthropods, namely the Roniviridae and Mesoniviridae (V'Kovski et al., 2015).
The ectodomain of nsp3, 3Ecto, is glycosylated in SARS-CoV at positions 1431 and 1434 (Harcourt et al., 2004) and the corresponding region of MHV (Kanjanahaluethai et al., 2007), and is predicted to be located on the luminal side of the membrane. Each copy of nsp3 is predicted span the membrane twice, placing the first 1395 residues of SARS-CoV nsp3 and the last 377 residues (Y and CoV—Y) on the cytosolic face of the membrane. Notably, the regions immediately before TM1 and after TM2, which would both have a cytosolic membrane topology are highly hydrophobic, and may serve to link the pre- and post-transmembrane regions of nsp3.
4. Nsp3 (Ub1 to PL1pro)
The N-terminal domain of all coronavirus nsp3 proteins containing Ub1, a hypervariable region (HVR) and PL1pro appears poorly conserved at first glance, showing less than 20% average amino acid identity between members of different coronavirus genera (Fig. 5 ), but secondary structure prediction (JPred; (Drozdetskiy et al., 2015)) suggests that the Ub1 and PL1pro domains adopt a conserved fold in all coronaviruses (data not shown). The NMR structure of the residues 1–112 of SARS-CoV nsp3 exhibits a globular ubiquitin-like fold with two additional helices which make the overall structure of the Ub1 domain somewhat more elongated than other ubiquitin-like proteins (Serrano et al., 2007b). In contrast, the following HVR was shown to be structurally disordered for SARS-CoV (Serrano et al., 2007b) and is dispensable for replication in MHV (Hurst et al., 2013).
Fig. 5.
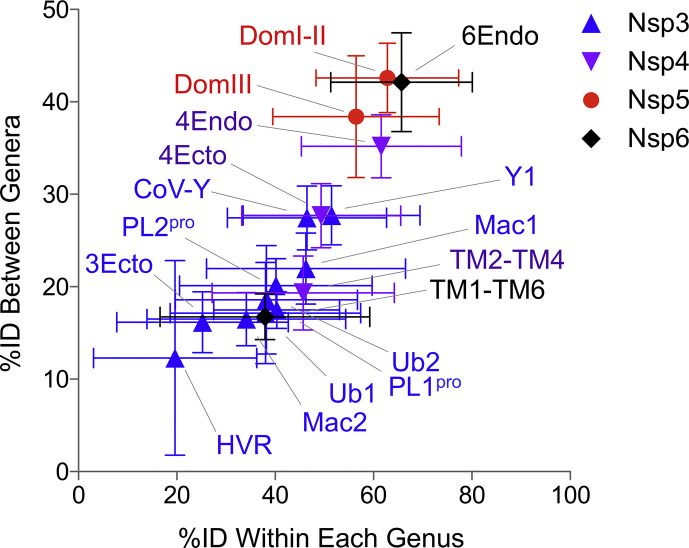
Variability in domain-level conservation in DMO-making proteins of the Coronavirinae. The thirty-three representative coronavirus protein sequences shown Fig. 4, Fig. 6 were aligned by Clustal Omega (Sievers et al., 2011). Percentage amino acid identity (%ID) was calculated between viruses in the same genus or in different genera. Average percent amino acid identity was calculated from 528 unique comparisons and plotted plus or minus standard deviation.
While the function of Ub1 has only been investigated in MHV, this domain has been found to play an essential role in initiating viral RNA synthesis, where it interacts with the viral nucleoprotein (N; (Hurst et al., 2013, Hurst-Hess et al., 2015)). This was demonstrated in experiments that attempted to delete nsp3 domains, or substitute with the corresponding domains from other coronaviruses. The interaction of Ub1 with N could effectively tether nsp3 to viral RNA during the replication process, but further experimentation is needed to better understand how the nsp3-N interaction leads to more efficient viral RNA synthesis.
Additionally, Ub1 has an extra alpha helix and a 310 helix that is unusual for ubiquitin folds in general (Serrano et al., 2007a). Among the closest structural matches to SARS-CoV Ub1 is one of the ubiquitin-like domains of ISG15, an interferon-induced protein constitutively present in higher eukaryotes. This has led to speculation that Ub1 may be involved in modulating the effects of intracellular immunity in a manner analogous to the immunomodulatory decoys of poxviruses (Johnston and McFadden, 2003). Some viruses have developed a mechanism to avoid the expression of ISG15. For example, Influenza B virus blocks its expression by means of NS1 protein in order to overcome the immune response. The PL2pro domain of SARS-CoV nsp3 also recognizes and cleaves ISG15 (Lindner et al., 2007), which could potentially modulate the intracellular response to infection (Morales et al., 2015). However, the comparison with ISG15 remains speculative because an immunomodulatory function for Ub1 has not yet been demonstrated experimentally. It is known that ISG15 is able to inhibit virus replication by abrogating nuclear processing of unspliced viral RNA precursors. However, some viruses have developed a mechanism to avoid the expression of ISG15. For example, influenza B virus blocks its expression by means of NS1 protein in order to overcome the immune response. It is possible that the PL2pro domain of nsp3 may also bind ISG15 and modulate the intracellular response to infection (Morales et al., 2015). However, the comparison with ISG15 remains speculative because an immunomodulatory function for Ub1 has not yet been demonstrated experimentally.
When SARS-CoV Ub1 was expressed in E. coli, it was found to bind tightly to a small RNA fragment that mass spectrometry analysis revealed to be consistent with the sequence GAUA or GUAA (Serrano et al., 2007a). While matching sequences can be found throughout the SARS-CoV, none are prominently located in regions of the 5′-UTR or 3′-UTR that are known to contain sequences essential for recognition of viral RNA by components of the replicase. The functional significance of RNA-binding by Ub1 therefore remains unknown, but may complement the Ub1-N interaction.
A papain-like protease domain (PL1pro) follows the HVR domain in some coronaviruses, but is absent in SARS-CoV and Middle east respiratory syndrome coronavirus (MERS-CoV). Where present, PL1pro generally cleaves at the N-terminal boundary of nsp3, but in viruses that have only one PLpro, this cleavage is carried out by PL2pro (Hilgenfeld, 2014). A transcription factor-like zinc finger is conserved in all complete coronavirus PLpro domains (Culver et al., 1993), which was taken as an early indication that nsp3 might be involved in coronavirus RNA synthesis. This hypothesis is supported by a report in which the Equine arteritis virus nonstructural protein 1, which is structurally and enzymatically similar to coronavirus PL1pro (Sun et al., 2009), was shown to be indispensable for viral subgenomic mRNA synthesis (Phizicky and Greer, 1993).
Some strains of Infectious bronchitis virus and Hipposideros pratti betacoronavirus-Zhejiang2013 contain only partial papain-like protease domains that are lacking one of the two catalytic domains (Fig. 2). It is not clear whether these domains are able to interact with the missing domains from PL2pro and remain functional, or whether these are inactive relict domains derived from an ancestral genome that are no longer necessary for viral growth and are in the process of being deleted.
5. Nsp3 (Mac1 to DPUP)
The next conserved domain is the first of up to three domains that adopt macrodomain folds similar to the histone H2A in SARS-CoV (Saikatendu et al., 2005). This domain was originally called an X-domain (Gorbalenya et al., 1991), an alternate name that is still used in some publications. The structure of Mac1 has been solved for several coronaviruses (Piotrowski et al., 2009, Saikatendu et al., 2005, Tan et al., 2009, Wojdyla et al., 2009, Xu et al., 2009). Purified SARS-CoV Mac1 was shown to have relatively weak ADP ribose 1″ phosphatase (ADRP) activity (Saikatendu et al., 2005). Enzymatic activity of Mac1 was predicted to be due to His45 based on comparison with the homologous protein from yeast Ymx7, Archaeoglobus fulgidus AF1521 and Er58 from E. coli. The active site was verified by site directed mutagenesis data in the Human coronavirus 229E (HCoV-229E) Mac1, which suggested that the corresponding residues Asn37, Asn40, His45, Gly44 and Gly48 of SARS-CoV Mac1 are part of the active site (Putics et al., 2005). Both the SARS-CoV ADRP and the HCoV-229E counterpart dephosphorylate ADP ribose 1‘’ phosphate to ADP-ribose in a highly specific manner, the enzyme having no detectable activity on several other nucleoside phosphates (Putics et al., 2005). Characterization of an ADP ribose 1″phosphatase-deficient HCoV-229E mutant revealed no significant effects on viral RNA synthesis and virus titer (Putics et al., 2005), but mutation of the Mac1 active site altered MHV pathogenicity, suggesting that this domain may play an immunomodulatory role by interacting with unknown host factors (Kuri et al., 2011).
The existence of an ADRP-like domain in all CoV nsp3s (as well as in several other RNA viruses) suggests that ADP ribose phosphatase activity confers some sort of advantage to coronaviruses. Egloff et al. suggested that Mac1 may primarily be a poly-ADP-ribose binding (PAR-binding) module, rather than an ADP-ribose cleaving enzyme (Egloff et al., 2006). PARylation occurs when PAR polymerases are activated, often in compromised cells, to trigger apoptosis or DNA repair. Perhaps more relevant to coronavirus infection, the formation of cytoplasmic stress granules, which are aggregates of cellular stalled translation complexes, appears to be regulated by PARylation (Leung, 2014). Further research is needed to determine whether coronavirus ADRP expression affects RNA granule formation or stability. However, the Infectious bronchitis virus Mac1 does not have detectable ADP-ribose binding activity suggesting that Mac1 domains may have other important functions that are unrelated to ADP-ribose (Piotrowski et al., 2009).
In SARS-CoV Mac1 is followed by two more macrodomains (Chatterjee et al., 2009, Tan et al., 2009) that form part of the region that was originally called the SARS-CoV Unique Domain (SUD). However, in light of more recent structural evidence that the SUD contains not one but three distinct structural domains, and phylogenetic evidence that some SUD domains are not unique to SARS-CoV, a revision to the name of this region is necessary (Chen et al., 2015). This region was originally divided into N-terminal and middle domains, which appear in the literature as SUD-N, SUD-M, but are here renamed as Mac2 and Mac3 to reflect their conservation outside SARS-CoV, followed by a small C-terminal domain known as DPUP for its position as the Domain Preceding Ub2 and PL2pro. The structure of both Mac2 and Mac3 is a close structural match for the SARS-CoV Mac1 domain despite a lack of detectable amino acid homology between these proteins (Tan et al., 2009). The presence of these additional macrodomain folds has also been confirmed by the NMR structure of the complete Mac2-Mac3-DPUP (Johnson et al., 2010) and the NMR structure of SUD-M (Chatterjee et al., 2009). The SARS-CoV DPUP domain contains a novel fold that consists of an antiparallel beta sheet (Johnson et al., 2010) that was surprisingly also found in the corresponding region of MHV despite negligible evidence of homology based on alignment of amino acid sequences (Chen et al., 2015).
All three of the domains that make up the SUD have been demonstrated to interact with nucleic acid in some way. Expressed Mac2-Mac3 has a high affinity for G-rich sequences and G-quadruplexes (Tan et al., 2009), while the Mac3-DPUP showed a general preference for purine nucleotides (Johnson et al., 2010). Notably, while Mac2 and Mac3 domains bear a close structural resemblance to the SARS-CoV Mac1 ADRP domain, neither domain has any demonstrable affinity for ADP-ribose (Tan et al., 2009). The amino acid residues responsible for Mac3 and DPUP RNA binding have been mapped, and appear to fall near the region of Mac3 that corresponds to the active site in the structurally similar Mac1 domain (Chatterjee et al., 2009). Together this suggests that the cluster of three macrodomains in SARS-CoV nsp3 arose through gene duplication and that additional macrodomains may contribute to the function of nsp3 as an accessory to the viral replication process (Neuman et al., 2008). Interestingly, in the context of a SARS-CoV infectious cDNA clone, Mac1 and Mac2 were dispensable but Mac3 was essential for RNA replication (Kusov et al., 2015).
6. Nsp3 (UB2 and PL2pro)
Unlike many coronaviruses that encode two papain-like proteases, SARS-CoV has a single copy of papain-like cysteine protease (PL2pro) that cleaves polyprotein 1a at three sites at the N-terminus to release nsp1, nsp2, and nsp3, respectively (Harcourt et al., 2004, Thiel et al., 2003). However, another important role for SARS-CoV PL2pro may be linked to its deubiquitinating activity; it efficiently disassembles diubiquitin and branched polyubiquitin chains, cleaves 7-amino-4-methylcoumarin-conjugated ubiquitin substrates, and has de-ISGylating activity (Chen et al., 2007, Lindner et al., 2005). Thus, PL2pro may have critical roles not only in proteolytic processing of the replicase complex but also in subverting cellular ubiquitination machinery to facilitate viral replication (Bailey-Elkin et al., 2014, Mielech et al., 2014, Mielech et al., 2015), as demonstrated for the arterivirus Equine arteritis virus (van Kasteren et al., 2013).
PL2pro is preceded by a second ubiquitin-like domain (Ratia et al., 2006). The protease catalytic domain adopts the canonical “thumb, palm and fingers” domain architecture. Two beta-hairpins at the fingertips region contain four cysteine residues, which coordinate a zinc ion. Mutational analysis of the zinc-coordinating cysteines of SARS-CoV PLpro showed that zinc-binding ability is essential for structural integrity and protease activity (Barretto et al., 2005). PL2pro has several structural homologues from the cysteine protease superfamily that are cellular deubiquitinating enzymes. The active site of PLpro consists of a catalytic triad of cysteine, histidine, and aspartic acid residues, consistent with catalytic triads found in many PLpro domains. Comparison of the SARS-CoV PL2pro structure with the structure of TGEV PL1pro demonstrates that the coronavirus-like PLpro folds have a common architecture (Wojdyla et al., 2010), and likely arose through gene duplication.
It has been demonstrated that an L/I X G G motif at the (pre-cleavage) P4—P1 positions of the substrate is essential for recognition and cleavage by Betacoronavirus PL2pro (Barretto et al., 2005, Han et al., 2005). There appear to be no preferences for the post-cleavage positions or for residues before P4. It is not surprising then that SARS-CoV PLpro is able to cleave after the four C-terminal residues of ubiquitin, LRGG. As predicted (Sulea et al., 2005), SARS-CoV PL2pro possesses de-ubiquitinating activity (Barretto et al., 2006, Lindner et al., 2005) in addition to cysteine protease activity involved in viral polyprotein processing. The specific deubiquitinating enzyme inhibitor, ubiquitin aldehyde, inhibited its activity at a Ki of 210 nM (Lindner et al., 2005).
Interestingly, a number of cellular deubiquitinases, including full-length USP14 and Ubp6, possess an N-terminal ubiquitin-like domain. Although the significance of this domain in these proteins is not well established, it has been demonstrated that the presence of the ubiquitin-like domain in USP14 and Ubp6 serves a regulatory function by mediating interactions between these deubiquitinases and specific components of the proteasome (Hu et al., 2005, Leggett et al., 2002). Comparisons of deubiquitinase activities between wild-type and mutant Ubp6 lacking the ubiquitin-like domain reveal that these associations are responsible for a 300-fold increase in catalytic rate and serve to activate the enzyme (Leggett et al., 2002). It is intriguing to consider whether the ubiquitin-like domain of PLpro may have a similar function.
While the role of PL2pro in polyprotein processing is well understood, the physiological significance of its deubiquitinating activity in the viral replication cycle is still not completely clear. However the conserved structural protein E is readily ubiquitinated in infected cells, suggesting that control of ubiquitination may be important for SARS-CoV assembly (Alvarez et al., 2010). Mounting evidence suggests that PL2pro interferes with interferon transcriptional activation pathways by inactivating TBK1, blocking NF-kappaB signaling and preventing translocation of IRF3 to the nucleus (Frieman et al., 2009, Wang et al., 2011, Zheng et al., 2008). In agreement with this proposed function, an arterivirus PL2pro was shown to play an important role in immune evasion (van Kasteren et al., 2013).
7. Nsp3 (NAB to CoV—Y)
The region between the PL2pro domain and the transmembrane region of nsp3 does not show obvious sequence similarity to any known domain that would give a clue to its function. However, a NMR structural study revealed that the nucleic acid binding (NAB) domain is an independently folded unit capable of binding RNA with relatively high affinity, and with duplex-unwinding activity reminiscent of an RNA chaperone (Neuman et al., 2008). The NAB, along with several other domains of nsp3 has been demonstrated to form homodimers upon incubation at 37 °C (Neuman et al., 2008) by perfluorooctanoic acid polyacrylamide gel electrophoresis. Little else is known about the function of NAB in the viral replication cycle, or about the structure and function of the betacoronavirus-specific marker (βSM) domain that follows or the conserved hydrophobic, non-transmembrane region that immediately precedes the first transmembrane region of nsp3.
The region from the first transmembrane helix to the carboxyl terminus of nsp3 was originally annotated as the Y-domain (Gorbalenya et al., 1991), in the sense that it followed the X-domain that we now refer to as Mac1. We have subdivided this region into two transmembrane regions, an ectodomain (3Ecto), a widely-conserved initial domain (Y1), and an apparently coronavirus-specific carboxyl-terminal domain (CoV—Y). No protein structures are available at this time for any part of the Y-domain, and the domain assignment in this region may change as new structures appear. A Fold and Function Annotation System search (FFAS; (Jaroszewski et al., 2005)) using the sequence from the SARS-CoV NAB to the end of nsp3 reveals three of seven significant hits (with expect values of −8 or better) to viral RNA-dependent RNA polymerase proteins, which may hint at the evolutionary origin of nsp3, which comprises nearly one fifth of most coronavirus genomes. The level of conservation in the Y1 domain in particular approaches levels consistent with the other enzymatic domains of nsp3, and exceeds the conservation of other domains that are believed to be non-enzymatic (Neuman et al., 2008), but the function and structure of this region remain to be explored (Fig. 5). The 3Ecto, Y1 and CoV—Y domains are highly conserved in all CoVs, but this region has not been structurally characterized yet. An Fold and Function Annotation System search (FFAS; (Jaroszewski et al., 2005)) using the sequence from the SARS-CoV NAB to the end of nsp3 reveals three of seven significant hits (with expect values of −8 or better) to viral RdRp proteins, which may hint at the evolutionary origin of nsp3, which comprises nearly one fifth of most coronavirus genomes. The level of conservation in the Y1 domain in particular approaches levels consistent with the other enzymatic domains of nsp3, and exceeds the conservation of other domains that are believed to be non-enzymatic (Neuman et al., 2008), but the function and structure of this region remain to be explored (Fig. 3).
It appears that domains from PL2pro to the CoV—Y domain have not undergone significant deletion or rearrangement during coronavirus evolution, while other nsps like nsp1, nsp2, and the N-terminal regions of nsp3 clearly have evolved by duplication and deletion of domains (Neuman et al., 2008, Neuman et al., 2014b). The C-terminal portion of nsp3 has been shown to change the localization of nsp4 (Hagemeijer et al., 2011), and cause a membrane proliferation phenotype in transfected cells (Angelini et al., 2013). The topology of nsp3 leaves only one domain, annotated here as the ectodomain of nsp3, or 3Ecto, on the luminal side of the membrane. If nsp3 participates directly in the membrane pairing exhibited in cells transfected with SARS-CoV nsp3 and nsp4 (Angelini et al., 2013), then the 3Ecto domain likely helps mediate membrane pairing directly. We previously noted that a cluster of cysteine and histidine residues in 3Ecto may coordinate a metal ion, but analysis of newly sequenced viruses shows that the Cys-His cluster is not conserved in all coronaviruses, hence this domain has been renamed in the annotation presented here.
8. Nsp4
Nsp4 is a transmembrane protein with four transmembrane helices and a cytosolic C-terminal domain (Oostra et al., 2007). Coronavirus nsp4 is approximately 500 amino acid residues in length, and is the only part of the viral polyprotein that is released after processing by both the PLpro and Mpro. The location and topology of the four transmembrane regions has been mapped (Oostra et al., 2007), but only the C-terminal portion of nsp4 appears to be conserved throughout the Nidovirales (Fig. 6 ).
Fig. 6.
Revised domain architecture and conservation of Coronavirinae nsp4, nsp5 and nsp6. (A) Previous annotations based on the ongoing work of Gorbalenya and collaborators (1991) and a new revised annotation (2016) are shown for comparison. Structures that were solved at the time of domain assignment are shown in red, and domain assignments based on amino acid alignments are shown in grey. (B) Annotation of nsp4-6 domains following the style of Fig. 4. Domain designations include nsp4 transmembrane regions (TM1, TM2-4), ectodomain (4Ecto) and endodomain (4Endo); nsp5 catalytic domains (Dom I and II) and dimerization domain (Dom III); and nsp6 transmembrane regions (TM1-4, TM5, TM6), ectodomain (6Ecto) and endodomain (6Endo). (C) Grey boxes show regions of apparent homology between coronavirus nsp4-6 and the equivalent proteins of Arteriviridae (Arteri), Mesoniviridae (Mesoni), Torovirinae (Toro), and Roniviridae (Roni). Regions of questionable homology based on predicted transmembrane regions or predicted protein secondary structure are marked with a question mark.
The C-terminal domain of nsp4 is conserved in all known coronaviruses, but deletion of this domain including the fourth transmembrane helix from a MHV infectious clone resulted in only slightly attenuated growth, consistent with a non-essential function (Sparks et al., 2007). It is somewhat surprising that the C-terminal domain of nsp4 is dispensible for replication since other nidoviruses with the exception of the Mesoniviridae contain a domain at this position with a similar predicted structure (V'Kovski et al., 2015). Mutation of the two glycosylation sites in nsp4, however, led to defective DMV formation and attenuation (Gadlage et al., 2010). The structure of the C-terminal 4Endo domain of Feline coronavirus has been reported. 4Endo consists of two small antiparallel beta-sheets and four alpha-helices (Manolaridis et al., 2009), and is highly conserved among coronaviruses at the amino acid level (Fig. 6).
SARS-CoV Nsp4 is an essential component for the formation of viral double-membrane vesicles (Angelini et al., 2013). Intracellular expression studies have demonstrated a biological interaction between the carboxyl-terminal region of MHV nsp3 and nsp4 (Hagemeijer et al., 2011), and co-expression of full-length SARS-CoV nsp3 and nsp4 results in extensive membrane pairing, in which the paired membranes are held at the same distance as observed in authentic DMVs (Angelini et al., 2013). Nsp4 has also been shown to interact with nsp2 in a yeast two-hybrid screen (von Brunn et al., 2007), and to interact with other nsp4 molecules in cells (Hagemeijer et al., 2011). Mutagenesis of the 4Ecto domain of nsp4 has been shown to cause aberrant DMV formation upon mutation, leading to a loss of nsp4 glycosylation (Gadlage et al., 2010, Sparks et al., 2007).
9. Nsp5
Coronavirus nsp5 is also known as the main protease (Mpro), a chymotrypsin-like protease related to the enteroviral 3C protease. It belongs to the C30 family of endopeptidases and is responsible for cleavage at 11 sequence specific sites within polyprotein 1a/1ab. The resultant “mature” protein products (nsp4 to nsp16) assemble into components of the replication complexes (reviewed in (Hilgenfeld et al., 2006, Ziebuhr et al., 2000)). Mpro is one of only three parts of the coronavirus replicase, along with the nsp12 polymerase and nsp13 helicase regions, that is conserved throughout the Nidovirales (Lauber et al., 2013).
Based on both structure and sequence characteristics, nsp5 can be divided into three domains: a two-domain active region (Dom I and II) and a third domain that plays a role in nsp5 dimerization (Dom III; (Yang et al., 2003)). The three-domain architecture is conserved in all coronaviruses, all nidoviral groups and several other RNA viruses that share a common polyprotein processing scheme (Ziebuhr et al., 2000). The sequence is related to chymotrypsin-like protease superfamily of endopeptidases (Murzin et al., 1995). The three-domain architecture is conserved in all coronaviruses, all nidoviral groups and several other RNA viruses that share a common polyprotein processing scheme (Ziebuhr et al., 2000). The sequence is related to chymotrypsin-like protease superfamily of endopeptidases.
The critical role of the first seven residues at the N terminus in dimerization and its close proximity to the active site results in this enzyme to be an obligate dimer (Anand et al., 2002), although modification of the termini appears to modulate higher order oligomerization (Zhang et al., 2010). Deletion of the first five amino acid residues results in complete inactivation of this enzyme. The helical C-terminal domain III mediates homodimerization of coronaviral Mpro proteases. This interaction is believed to be important for its trans-proteolytic activity. The active site is located at the interface of the two beta-barrels with the catalytic residues H51 and C145 being contributed by domains 1 and 2 respectively in the SARS-CoV Mpro.
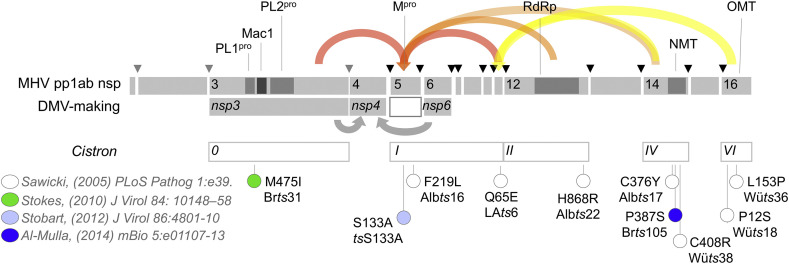
As part of its proteolytic activity, Mpro is destined to interact with all the non-structural proteins from nsp4 to nsp16, presumably near its catalytic site. Interestingly, comparison of all the known sites where mutation can lead to a temperature-sensitive, RNA-negative phenotype suggests a close connection to Mpro (Fig. 7 ). To date, the only proteins where temperature-sensitive mutations have been discovered are nsp3, nsp5, nsp10, nsp12, nsp14 and nsp16 (Al-Mulla et al., 2014, Sawicki et al., 2005, Stokes et al., 2010), all of which make fewer or smaller DMVs compared to wild-type virus (Al-Mulla et al., 2014). Nsp10 interacts with nsp5 directly (Imbert et al., 2008), and both nsp10 and nsp3 mutations paradoxically inhibit Mpro activity (Donaldson et al., 2007, Stokes et al., 2010). One interactome study showed that nsp12 and nsp14 also directly interact with nsp5 (Pan et al., 2008), and nsp14 and 16 would also indirectly interact with nsp5 as part of the nsp10-14-16 complex (Bouvet et al., 2010, Decroly et al., 2011, Imbert et al., 2008, Pan et al., 2008). Together this suggests that nsp5 plays a critical role in both RNA replication and DMV formation, likely by proteolytically releasing nsp4 and nsp6.
Fig. 7.
Functional map of potential connections between DMO making and RNA synthesis based on MHV ts mutants. Mutations linked to temperature-sensitive phenotypes, publications describing those mutations and their position in the system of cis-acting “cistrons” established by the complementation studies of Sawicki and Siddell are shown. Functional domain abbreviations include two papain-like proteases (PL1pro, PL2pro), a macrodomain (Mac1), main protease (Mpro), RNA-dependent RNA polymerase (RdRp), and RNA cap-methyltransferases (NMT, OMT). Temperature-sensitive mutations known to inhibit polyprotein processing by nsp5 Mpro or in proteins that interact with Mpro are shown in red, numbered from the start of the nonstructural protein where each is found. Mpro interactions based on mammalian two-hybrid data (Pan et al., 2008) are shown in orange. Interactions that indirectly link to Mpro via nsp10 are shown in yellow. Proposed interactions among DMV-making proteins are shown with grey arrows.
10. Nsp6
Nsp6 has six transmembrane regions, with both termini located on the cytosolic side of the membrane (Oostra et al., 2008). Although most coronavirus nsp6 proteins are predicted by TMHMM2.0 (Krogh et al., 2001) to contain seven transmembrane regions, only six of these function as membrane-spanning helices. The presence of additional non-transmembrane hydrophobic domains near authentic transmembrane domains is a common theme running through the DMO making proteins. IBV and SARS-CoV nsp6 have been shown to activate autophagy, inducing vesicles containing Atg5 and LC3-II (Cottam et al., 2011). MHV Nsp6 colocalizes with nsp4 when co-expressed (Hagemeijer et al., 2012), suggesting that the two proteins may interact. SARS-CoV nsp6 has also been shown to interact with nsp2, nsp8, nsp9 and accessory protein 9b via yeast two-hybrid assays (von Brunn et al., 2007). It is notable that both the 4Endo and 6Endo domains are nearly as well conserved in coronaviruses as the catalytic domain of Mpro (Fig. 5).
Overexpression of nsp6 disturbs intracellular membrane trafficking (Cottam et al., 2011), resulting in an accumulation of single-membrane vesicles around the microtubule organization complex (Angelini et al., 2013). However, coexpression with nsp4 prevents vesicle accumulation due to nsp6 expression, suggesting that nsp4 and 6 interact. While nsp6 was not necessary for membrane pairing, it was essential for forming the DMVs that are characteristic of coronavirus replicative organelles.
After serial passage of Human coronavirus 229E virus in the presence of the experimental antiviral K22, resistant viruses carrying mutations in nsp6 were selected (Lundin et al., 2014). K22 showed antiviral effects in the high nanomolar to low micromolar range, requiring higher amounts to achieve an antiviral effect than would normally be considered practical for wider clinical use. Time of addition and removal studies showed that K22 was most effective early in infection, after entry, consistent with effects on establishment of viral replication complexes or direct interference with the process of RNA replication (Lundin et al., 2014). Surprisingly, two independently isolated resistance mutations mapped to opposite ends of transmembrane helices in nsp6 at positions H121L and M159V. The resistant viruses released similar amounts of new progeny compared to wt, but produced only about half as many DMVs per infected cell, confirming the importance of nsp6 for authentic DMV formation. In addition, the DMVs induced by resistance mutants appeared structurally impaired. Similarly to MHV nsp4 mutants (Beachboard et al., 2015, Gadlage et al., 2010), K22 escape mutants induced DMV with partially collapsed inner membranes, even when K22 was not present. Moreover, the specific infectivity of those newly released virions was about ten-fold lower for nsp6 mutants than for wt. This suggested that the mutations in nsp6 conferred resistance to K22 at a cost of impairing an early intracellular step in the establishment of infection.
11. DMOs and viral replication fitness
While the structure of coronavirus DMOs has become increasingly clear, their purpose remains somewhat mysterious. Mutations in nsp4 of MHV resulted in decreased replication and competitive fitness (Beachboard et al., 2015). MHV mutants that produced fewer DMVs, but equal or greater amounts of genomic and subgenomic viral RNA have been described (Al-Mulla et al., 2014), suggesting that coronavirus replication is not strictly dependent on high numbers of DMVs or the size of the DMV interior. Notably, when equal infectivities of two viruses were added to the same flask at a temperature where both viruses could grow normally, several mutants with small DMV and low-DMV phenotypes did not appear to be at a competitive disadvantage compared to wild-type virus (Al-Mulla et al., 2014). This result was replicated in several continuous cell lines and primary cells. Furthermore, a survey of Infectious bronchitis virus revealed that one high-pathogenicity strain produced abnormally low numbers of double-membrane spherules, while still producing an equivalent amount of RNA to a vaccine strain (Maier et al., 2016). These results suggest that whatever their purpose, coronavirus replicative organelles display a surprising degree of structural plasticity without necessarily impairing RNA production, pathogenicity or competitive fitness.
References
- Al-Mulla H.M., Turrell L., Smith N.M., Payne L., Baliji S., Zust R., Thiel V., Baker S.C., Siddell S.G., Neuman B.W. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. MBio. 2014;5:e01107–01113. doi: 10.1128/mBio.01107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Marcos-Villar L., Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524–00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. Deubiquitinating activity of the SARS-CoV papain-like protease. Adv. Exp. Med. Biol. 2006;581:37–41. doi: 10.1007/978-0-387-33012-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachboard D.C., Anderson-Daniels J.M., Denison M.R. Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications. J. Virol. 2015;89:2080–2089. doi: 10.1128/JVI.02776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R., Galli C., Noack J., Bianchi S., de Haan C.A., Reggiori F., Molinari M. Role of the SEL1L: LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol. Cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calì T., Galli C., Olivari S., Molinari M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008;371:405–410. doi: 10.1016/j.bbrc.2008.04.098. [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J. Virol. 2009;83:1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Savinov S.N., Mielech A.M., Cao T., Baker S.C., Mesecar A.D. X-ray structural and functional studies of the three tandemly linked domains of non-structural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J. Biol. Chem. 2015;290:25293–25306. doi: 10.1074/jbc.M115.662130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A.D., Wilkinson K.D., Baker S.C. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M., McCraith S.M., Zillmann M., Kierzek R., Michaud N., LaReau R.D., Turner D.H., Phizicky E.M. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″-2″ cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2'-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Graham R.L., Sims A.C., Denison M.R., Baric R.S. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 2007;81:7086–7098. doi: 10.1128/JVI.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage M.J., Sparks J.S., Beachboard D.C., Cox R.G., Doyle J.D., Stobart C.C., Denison M.R. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 2010;84:280–290. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J., de Haan C.A. Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 2011;85:4572–4577. doi: 10.1128/JVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Verheije M.H., Ulasli M., Shaltiel I.A., de Vries L.A., Reggiori F., Rottier P.J., de Haan C.A. Dynamics of coronavirus replication-transcription complexes. J. Virol. 2010;84:2134–2149. doi: 10.1128/JVI.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.S., Chang G.G., Juo C.G., Lee H.J., Yeh S.H., Hsu J.T., Chen X. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry. 2005;44:10349–10359. doi: 10.1021/bi0504761. [DOI] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Anand K., Mesters J.R., Rao Z., Shen X., Jiang H., Tan J., Verschueren K.H. Structure and dynamics of SARS coronavirus main proteinase (Mpro) Adv. Exp. Med. Biol. 2006;581:585–591. doi: 10.1007/978-0-387-33012-9_106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst-Hess K.R., Kuo L., Masters P.S. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J. Virol. 2015;89:6033–6047. doi: 10.1128/JVI.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lecine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Chatterjee A., Neuman B.W., Wuthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J. Mol. Biol. 2010;400:724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.B., McFadden G. Poxvirus immunomodulatory strategies: current perspectives. J. Virol. 2003;77:6093–6100. doi: 10.1128/JVI.77.11.6093-6100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Barcena M., Limpens R.W., Koster A.J., Mommaas A.M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuri T., Eriksson K.K., Putics A., Zust R., Snijder E.J., Davidson A.D., Siddell S.G., Thiel V., Ziebuhr J., Weber F. The ADP-ribose-1'-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. General Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9:e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett D.S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R.T., Walz T., Ploegh H., Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Leung A.K. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 2014;205:613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Menard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Archives Biochem. Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergstrom T., Kann N., Adamiak B., Hannoun C., Kindler E., Jonsdottir H.R., Muth D., Kint J., Forlenza M., Muller M.A., Drosten C., Thiel V., Trybala E. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10:e1004166. doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4:e00801–00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Neuman B.W., Bickerton E., Keep S.M., Alrashedi H., Hall R., Britton P. Extensive coronavirus-induced membrane rearrangements are not a determinant of pathogenicity. Sci. Rep. 2016;6:27126. doi: 10.1038/srep27126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolaridis I., Wojdyla J.A., Panjikar S., Snijder E.J., Gorbalenya A.E., Berglind H., Nordlund P., Coutard B., Tucker P.A. Structure of the C-terminal domain of nsp4 from feline coronavirus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:839–846. doi: 10.1107/S0907444909018253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Deng X., Chen Y., Kindler E., Wheeler D.L., Mesecar A.D., Thiel V., Perlman S., Baker S.C. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J. Virol. 2015;89:4907–4917. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Kilianski A., Baez-Santos Y.M., Mesecar A.D., Baker S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology. 2014;450–451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D.J., Monte K., Sun L., Struckhoff J.J., Agapov E., Holtzman M.J., Stappenbeck T.S., Lenschow D.J. Novel mode of ISG15-mediated protection against influenza A virus and Sendai virus in mice. J. Virol. 2015;89:337–349. doi: 10.1128/JVI.02110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Angelini M.M., Buchmeier M.J. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22:642–647. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Chamberlain P., Bowden F., Joseph J. Atlas of coronavirus replicase structure. Virus Res. 2014;194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M., Greer C.L. Pre-tRNA splicing: variation on a theme or exception to the rule? Trends Biochem. Sci. 1993;18:31–34. doi: 10.1016/0968-0004(93)90085-2. [DOI] [PubMed] [Google Scholar]
- Piotrowski Y., Hansen G., Boomaars-van der Zanden A.L., Snijder E.J., Gorbalenya A.E., Hilgenfeld R. Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 2009;18:6–16. doi: 10.1002/pro.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1”-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Cali T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1'-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wuthrich K. NMR structure of the N-terminal domain of the nonstructural protein 3 from the SARS coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Schiller J.J., Kanjanahaluethai A., Baker S.C., Oh J.W., Lai M.M. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J. Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E.L., Hegde R.S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. vol. 6. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences; pp. 175–182. (Proceedings/… International Conference on Intelligent Systems for Molecular Biology; ISMB). International Conference on Intelligent Systems for Molecular Biology. [PubMed] [Google Scholar]
- Sparks J.S., Lu X., Denison M.R. Genetic analysis of Murine hepatitis virus nsp4 in virus replication. J. Virol. 2007;81:12554–12563. doi: 10.1128/JVI.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F., Kincaid A., Gordon M., Porter B.F., Baumgartner W., Stahl S., Kelley K., Towner J.S., DeRisi J.L. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in Python regius. MBio. 2014;5 doi: 10.1128/mBio.01484-14. e01484–01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes H.L., Baliji S., Hui C.G., Sawicki S.G., Baker S.C., Siddell S.G. A new cistron in the murine hepatitis virus replicase gene. J. Virol. 2010;84:10148–10158. doi: 10.1128/JVI.00901-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Xue F., Guo Y., Ma M., Hao N., Zhang X.C., Lou Z., Li X., Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J. Virol. 2009;83:10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5:e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. General Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- V'Kovski P., Al-Mulla H., Thiel V., Neuman B.W. New insights on the role of paired membrane structures in coronavirus replication. Virus Res. 2015;202:33–40. doi: 10.1016/j.virusres.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J., Locker J.K. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., Bailey-Elkin B.A., James T.W., Ninaber D.K., Beugeling C., Khajehpour M., Snijder E.J., Mark B.L., Kikkert M. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS One. 2011;6:e17192. doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., Snijder E.J., Gorbalenya A.E., Coutard B., Piotrowski Y., Hilgenfeld R., Tucker P.A. Structure of the X (ADRP) domain of nsp3 from feline coronavirus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:1292–1300. doi: 10.1107/S0907444909040074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., van Kasteren P.B., Kikkert M., Snijder E.J., Gorbalenya A.E., Tucker P.A. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J. Virol. 2010;84:10063–10073. doi: 10.1128/JVI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Cong L., Chen C., Wei L., Zhao Q., Xu X., Ma Y., Bartlam M., Rao Z. Crystal structures of two coronavirus ADP-ribose-1'-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain. J. Virol. 2009;83:1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhong N., Xue F., Kang X., Ren X., Chen J., Jin C., Lou Z., Xia B. Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease. Protein Cell. 2010;1:371–383. doi: 10.1007/s13238-010-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 2001;276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]