Highlights
-
•
Fluorescent RT-LAMP assays using quenching probes for MERS-CoV were developed.
-
•
Quenching probe (QProbe) can solve the problem in turbidity monitoring mechanism.
-
•
Only primer-derived signal can be monitored specifically by QProbes.
-
•
Two primer sets were developed to enable to confirm MERS case by RT-LAMP only.
-
•
Both sets were highly specific and sensitive in comparison with real-time RT-PCR.
Abbreviations: ADV, adenovirus; ATCC, American Type Culture Collection; BIP, backward inner primer; CoV, coronavirus; FFU, focus forming unit; FIP, forward inner primer; HBoV, human bocavirus; HCoV, human coronavirus; MERS, Middle East respiratory syndrome; MPV, metapneumovirus; N, nucleocapsid; ORF, open reading frame; PBS, phosphate-buffered saline; PIV, parainfluenza virus; PFU, plaque forming unit; QProbe or QP, quenching probe; RSV, respiratory syncytial virus; RT-LAMP, reverse transcription-loop-mediated isothermal amplification; TCID50, 50% tissue culture infectious dose; upE, upstream E
Keywords: Middle East respiratory syndrome, MERS coronavirus, Quenching probe, RT-LAMP
Abstract
Clinical detection of Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) in patients is achieved using genetic diagnostic methods, such as real-time RT-PCR assay. Previously, we developed a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay for the detection of MERS-CoV [Virol J. 2014. 11:139]. Generally, amplification of RT-LAMP is monitored by the turbidity induced by precipitation of magnesium pyrophosphate with newly synthesized DNA. However, this mechanism cannot completely exclude the possibility of unexpected reactions. Therefore, in this study, fluorescent RT-LAMP assays using quenching probes (QProbes) were developed specifically to monitor only primer-derived signals. Two primer sets (targeting nucleocapsid and ORF1a sequences) were constructed to confirm MERS cases by RT-LAMP assay only. Our data indicate that both primer sets were capable of detecting MERS-CoV RNA to the same level as existing genetic diagnostic methods, and that both were highly specific with no cross-reactivity observed with other respiratory viruses. These primer sets were highly efficient in amplifying target sequences derived from different MERS-CoV strains, including camel MERS-CoV. In addition, the detection efficacy of QProbe RT-LAMP was comparable to that of real-time RT-PCR assay using clinical specimens from patients in Saudi Arabia. Altogether, these results indicate that QProbe RT-LAMP assays described here can be used as powerful diagnostic tools for rapid detection and surveillance of MERS-CoV infections.
1. Introduction
Middle East respiratory syndrome (MERS) is an emerging respiratory disease caused by the MERS coronavirus (MERS-CoV). MERS has been endemic mainly in Saudi Arabia since 2012 (Assiri et al., 2013; Azhar et al., 2014). As of 15 March 2018, there have been 2144 confirmed cases, with 750 deaths, reported from 27 countries [The World Health Organization (WHO), Global Alert and Response (GAR), Coronavirus infections, updated on 15 March 2018, http://www.who.int/csr/don/15-march-2018-mers-oman/en/].
According to the case definition of the WHO, at least two distinct genomic targets are required for a positive diagnosis [WHO, GAR, Revised interim case definition for reporting to WHO – Middle East respiratory syndrome coronavirus (MERS-CoV), updated on 3 July 2013, http://www.who.int/csr/disease/coronavirus_infections/case_definition/en/index.html]. Therefore, many genetic diagnostic methods have been developed for the stable and reliable diagnosis of MERS-CoV infections. Currently, the main diagnostic method of MERS-CoV is real-time RT-PCR assays, and the primer/probe sets [upE and open reading frame (ORF) 1a] developed by Corman et al. are widely used as standard assays (Corman et al., 2012a, Corman et al., 2012b).
The loop-mediated isothermal amplification (LAMP) method amplifies specific nucleotide sequences using a set of four or six unique primers (Nagamine et al., 2002; Notomi et al., 2000). This method is relatively quick and user-friendly; amplification signals can be readily detected within an hour, and it only requires a single incubation temperature. As such, various LAMP assays have been developed for the detection of a wide range of pathogens, such as bacteria (Adhikari et al., 2009; Geojith et al., 2011; Ueda and Kuwabara, 2009), parasites (Arimatsu et al., 2012; Wang et al., 2010), and viruses (Hong et al., 2004; Imai et al., 2006; Mahony et al., 2013; Shirato et al., 2007; Ushio et al., 2005) including MERS-CoV (Bhadra et al., 2015; Lee et al., 2016; Shirato et al., 2014).
A reverse transcription (RT)-LAMP assay for the detection of MERS-CoV was developed by our group recently (Shirato et al., 2014), which employs a primer set targeting the viral nucleocapsid (N) sequence, comparable to standard real-time RT-PCR assays. In the LAMP assay, positive signals are indicated by turbidity that results from magnesium pyrophosphate precipitation following LAMP reaction. However, in this mechanism, the possibility of unexpected signals derived from primer dimer and/or non-primer reactions cannot be excluded (Njiru, 2012). There is also a possibility of detecting turbidity if the host-derived DNA makes LAMP product non-specifically. Thus, if unexpected signals are detected, it is very difficult to identify the origin of the signal. The validity of MERS-CoV detection by previous RT-LAMP assay has been confirmed (Shirato et al., 2014), but the mechanism of turbidity detection can be improved.
Florescence dye (calcein, etc.) or DNA intercalator can be added for fluorescence monitoring (http://loopamp.eiken.co.jp/e/tech/detect_index.html), which may help to improve turbidity detection. The addition of DNA intercalator was used in Zika virus detection (Kurosaki et al., 2017). However, the detection principle of these methods is the same as turbidity detection. Using fluorescence labeled primer can solve the problem of non-primer-derived signals. Recently, Fowler et al. (2016) reported RT-LAMP assays for detection of vesicular stomatitis, foot and mouth diseases, and swine vesicular disease viruses using fluorescence labeled forward inner primers (FIPs) or backward inner primers (BIPs). However, this study used 5´ end-labeled primers. Therefore, if the fluorescent primer causes non-specific extension at the 3′ end, unexpected signals will be detected. To avoid non-specific signals, melting curve analysis of the LAMP amplicon is useful to confirm amplification of the targeted sequence (Fowler et al., 2016; Kurosaki et al., 2017). However, melting curve analysis requires incubation of at a higher temperature than that LAMP, and requires additional time after amplification, which negates the main advantage of LAMP.
In this study, to address these problems, a quenching probe 3G (QProbe) was used for monitoring RT-LAMP. In QProbe, the fluorescence dye is labeled at the 3´ end of the primer. Therefore, the extension of the primer sequence is blocked by dye even if the primer anneals non-specifically at its 3´ end. Use of QProbe can detect primer-derived signals only, and thus can avoid detecting non-specific amplification caused by fluorescent primer. In addition, to validate a positive MERS-CoV diagnosis, an additional primer set (targeting the ORF1a region) for use in QProbe RT-LAMP assays was developed to enable to confirm MERS cases only by QProbe RT-LAMP.
2. Materials and methods
2.1. Viruses
MERS-CoV EMC strain was kindly provided by Ron A. M. Fouchier, Erasmus Medical Center, Rotterdam, the Netherlands. MERS-CoV was propagated and titrated using Vero cells. Human respiratory syncytial viruses (RSV; Long, A2, B WV/14617/85 and 18537) were obtained from the American Type Culture Collection (ATCC). Human metapneumovirus (HMPV; Sendai-H/2404/2003) was obtained from the Virus Research Center, Sendai Medical Center, Japan. Human coronavirus (HCoV)-229E isolates ATCC VR-740 and Sendai-H/1121/04 (Shirato et al., 2012) were used. HCoV-NL63 was supplied by Dr. Lia van der Hoek, University of Amsterdam, the Netherlands. HCoV-OC43 isolate ATCC VR-1558 was used. SARS coronavirus (Frankfurt strain) was supplied by Dr. J. Ziebuhr, University of Würzburg, Germany. Human parainfluenza viruses (PIV) 1 (strain C35) and 3 (strain C243) were obtained from ATCC. Adenoviruses (ADVs) (serotype 3, strain G.B.; serotype 4, strain RI-67; and serotype 7, strain Gomen) were obtained from ATCC. Viruses were propagated and titrated using HEp-2, HeLa, RD, Vero cells, or LLC-Mk2 cells (Shirogane et al., 2008). Influenza viruses [Flu; A/California/7/2009 (H1N1pdm), A/Victoria/210/2009 (H3N2), and B/Brisbane/60/2008] were propagated and titrated using MDCK cells. Clinical isolates of HCoV-OC43 (Tokyo/SGH-36/2014, LC315646: Tokyo/SGH-61/2014, LC315647: Tokyo/SGH-06/2015, LC315648) and HKU1 (Tokyo/SGH-15/2014, LC315050: Tokyo/SGH-18/2016, LC315051) were isolated and propagated using human bronchial tracheal epithelial cells (Lifeline Cell Technology, Frederick, MD, USA) that were cultured and differentiated at the air–liquid interface.
2.2. Construction of primers for QProbe RT-LAMP
For amplification of the N sequences, the primer set reported previously was utilized (Shirato et al., 2014). The primer set for the amplification of the ORF1a region was constructed using the online LAMP primer design software (PrimerExplorer V4; http://primerexplorer.jp/e/) based on the sequence of the MERS-CoV EMC strain (GenBank JX869059.2). The nucleotide sequence and concentration of primers used in each reaction are listed in Table 1 . For the detection of the RT-LAMP reaction by fluorescent signals, the QProbe was used (Nippon Steel & Sumikin Eco-Tech Corp., Tsukuba, Japan) (Tani et al., 2009). For primer sets targeting N and ORF1a, QProbes were constructed based on LB primers, and several nucleotides were added to LB primers (Table 1). The final reaction mixture contained 1 pmol of QProbe-LBs and the six general MERS-CoV primers.
Table 1.
Primer sets for MERS-CoV QProbe RT-LAMP assay.
| N | Primer sequence (5′–3′, EMC, JX869059.2) | Position | Volume (pmol/test) | Number of matched sequences on GenBank | Percentage of matched sequences |
|---|---|---|---|---|---|
| N-F3 | GCTCCCAGGTGGTACTTCT | 28848–28866 | 5 | 293/300 | 97.7 |
| N-B3 | cagtcccctcaatgtggaag | 29061–29042 | 5 | 300/300 | 100.0 |
| N-FIP | tcatggacccaaacgatgccatACTGGAACTGGACCCGAAG | 28939–28918 | 40 | 299/300 | 99.7 |
| +28872–28890 | 245/300 | 81.7 | |||
| N-BIP | GCTCCTTCAACTTTTGGGACGCtagtaccgggcgcgaatt | 28956–28977 | 40 | 291/300 | 97.0 |
| +29028–29011 | 293/300 | 97.7 | |||
| N-LF | cggaatgggagtgctg | 28906–28891 | 20 | 300/300 | 100.0 |
| N-LB | GGAACCCTAACAATGATTCAGCT | 28978–29000 | 10 | 285/300 | 95.0 |
| N-LB-QP | GGAACCCTAACAATGATTCAGCTATTGTTACAC | 28978–29010 | 1 |
| ORF1a | Primer sequence (5′–3′, EMC, JX869059.2) | Position | Volume (pmol/test) | Number of matched sequences on GenBank | Percentage of matched sequences |
|---|---|---|---|---|---|
| ORF1a-F3 | GCCTACTTTGGATGTGAGG | 1572–1590 | 5 | 278/278 | 100.0 |
| ORF1a-B3 | acaacgaactctcccaca | 1753–1736 | 5 | 279/279 | 100.0 |
| ORF1a-FIP | taaagatggagtctccaatccttgaAAGGTACTATGTACTTTGTGCC | 1656–1632 | 40 | 264/278 | 95.0 |
| +1591–1612 | 239/278 | 86.0 | |||
| ORF1a-BIP | GTACTGGCTCTTGGAACAAGGagttaagggaatgctgagt | 1663–1683 | 40 | 278/278 | 100.0 |
| +1734–1716 | 278/279 | 99.6 | |||
| ORF1a-LF | acaacagacttagctctag | 1612–1630 | 20 | 278/278 | 100.0 |
| ORF1a-LB | GGTCACTCAAATTGCTAACATG | 1682–1703 | 20 | 253/279 | 90.7 |
| ORF1a-LB-QP | GGTCACTCAAATTGCTAACATGTTCTTGGAACAGAC | 1682–1717 | 1 |
Capital letters indicate the sense strand; lowercase letters indicate the antisense strand.
QP: Quenching probe
2.3. Extraction of nucleic acids from virus stocks
RNA was extracted from viral stocks using TRIzol LS, TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA), QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) or MagnaPure Compact Nucleic Acid Isolation kit (Roche, Basel, Switzerland), according to the manufacturer’s instructions. Viral DNA was extracted using the SimplePrep Reagent for DNA (Takara-Bio Inc., Shiga, Japan), according to the manufacturer’s instructions. Total RNA and genomic DNA were quantified using standard methods of measuring the OD value. For sensitivity assays, to isolate RNA from virion only, Vero cells were infected with MERS-CoV, and incubated for 4 days. Cell supernatants were then collected and centrifuged at 1500×g for 30 min at 4 °C, and the supernatants were treated with RNaseA (Nippongene, Tokyo Japan) at a concentration of 10 μg/mL for 30 min at 37 °C to exclude non-viral RNA as previously reported (Shirato et al., 2014). The MERS-CoV RNA copy number was calculated based upon the standard curve generated by real-time RT-PCR assay using the upE primer set (Corman et al., 2012a) and a positive control RNA template. Total RNAs were diluted in ribonuclease-free water containing 10 μg/mL of ribonucleic acid from baker’s yeast (R6750; Sigma-Aldrich, St. Louis, MO, USA) as carrier RNA.
2.4. RT-LAMP assay
The QProbe RT-LAMP assay was performed in a 25-μl (total) reaction mixture containing the appropriate amount of primer sets (see Table 1), 1.4 mM of each deoxynucleoside triphosphates, 0.5% Tween 20, 8 mM MgSO4, 30 mM KCl, 20 mM Tricine (pH 8.6), 16 U of Bst DNA polymerase (New England Biolabs, Ipswich, MA, USA), 1 U of avian myeloblastosis virus reverse transcriptase (Thermo Fisher Scientific), and the extracted RNA. As a negative control, PCR-graded water containing carrier RNA only was utilized. The reaction mixture was incubated at 63 °C for 30 min in a thermostatic fluorometer capable of detecting FAM dye, which included LightCycler 480 (Roche), ABI 7500 Fast (Thermo Fisher Scientific), or ESEQuant TS2 tube scanner (Qiagen).
As the positive control for amplification of the viral N region, a previously synthesized RNA was utilized (Shirato et al., 2014). To synthesize the control RNA for amplification of ORF1a region, the EMC strain sequence (1000–2000) was subcloned into pGEM-T Easy vector and subsequently amplified using PrimeSTAR MAX (Takara-Bio Inc.) and the following primers: 5′-TAATACGACTCACTATAGGGTCATCACATTAAAGAACAATCTATA-3′, and 5′-GGTTGCAACTTTCTTAAAGGACTCAC-3′. The amplicons were gel-purified and used as templates for RNA transcription using the MEGAscript T7 Transcription Kit (Thermo Fisher Scientific). The resultant RNA transcripts were quantified based on the OD value, and the copy number was calculated. The RNA was diluted in ribonuclease-free water containing 10 μg/mL of yeast RNA.
To evaluate the sensitivities of each primer set for detection of various target sequences, point mutations were introduced into the N and ORF1a sequences on plasmid by site-direct mutagenesis. The control RNA transcripts with the incorporated mutations were generated as described above.
2.5. Real-time RT-PCR
Real-time RT-PCR assays using upE and ORF1a primer sets (Corman et al., 2012a, Corman et al., 2012b) were performed using a QuantiTect Probe RT-PCR kit (Qiagen) and a LightCycler 480 or LightCycler96 Instrument (Roche) as per the manufacturers' instruction. The amplification conditions as previously reported were utilized (Corman et al., 2012a, Corman et al., 2012b).
2.6. Processing of clinical specimens
All experiments using human clinical specimens were approved by the Research and Ethical Committee for the Use of Human Subjects of the National Institute of Infectious Diseases, Japan (Approval #746) ; the Ethical Committee of Showa General Hospital (Approval #REC-094); and the Research Ethics Committee, Faculty of Medicine, King Abdulaziz University, Kingdom of Saudi Arabia (Approval #121-16). Clinical specimens diagnosed to be positive for other respiratory pathogens were used for the evaluation of non-specific reaction in MERS-CoV-negative specimens. From January 2014 to February 2016, 19 nasal aspirates, secretions, or swabs were collected from patients presenting with influenza-like illnesses at the outpatient pediatrics clinic of Showa General Hospital. Parents or legal guardians of all children/minor participants provided written informed consent. Specimens were collected in 1 ml of universal transport medium (Copan Italia, Brescia, Italy), and RNA extraction was performed as described above. Detection of other respiratory pathogens was confirmed by real time RT-PCR as previously described (Do et al., 2010; Kaida et al., 2014). The presence of MERS-CoV was determined by QProbe RT-LAMP, using the protocol described above.
The QProbe RT-LAMP assay using MERS-CoV positive specimens was performed in the Special Infectious Agents Unit, King Abdulaziz University, Jeddah, Saudi Arabia, using lyophilized reaction mixtures in 12 stripe tubes and an ESEQuant TS2 tube scanner (Qiagen). Specimens used for validation were archived specimens collected from MERS cases since 2014. These were stored at −80 °C until testing. Total RNA (5 μL) extracted from MERS-CoV-positive specimens that were pre-tested by real-time RT-PCR was mixed with 20 μL of RT-PCR-grade water (Thermo Fisher Scientific), and was subsequently added to each well in the tube strip, and then used for MERS-CoV detection. Quenching signals were detected using the ESEQuant TS2 tube scanner at 63 °C for 30 min.
3. Results
3.1. Sensitivity of the QProbe RT-LAMP assay
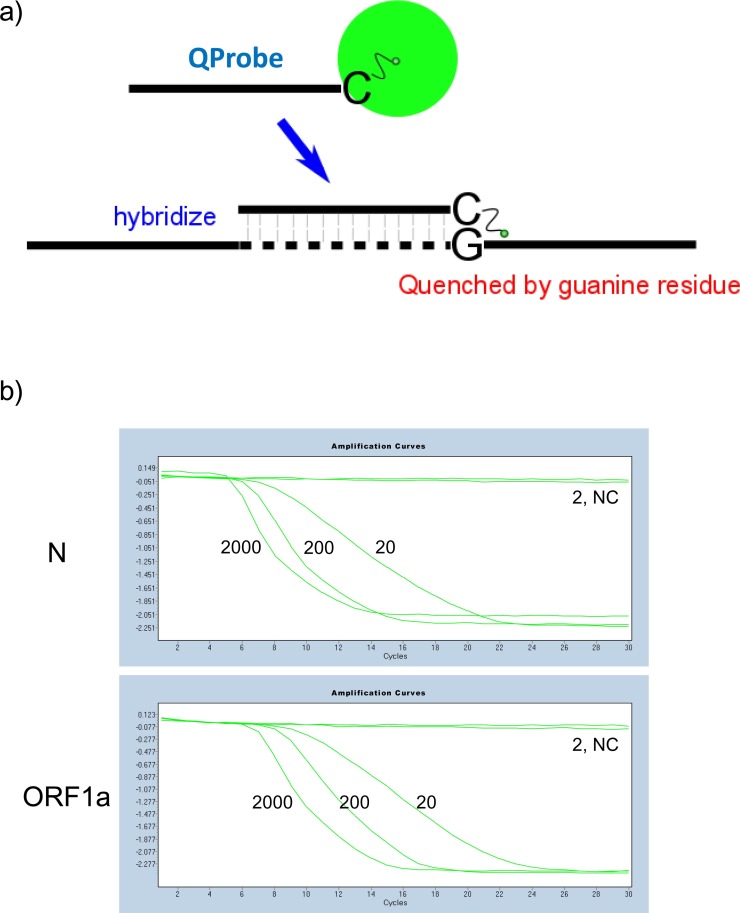
The detection principle of QProbe is shown as in the schematic diagram of Fig.1 ; fluorescence from the fluorophore bound to the cytosine residue at the 3′ end of the QProbe is quenched by the guanine residue present in the target sequence during hybridization (Fig. 1a). The positive signal is shown as quenching of fluorescence, which generates a reverse sigmoid curve (Fig. 1b). In contrast, negative signals due to the lack of fluorescence quenching generate a straight line (Fig. 1b). The detection limit of the QProbe RT-LAMP assay was determined using serially diluted MERS-CoV RNA templates and was evaluated in comparison to those of real-time RT-PCR (upE and ORF1a) and RT-LAMP (turbidity) assays (Table 2 ). Although target regions of QProbe RT-LAMP assays were different from real-time RT-PCR assays, the validation was performed using copy number-determined viral RNA, and each amplification was performed using the same samples. As reported previously, both real-time RT-PCR and RT-LAMP assays are capable of detecting MERS-CoV RNA at a copy level as low as 20 (Corman et al., 2012a, Corman et al., 2012b; Shirato et al., 2014). As shown in Table 2, QProbe RT-LAMP assays, which targeted N and ORF1a sequences, were able to detect MERS-CoV RNA at a similar level, comparable to real-time RT-PCR and RT-LAMP. These data indicate that the sensitivity of QProbe RT-LAMP assays is similar to that of existing genetic diagnostic methods.
Fig. 1.
a) Schematic representation of quenching probe (QProbe). QProbe is labeled with fluorescent dye at the cytosine residue at the 3′ end. When the QProbe hybridizes with the target, fluorescence is quenched by the guanine residue present in the target sequence. b) Images of detecting fluorescence quenching. Fluorescence RT-LAMP (N and ORF1a) was performed with serially diluted MERS-CoV viral RNA using the LightCycler480 instrument. The wavelength used for signal detection is the same as FAM. Negative signal is represented by an upper line. Positive signal is represented by a reverse S-shaped curve. NC, negative control.
Table 2.
Sensitivity of QProbe RT-LAMP assays.
| Copies/reaction | 2000 | 200 | 20 | 2 | 0.2 | Sensitivity (copies/reaction) | Time required (h) |
|---|---|---|---|---|---|---|---|
| Real-time RT-PCR | Positive/Number | ||||||
| upE | 6/6 | 6/6 | 5/6 | 1/6 | 0/6 | 6.3 | 2 |
| ORF1a | 6/6 | 6/6 | 4/6 | 0/6 | 0/6 | 13.6 | 2 |
| Copies/reaction | 2000 | 200 | 20 | 2 | 0.2 | ||
| RT-LAMP | Positive/Number | ||||||
| N (turbidity) | 6/6 | 6/6 | 2/6 | 1/6 | 0/6 | 20 | 0.5 |
| N (QP) | 8/8 | 8/8 | 3/8 | 1/8 | 0/8 | 20 | 0.5 |
| ORF1a (QP) | 8/8 | 8/8 | 5/8 | 0/8 | 0/8 | 15 | 0.5 |
QP: Quenching probe.
3.2. Specificity of the QProbe RT-LAMP assays
Next, the specificity of QProbe RT-LAMP was determined using various respiratory virus isolates (Table 3 ). For both N and ORF1a primer sets, no cross reaction was detected with other respiratory pathogens included in this study. Similarly, no cross-reactivity was observed in the QProbe RT-LAMP assay where clinical specimens positive for other respiratory pathogens (determined by real-time RT-PCR) were utilized (Table 4 ). These data demonstrate that QProbe RT-LAMP possessed a high specificity for the diagnosis of MERS-CoV.
Table 3.
Specificity of QProbe RT-LAMP assays.
| Primer set |
||||
|---|---|---|---|---|
| Strain | Name of isolate | Amount | N | ORF1a |
| MERS-CoV | EMC | 1 × 105 copies | + | + |
| HCoV-229E | ATCC VR-740 | 2.5 × 104 PFU | – | – |
| Sendai-H/1121/04 | 5 × 103 PFU | – | – | |
| Niigata/01/08 | 4 × 102 PFU | – | – | |
| HCoV-NL63 | 1 × 102 FFU | – | – | |
| HCoV-HKU1 | Tokyo/SGH-15/2014 | 5 × 104 copies | – | – |
| Tokyo/SGH-18/2016 | 6 × 102 copies | – | – | |
| HCoV-OC43 | ATCC VR-1558 | 2.5 × 102 TCID50 | – | – |
| Tokyo/SGH-36/2014 | 2 × 105 copies | – | – | |
| Tokyo/SGH-61/2014 | 1 × 106 copies | – | – | |
| Tokyo/SGH-06/2016 | 1 × 105 copies | – | – | |
| SARS-CoV | Frankfurt | 2 × 106 TCID50 | – | – |
| Other respiratory pathogens | ||||
| ADV 3 | G.B. | 2 × 106 TCID50 | – | – |
| ADV 4 | RI-67 | 2 × 106 TCID50 | – | – |
| ADV 7 | Gomen | 2 × 106 TCID50 | – | – |
| PIV1 | C-35 | 1.2 × 103 PFU | – | – |
| PIV3 | C-243 | 1 × 105 PFU | – | – |
| RSV A | Long | 5 × 107 copies | – | – |
| RSV A | A2 | 5 × 105 copies | – | – |
| RSV B | CH/18537 | 5 × 107 copies | – | – |
| RSV B | B1 | 5 × 106 copies | – | – |
| HMPV | Sendai-H/2404/2003 | 1.2 × 106 PFU | – | – |
| Influenza | ||||
| A(H1N1)pdm09 | A/California/7/2009 | 4 × 103 TCID50 | – | – |
| A(H3N2) | A/Victoria/210/2009 | 1.25 × 106 TCID50 | – | – |
| B | B/Brisbane/60/2008 | 1.25 × 104 TCID50 | – | – |
PFU: plaque forming unit.
FFU: focus forming unit.
TCID50: median tissue culture infectious dose.
Table 4.
QProbe RT-LAMP assays using clinical specimens positive for other respiratory viruses.
| Specimen | Type | Detected viruses |
Primer set |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Name | Cq | Name | Cq | Name | Cq | N | ORF1a | |
| F14-15 | Nasal secretion | HCoV-HKU1 | 23.7 | – | – | ||||
| F14-61 | Nasal aspiration | HCoV-OC43 | 18.3 | ADV2 | 33.1 | Rhino | 31.6 | – | – |
| F16-18 | Nasal secretion | HCoV-HKU1 | 25.1 | – | – | ||||
| F16-65 | Nasal aspiration | HCoV-OC43 | 24.0 | RSV A | 34.3 | – | – | ||
| F14-56 | Nasal aspiration | RSV A | 22.0 | ADV4 | 24.8 | – | – | ||
| F15-25 | Nasal aspiration | HBoV | 25.5 | Rhino | 26.9 | – | – | ||
| F15-35 | Nasal aspiration | PIV3 | 28.9 | – | – | ||||
| F15-42 | Nasal aspiration | RSV B | 21.0 | HBoV | 31.9 | – | – | ||
| F15-47 | Nasal aspiration | ADV2 | 28.3 | Rhino | 19.7 | – | – | ||
| F15-50 | Nasal aspiration | PIV4 | 32.0 | – | – | ||||
| F15-52 | Nasal aspiration | RSV B | 19.5 | – | – | ||||
| F16-55 | Nasal secretion | HMPV | 25.0 | – | – | ||||
| F15-56 | Nasal aspiration | RSV A | 27.2 | ADV2 | 22.5 | – | – | ||
| F15-7 | Nasal secretion | FluA, H3 | 19.1 | – | – | ||||
| F16-9 | Nasal swab | FluA, H1pdm | 22.2 | – | – | ||||
| F16-17 | Nasal secretion | FluA, H3 | 18.6 | – | – | ||||
| F16-26 | Nasal secretion | FluA, H1pdm | 20.6 | – | – | ||||
| F16-44 | Nasal secretion | FluB | 21.0 | – | – | ||||
| F16-56 | Nasal secretion | FluB | 18.6 | – | – | ||||
| Positive control (viral RNA) | + | + | |||||||
| Negative control | – | – | |||||||
Cq: quantification cycle value.
To evaluate the accuracy of QProbe RT-LAMP in detecting MERS-CoV from human specimens, QProbe RT-LAMP assays were performed using seven total RNAs extracted from clinical specimens that were initially confirmed to be MERS-CoV-positive by real-time RT-PCR (upE) (Table 5 ). Two MERS-CoV negative specimens were used as negative controls. Taking into consideration the recent MERS case occurrence rate, it was difficult to obtain fresh specimens; therefore, stored specimens were used for validation. Specimens deemed to be positive had quantification cycle values of 20.2–30.9 for the upE set. Using the N and ORF1a primer sets, QProbe RT-LAMP confirmed a positive diagnosis for all seven positive samples and a negative diagnosis for the other two. In short, the QProbe RT-LAMP assays developed in this study were capable of detecting MERS-CoV from human clinical specimens.
Table 5.
QProbe RT-LAMP assays using clinical specimens positive for MERS-CoV viruses.
| Specimen | Real-time RT-PCR |
QProbe RT-LAMP |
||
|---|---|---|---|---|
| No. | upE | Cq value | N | ORF1a |
| 1 | + | 20.2 | + | + |
| 2 | + | 26.3 | + | + |
| 3 | + | 23.4 | + | + |
| 4 | + | 30.6 | + | + |
| 5 | + | 30.9 | + | + |
| 6 | + | 22.7 | + | + |
| 7 | + | 25.8 | + | + |
| 8 | – | >40 | – | – |
| 9 | – | >40 | – | – |
Cq: quantification cycle value.
3.3. Validations for mismatched sequences
The primer sets utilized in this study were constructed based on the conserved region of the N protein and ORF1a from the MERS-CoV EMC strain. However, significant genetic variations are present in these viral genomic regions as demonstrated by the large amount of sequences registered in GenBank. As shown in Table 1, 300 and 278/9 variations of nucleotide sequences for N and ORF1a have been registered in the database, with the majority of them showing a complete match to the corresponding primer set. Mismatches (1–3 base-pairs) were identified in several sequences when aligned with our primers (see Supplemental figure). In particular, the FIP primers had a high mismatch rate in the F2 region; the identities were 81.7% for N set and 86.0% for ORF1a set (Table 1). To determine whether QProbe RT-LAMP could amplify target sequences with mismatches to our primers, a panel of N and ORF1a RNA templates were synthesized by in vitro transcription, and QProbe RT-LAMP assays were performed using the appropriate primer sets (Table 6 for N, Table 7 for ORF1a). The detection limit of QProbe RT-LAMP for the EMC isolate sequence was 7.3–15.8 copies. QProbe RT-LAMP assays showed similar levels of amplification among all RNA template sequences, compared with that of the EMC isolate. The MERS-CoV sequences with mismatches were either of camel and/or human origin. Regardless of the target sequence origin, QProbe RT-LAMP well tolerated the nucleotide mismatches (1–3 base-pairs) without affecting the overall assay performance. In the previous study, mismatch in the B2 region (G29018T) slightly altered the amplification efficiency of RT-LAMP, leading to a five-fold decrease in detection sensitivity (Shirato et al., 2014). In contrast, the amplification efficiency in the QProbe RT-LAMP assays was not affected by mismatch in this region (Table 6). These findings indicate that the QProbe RT-LAMP assays could be used for the detection of all MERS-CoV isolates reported thus far, including for camels and humans.
Table 6.
Sensitivity of QProbe RT-LAMP using sequence with mismatches to the N primer set.
Camel MERS-CoV sequences are indicated in bold.
Based on EMC isolate (JX869059.2).
Table 7.
Sensitivity of QProbe RT-LAMP using sequences with mismatches to the ORF1a primer set.
| Positiona | Accession No. | Sensitivity (copies) |
|---|---|---|
| C1604T | KX108942, KX108941, KX108940, KX108939, KX108938, KX108937, KU242424, KU242423, KT751244, KT156561, KT156560, KP719933, KP719932, KP719931, KP719930, KP719929, KP719928, KP719927, KP209313, KP209312, KP209311, KP209310, KP209309, KP209308, KP209307, KP209306, KJ650297, KJ650296, KJ650295, KJ361503, KJ361502, KJ361501, KJ361500, KJ361499, KJ156896, KJ156863, KF745068 | 7.3 |
| A1650G | KX154687 | 1.6 |
| A1650G, C1685T | KX108944, KT368875, KT368832. KT368831, KT368830, KT368829, KR011266, KR011265, KR011264, KR011263, KJ713299, KJ713297, KJ713296, KJ713295 | 3.4 |
| A1650G, C1685T, T1694C | KJ713298 | 7.3 |
| C1685T | KT861628, KT368824, KM027257, KJ556336, KJ156949, KJ156944, KJ156938, KJ156881, KF958702, KF917527 | 15.8 |
| C1696T | KT368826 | 15.8 |
| T1718C | KX108943 | 1.6 |
| JX869059 (QProbe) | 7.3 |
Camel MERS-CoV sequences are indicated in bold.
Based on EMC isolate (JX869059.2).
4. Discussion
Real-time RT-PCR is the most commonly used technique for the detection and confirmation of MERS-CoV infection. According to the case definition outlined by the WHO, positive amplification of at least two different virus-specific genomic targets is required for case confirmation. Two real-time RT-PCR assays were developed by Corman et al., using primer sets targeting upE and ORF1a region. These assays have been proven to be highly sensitive and specific; therefore, they are used as the standard diagnostic method for MERS-CoV (Corman et al., 2012a, Corman et al., 2012b). However, PCR amplification involves a relatively long running process and may be unsuitable for field-based studies. Recently, other genetic diagnostic methods using different mechanisms have been developed, which include RT-LAMP (Bhadra et al., 2015; Shirato et al., 2014) and reverse transcription isothermal recombinase polymerase amplification (RT-RPA) (Wahed et al., 2013). In this study, the QProbe RT-LAMP targeted different positions in the MERS-CoV genome [ORF1a (nt 1572–1753) and N] from Corman’s assays [ORF1a (nt 18265–18314) and upE]. Two positives in the QProbe RT-LAMP or real-time PCR assays are enough, to confirm the presence of MERS-CoV. However, this means if the specimen is positive in two of four sets, it can be considered positive for MERS-CoV; if one of the real-time RT-PCR assay is negative, one positive QProbe RT-LAMP is sufficient for case confirmation, and vice versa. Thus, these techniques have improved the sensitivity and diagnostic outcomes of MERS-CoV by increasing the number of viral genomic targets available for amplification.
The results of RT-LAMP can be detected at the endpoint by checking magnesium pyrophosphate precipitation or fluorescent signal generated by DNA intercalators under ultraviolet light (Mori et al., 2001). Because they are easy to use, and they do not require large equipment for processing, RT-LAMP assays are more suitable for field-based studies. However, in turbidity monitoring, the salt accumulation accompanied by the LAMP reaction can be induced by primer dimers and/or non-primer reactions (Njiru, 2012). It is possible to detect unexpected increase in turbidity derived from non-primer signal, such as fragments of host DNA. Therefore, in this study, we developed a fluorescent RT-LAMP method with the addition of QProbes, in which only fluorescence quenching-derived from the probes was measured as a positive signal (Kurata et al., 2001). As such, QProbes provide additional specificity for detection as they bind to unique nucleotides that are only present in the target sequence and amplicon by LAMP primers. This means that a positive signal in the QProbe RT-LAMP assay is dependent on the primer reaction only. In addition, labeling the 3′ end with fluorescence dye abrogates non-specific signals derived from primers because extension of the QProbe is physically blocked by the dye. Thus, QProbe allows for highly specific detection under isothermal conditions and in a short time without melting curve analysis. Furthermore, materials used in QProbe RT-LAMP assays can be prepared as lyophilized form and packaged into diagnostic kits, increasing product integrity during shipment and handling. As demonstrated using clinical specimens (Table 5), this assay could be run in a portable device (e.g., ESEQuant TS2) and be completed in 30 min or less for accurate diagnosis of MERS-CoV, making it more suitable for field-based studies. We also showed that the two primer sets constructed in the study (targeting the viral N and ORF1a sequences) were equally capable of detecting MERS-CoV RNA. This means that a positive diagnosis can be confirmed by QProbe RT-LAMP alone without the need for other confirmation methods.
To determine the homology between the QProbe RT-LAMP primers and the generic variants of N and ORF1a, nucleotide sequence alignment was performed using 300 N and 279 ORF1a sequences available on GenBank. Most primer sets described in this study matched 90–100% of sequence variants, except for the FIP primers, which had matched 81.7% and 86.0% of N and ORF1a sequences, respectively. Our data indicate that these mismatches did not affect the ability of QProbe RT-LAMP assays to amplify these target sequences. In fact, these assays demonstrated comparable levels of sensitivity and specificity in detecting MERS-CoV genetic variants as the EMC strain, which was used for primer construction. When using LAMP, mismatches in primer sequences seem to be tolerated, but mismatches can occur in the primer’s 3′ end and the BIP primer should be avoided (Peyrefitte et al., 2008; Wang, 2016). In previous report, a mismatch in the BIP primer of the MERS-CoV N set slightly affected reactivity for the Riyadh-3 clade of MERS-CoV (Shirato et al., 2014). However, this decrease in sensitivity was not seen in QProbe RT-LAMP. This difference might be due to the difference in detection mechanism.
It seems that a significant amount of the newly registered MERS-CoV sequences on GenBank are of dromedary origin. The QProbe RT-LAMP assays were also able to detect target sequences (synthesized RNA) derived from dromedary MERS-CoV. These data suggest that QProbe RT-LAMP assays can be used as an easy, rapid and reliable surveillance technique for MERS-CoV in the field-based studies for both humans and dromedaries.
5. Conclusions
In this study, QProbe RT-LAMP assays were developed for the detection of MERS-CoV. Quenching of fluorescence from labeled probe-specific reactions is measured as positive signals. These assays are capable of detecting MERS-CoV RNA at a level similar to that of standard real-time RT-PCR assays and the previously reported RT-LAMP, with no cross-reactivity observed with other respiratory viruses. QProbe RT-LAMP assays were demonstrated to be rapid, simple, and convenient as they employed a dry form of reagents and a portable fluorometer. QProbe RT-LAMP assays thus offer a reliable alternative for the diagnosis of MERS-CoV in humans and dromedaries. Altogether, these results indicate that the QProbe RT-LAMP assay can be used as a powerful tool for the diagnosis and surveillance of MERS-CoV infection in the field.
Funding
This work was supported by a Grant-in-Aid (Research Program on Emerging and Re-emerging Infectious Diseases, No. 16fk0108213j0102, 16fk0108303j0303, 17fk0108313j0203, and 17fk0108103j0301) from the Japan Agency for Medical Research and Development (AMED), and a Grant-in-Aid for Scientific Research (B: 17H04642) from the Japan Society for the Promotion of Science.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
We thank Dr. Ron A. M. Fouchier, Erasmus Medical Center, Rotterdam, the Netherlands for providing the MERS-CoV EMC isolate. We thank Dr. Kunihiro Oba, Showa General Hospital, Japan for providing clinical specimens from patients presenting with flu-like symptoms.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2018.05.006.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Adhikari B.R., Pandey B.D., Ghimire P., Shrestha B., Khadka M., Yoda T., Suzuki Y. Loop-mediated isothermal amplification (LAMP) for the direct detection of human pulmonary infections with environmental (nontuberculosis) mycobacteria. Jpn. J. Infect. Dis. 2009;62:212–214. [PubMed] [Google Scholar]
- Arimatsu Y., Kaewkes S., Laha T., Hong S.J., Sripa B. Rapid detection of Opisthorchis viverrini copro-DNA using loop-mediated isothermal amplification (LAMP) Parasitol. Int. 2012;61:178–182. doi: 10.1016/j.parint.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A., Team K.M.-C.I. Hospital outbreak of Middle East respiratory syndrome coronavirus. New Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. New Engl. J. Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS One. 2015;10:e0123126. doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T., Muth D., Muller M., Drexler J., Zambon M., Osterhaus A., Fouchier R., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Muller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M., Nitsche A., Bleicker T., Landt O., Schweiger B., Drexler J.F., Osterhaus A.D., Haagmans B.L., Dittmer U., Bonin F., Wolff T., Drosten C. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- Do D.H., Laus S., Leber A., Marcon M.J., Jordan J.A., Martin J.M., Wadowsky R.M. A one-step, real-time PCR assay for rapid detection of rhinovirus. J. Mol. Diagn. 2010;12:102–108. doi: 10.2353/jmoldx.2010.090071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V.L., Howson E.L., Madi M., Mioulet V., Caiusi C., Pauszek S.J., Rodriguez L.L., King D.P. Development of a reverse transcription loop-mediated isothermal amplification assay for the detection of vesicular stomatitis New Jersey virus: use of rapid molecular assays to differentiate between vesicular disease viruses. J. Virol. Methods. 2016;234:123–131. doi: 10.1016/j.jviromet.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Geojith G., Dhanasekaran S., Chandran S.P., Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J. Microbiol. Methods. 2011;84:71–73. doi: 10.1016/j.mimet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Hong T.C., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T., Hasebe F., Morita K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Tashiro M., Odagiri T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine. 2006;24:6679–6682. doi: 10.1016/j.vaccine.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Kaida A., Kubo H., Takakura K., Sekiguchi J., Yamamoto S.P., Kohdera U., Togawa M., Amo K., Shiomi M., Ohyama M., Goto K., Hase A., Kageyama T., Iritani N. Associations between co-detected respiratory viruses in children with acute respiratory infections. Jpn. J. Infect. Dis. 2014;67:469–475. doi: 10.7883/yoken.67.469. [DOI] [PubMed] [Google Scholar]
- Kurata S., Kanagawa T., Yamada K., Torimura M., Yokomaku T., Kamagata Y., Kurane R. Fluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY((R)) FL-labeled probe or primer. Nucleic Acids Res. 2001;29:E34. doi: 10.1093/nar/29.6.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Y., Martins D.B.G., Kimura M., Catena A.D.S., Borba M., Mattos S.D.S., Abe H., Yoshikawa R., de Lima Filho J.L., Yasuda J. Development and evaluation of a rapid molecular diagnostic test for Zika virus infection by reverse transcription loop-mediated isothermal amplification. Sci. Rep. 2017;7:13503. doi: 10.1038/s41598-017-13836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Baek Y.H., Kim Y.H., Choi Y.K., Song M.S., Ahn J.Y. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2016;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Chong S., Bulir D., Ruyter A., Mwawasi K., Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J. Clin. Virol. 2013;58:127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- Njiru Z.K. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl. Trop. Dis. 2012;6:e1572. doi: 10.1371/journal.pntd.0001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrefitte C.N., Boubis L., Coudrier D., Bouloy M., Grandadam M., Tolou H.J., Plumet S. Real-time reverse-transcription loop-mediated isothermal amplification for rapid detection of rift valley Fever virus. J. Clin. Microbiol. 2008;46:3653–3659. doi: 10.1128/JCM.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Watanabe O., Hirokawa C., Matsuyama S., Nishimura H., Taguchi F. Differences in neutralizing antigenicity between laboratory and clinical isolates of HCoV-229E isolated in Japan in 2004-2008 depend on the S1 region sequence of the spike protein. J. Gen. Virol. 2012;93:1908–1917. doi: 10.1099/vir.0.043117-0. [DOI] [PubMed] [Google Scholar]
- Shirato K., Nishimura H., Saijo M., Okamoto M., Noda M., Tashiro M., Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2007;139:78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Yano T., Senba S., Akachi S., Kobayashi T., Nishinaka T., Notomi T., Matsuyama S. Detection of Middle East respiratory syndrome coronavirus using reverse transcription loop-mediated isothermal amplification (RT-LAMP) Virol. J. 2014;11:139. doi: 10.1186/1743-422X-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Miyata R., Ichikawa K., Morishita S., Kurata S., Nakamura K., Tsuneda S., Sekiguchi Y., Noda N. Universal quenching probe system: flexible, specific, and cost-effective real-time polymerase chain reaction method. Anal. Chem. 2009;81:5678–5685. doi: 10.1021/ac900414u. [DOI] [PubMed] [Google Scholar]
- Ueda S., Kuwabara Y. The rapid detection of salmonella from food samples by loop-mediated isothermal amplification (LAMP) Biocontrol Sci. 2009;14:73–76. doi: 10.4265/bio.14.73. [DOI] [PubMed] [Google Scholar]
- Ushio M., Yui I., Yoshida N., Fujino M., Yonekawa T., Ota Y., Notomi T., Nakayama T. Detection of respiratory syncytial virus genome by subgroups-A, B specific reverse transcription loop-mediated isothermal amplification (RT-LAMP) J. Med. Virol. 2005;77:121–127. doi: 10.1002/jmv.20424. [DOI] [PubMed] [Google Scholar]
- Wahed A.A.E., Patel P., Heidenreich D., Hufert F.T., Weidmann M. Reverse transcription recombinase polymerase amplification assay for the detection of Middle East respiratory syndrome coronavirus. PLOS Curr. Outbreaks Ed. 2013;1 doi: 10.1371/currents.outbreaks.62df1c7c75ffc96cd59034531e2e8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Effect of internal primer–template mismatches on loop-mediated isothermal amplification. Biotechnol. Biotechnol. Equip. 2016;30:314–318. [Google Scholar]
- Wang L.X., He L., Fang R., Song Q.Q., Tu P., Jenkins A., Zhou Y.Q., Zhao J.L. Loop-mediated isothermal amplification (LAMP) assay for detection of Theileria sergenti infection targeting the p33 gene. Vet. Parasitol. 2010;171:159–162. doi: 10.1016/j.vetpar.2010.02.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.