Highlights
-
•
DDX5 acts as a positive regulator for JEV RNA replication.
-
•
DDX5 interacts with several viral proteins.
-
•
DDX5 is recruited to the cytoplasm and colocalizes with viral proteins and viral RNA.
-
•
DDX5 can only bind to the JEV 3′ UTR.
Keywords: Japanese encephalitis virus, DEAD-box RNA helicase DDX5, Replication, Non-structural protein, Untranslated region
Abstract
Japanese encephalitis virus (JEV), one of the causes for epidemic encephalitis, belongs to the family of Flaviviridae. In this study, we demonstrated that cellular DEAD-box RNA helicase DDX5 plays an important role in JEV replication. The knockdown of DDX5 was able to decrease JEV replication, and overexpression of DDX5 mutants lacking the helicase activity also reduced JEV replication, suggesting the helicase activity is essential for JEV replication. DDX5 knockdown did not affect virus assembly and release. GST-pulldown and co-immunoprecipitation experiments demonstrated that DDX5 could interact with JEV core protein, non-structural protein 3 (NS3) and 5 (NS5-MTase and NS5-RdRp domains). Meanwhile, we also confirmed that DDX5 interacts with these viral proteins during JEV infection. Confocal microscopy analysis showed that endogenous DDX5 is recruited to the cytoplasm and colocalizes with these viral proteins and viral RNA. RNA-pulldown experiment showed that DDX5 only binds to the JEV 3′ untranslated region (UTR). Finally, we confirmed the role of DDX5 in JEV RNA replication using JEV-replicon system. In conclusion, we identified DDX5 as a positive regulator for JEV replication.
1. Introduction
Japanese encephalitis virus (JEV), one of the causes for epidemic encephalitis, belongs to the family of Flaviviridae. It is widely distributed in Asia (Harris et al., 2006, Solomon, 2003, Tsai, 2000). The outbreaks of emerging flaviviruses have made the prevention and treatment of flavivirus infection a global public health concern (Hanna et al., 1996, Mackenzie et al., 2004, Wang et al., 2007). The genome of JEV is a single-stranded and plus-sense RNA approximately 11 kb in length, encoding 3 structural and 7 non-structural proteins in the order of 5′-C-prM-E-NS1-NS2a-NS2b-NS3-NS4a-NS4b-NS5-3′ (Chambers et al., 1990).
The cellular DEAD-box helicases family of RNA helicase utilizes the energy derived from nucleotide triphos phate (NTP) hydrolysis to modulate the unwinding of RNA. They are highly conserved from bacteria to humans and play important roles in all biological steps of RNA, such as RNA transcription, splicing, transport and decay, ribosome assembly and translation (Gustafson and Wessel, 2010, Lee et al., 2008). DDX5, one of the cellular DEAD helicase, acts as an important co-activator for a range of transcription factors (Endoh et al., 1999, Fuller-pace and Ali, 2008, Steimer and Klostermeier, 2012). Moreover, DDX5 has been shown to be recruited to the promoters of responsive genes when these transcription factors are activated, consistent with its role in transcription initiation (Clark et al., 2008).
Theoretically, a drug targeting the cellular factors important for the viral life cycle can be used to inhibit the virus infection. DDX5 attracts great interests in antiviral researches because several studies showed it is involved in the replication of several viruses such as HIV-1 (Zhou et al., 2013), SARS-CoV, influenza virus and Hepatitis C virus (HCV) (Chen et al., 2009, Goh et al., 2004). DDX5 interacts with SARS-CoV helicase and SARS-CoV replication is significantly inhibited when expression of DDX5 was silenced by small interfering RNA (Chen et al., 2009). DDX5 also colocalizes with influenza virus RNP in the nucleus and interacts with influenza virus polymerase (Bortz et al., 2011). HCV RNA-dependent RNA polymerase can interact with DDX5 and transient expression of RdRp causes the redistribution of endogenous DDX5 from the nucleus to the cytoplasm (Goh et al., 2004).
Meanwhile, other cellular helicases such as DDX1, DDX3 and DDX6 can act as the cofactors to promote HIV and HCV replication (Fang et al., 2005, Fang et al., 2004, Reed et al., 2012). DDX3 inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids (Wang et al., 2009b). DDX1 can also stimulate the replication of IBV (Xu et al., 2010). It has been reported that DDX6, DDX28, DDX42, DHX15 and DDX56 are important for infectivity of West Nile virus (Chahar et al., 2013, Krishnan et al., 2008, Xu et al., 2011, Xu and Hobman, 2012). DDX6 is also involved in DENV, HCV, foamy virus and adenovirus replication (Greer et al., 2011, Jangra et al., 2010, Ward et al., 2011, Yu et al., 2011).
In this study, we demonstrated that DDX5 is required for JEV replication, and the helicase activity of DDX5 is crucial for JEV replication. DDX5 interacted and colocalized with JEV core, NS3, NS5 (MTase and RdRp domain) in the cytoplasm of the infected cells. DDX5 was recruited from the nucleus to cytoplasm during virus infection. DDX5 can bind to the JEV 3′ UTR and colocalizes with viral RNA. Finally we used a JEV replicon system to confirm that DDX5 plays an important role in JEV RNA replication.
2. Materials and methods
2.1. Viruses, cells and transfection
Baby hamster kidney (BHK)-21 cells (C-13, American Type Culture Collection) was maintained in Dulbecco’s minimal essential medium supplemented with 10% fetal bovine serum and penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37 °C in 5% CO2. JEV SA14-14-2 strain was propagated in the BHK-21 cells, and the viral titers were determined in BHK-21 cells by plaque assay. Cells were transfected using polyethylenimine (25 KD, Sigma–Aldrich). The amounts of plasmid DNA with which cells were transfected (1 μg per 15.4 mm and 10 μg per 80 mm) were kept constant by the inclusion of DNA vector. Transfection efficiencies of over 70% were routinely obtained when using the pEGFP-N2 (Novagen) plasmid to monitor the transfection efficiencies.
2.2. Plasmids
To construct HA-tagged DDX5 pcDNA3.1-DDX5 (1–614), pCold-I-DDX5 or pGEX-4T-3-DDX5 plasmids, the gene fragments were amplified using DDX5-HA plasmid (a gift from Prof. Ralf Janknecht) as the template by PCR using specific primers (Table 1 ) and were cloned in frame into pcDNA3.1(+) (Invitrogen), pGEX-4T-3(GE Healthcare) or pCold-I (Takara) vector respectively. JEV core, NS3, NS5-MTase (1–268) and NS5-RdRp (318–905) were also subcloned into pcDNA3.1 (+) vector, the MYC tag was introduced during PCR by inclusion of the tag coding sequence in the antisense primer. Kozak sequence was added in the sense primers (Table 1). To express JEV viral proteins, the cDNA fragments of JEV SA14-14-2 strain encoding core, NS3, NS5-MTase and NS5-RdRp were all subcloned into pET-24a-(+) vector (Novagen) and pGEX-4T-3(GE Healthcare) by PCR using specific primers (Table 1). pcDNA3.1-DDX5-K144E-HA, pcDNA3.1-DDX5-S279L -HA mutants were made with Quick-Change site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions using pcDNA3.1-DDX5-HA as the template by PCR using specific primers (Table 1). Oligonucleotides with the following sequence: CTCTAATGTGGAGTGCGAC was used to clone short hairpin RNA (shRNA)-encoding sequences against DDX5 in the pGPU6/Neo (Genepharma, China) vector, the Oligonucleotides encoding non-targeting shRNA was cloned into pGPU6/Neo vector and the sequence is as follows: GTTCTCCGAACGTGTCACGT which was used as the control shRNA.
Table 1.
The primers of the plasmids used in the study (the restriction endonucleases sites were underlined and italic, T7 promoter and tobramycin aptamer tag sequences were underlined in the forward and reverse primers).
| Plasmids | Primers |
|---|---|
| pCold-I-DDX5/pGEX-4T-3-DDX5 | Forward primer: GATACTAGGATCCATGTCGGGTTATTCGAG |
| Reverse primer: GAGCTCGGTCGACTTATTGGGAATATCCTG | |
| pcDNA3.1-DDX5 (K144E and S279L)-HA |
Forward primer: GTCATGCAAGCTTGCCACCATGTCGGGTTA TTCGAGTGAC |
| Reverse primer: GTAGTTAGGATCCCTAGGCGTAGTCGGGGA CGTCGTAGGGGTATTGGGAATATCCTGTT | |
| pcDNA3.1-core-MYC | Forward primer: GCCGCCAAGCTTGCCACCATGATGACTAAA AAACCAGGAGGGCC |
| Reverse primer: GCGGGCGGGATCCCTACAAATCTTCTTCAG AAATCAACTTCTGTTCGGCTCCTGCACAA | |
| pcDNA3.1-NS3-MYC | Forward primer: GTATTATGGTACCGCCACCATGGGGGGCGT GTTTTGGGAC |
| Reverse primer: GCGGGCGCTCGAGCTACAAATCTTCTTCAG AAATCAACTT CTGTTCTCTCTTCCCTGCT | |
| pcDNA3.1-NS5 (Mtase) -MYC |
Forward primer: GATATATGGTACCGCCACCATGGGAAGGCC TGGGGG |
| Reverse primer: GCGGGCGCTCGAGCTACAAATCTTCTTCAG AAATCAACTTCTGTTCTCCCACGGCTCTT | |
| pcDNA3.1-NS5 (RdRp) -MYC |
Forward primer: GACATATGGTACCGCCACCATGTCAGCC AGCTCTCTC |
| reverse primer: GCGGGCGCTCGAGCTACAAATCTTCTTCAG AAATCAACTTCTGTTCGATGACCCTGTCT | |
| DDX5-K144E (Quikchange) | Forward primer: GTGGCACAGACTGGATCTGGGGAGACATT GTCTTATTTGCTTCCT |
| Reverse primer: AGGAAGCAAATAAGACAATGTCTCCCCA GATCCAGTCTGTGCCAC | |
| DDX5-S279L (Quikchange) |
Forward primer: GATAGGCAAACTCTAATGTGGCTAGCGAC TTGGCCAAAAG AAGTA |
| Reverse primer: TACTTCTTTTGGCCAAGTCGCTAGCCACA TTAGAGTTTGCCTATC | |
| pET-24a-core | Forward primer: GCCGCCGAATTCATGACTAAAAAACCAG |
| Reverse primer: ATAATACTCGAGGGCTCCTGCACAAGCTAT | |
| pET-24a-NS3 | Forward primer: GACGACACATATGGGGGGCGTGTTTTGGGA |
| Reverse primer: GCGCGCGCTCGAGTCTCTTCCCTGCTGCAA | |
| pET-24a-NS5-MTase | Forward primer: GATTTACATATGGGAAGGCCTGGGGGCAG |
| Reverse primer: GCTCATACTCGAGTCCCACGGCTCTTGTTCC | |
| pET-24a-NS5-RdRP | Forward primer: GATATGCGGATCCTCAGCCAGCTCTCTCGT |
| Reverse primer: GGCCGACTCGAGGATGACCCTGTCTTCCTGG ATCAAGACG | |
| pcDNA3.1-DDX5r, pcDNA3.1-DDX5r- (K144E,S279L)-HA (Quikchange) | Forward primer: TAAGACCTGATAGGCAAACCTTGATGTGGTC CGCTACTTGGCCAAAAGAAGTAAGA |
| Reverse primer: TCTTACTTCTTTTGGCCAAGTAGCGGACCAC ATCAAGGTTTGCCTATCAGGTCTTA | |
| JEV 5′ UTR | Forward primer: TAATACGACTCACTATAGGGAGAAGTTTATC TGTGTGAACTTCTTGGCTTA |
| Reverse primer: GGCTCAGCACGAGTGTAGCTAAACCTCGCTA TACTAAGCCGGTTATCTTCCGTTCTAA | |
| JEV 3′ UTR | Forward primer: TAATACGACTCATAGGGTAGTGTGATTTAAG GTAGAAAAGTAG |
| Reverse primer: GGCTCAGCACGAGTGTAGCTAAACCTCGCTA TACTAAGCCAGATCTTGTGTTCTTCCT | |
| Ampicillin resistance gene (1–250 bp) |
Forward primer: TAATACGACTCACTATAGGGATGAGTATTCA ACATTTCCGTGTCG |
| Reverse primer: GGCTCAGCACGAGTGTAGCTAAACCTCGCTA TACTAAGCCCGTCAATACGGGATAATAC | |
| pGEX-4T-3-core | Forward primer: GCCGCCGGATCCATGACTAAAAAACCAG |
| Reverse primer: ATAATACTCGAGGGCTCCTGC ACAAGCTAT | |
| pGEX-4T-3-NS3 | Forward primer: GACGACAGAATTCGGGGGCGTGTTTTGGGA |
| Reverse primer: GCGCGCGCTCGAGTCTCTTCCCTGCTGCAA | |
| pGEX-4T-3-NS5-MTase | Forward primer: GATTTAGAATTCGGAAGGCCTGGGGGCAG |
| Reverse primer: GCTCATACTCGAGTCCCACGGCTCTTGTTCC | |
| pGEX-4T-3-NS5-RdRP | Forward primer: GATATGCGGATCCTCAGCCAGCTCTCT CGT |
| Reverse primer: GGCCGACTCGAGGATGACCCTGTCTTCCTGG ATCAAGACG |
To construct RNAi resistant pcDNA3.1-DDX5r, pcDNA3.1-DDX5r-K144E-HA and pcDNA3.1-DDX5r-S279L-HA mutants, the shRNA target sequence in DDX5 gene was changed into CCTTGATGTGGTCCGCTAC without introducing any residue change using QuikChange site-directed mutagenesis kit (Stratagene) using pcDNA3.1-DDX5-HA as the template by PCR using specific primers (Table 1). All the nucleotide sequence was confirmed by using DNA sequence analysis.
For construct of templates for JEV 5′ and 3′ UTR RNA transcription, the JEV 5′ and 3′ UTR gene was amplified by PCR with specific primers. A control RNA was generated by PCR amplification of part of the ampicillin resistance gene (1–250 bp) using the primers (Vashist et al., 2012) (Table 1). T7 promoter and tobramycin aptamer tag (Hartmuth et al., 2004) sequences were included in sense and anti-sense primer respectively. The PCR products were subcloned into pMD-18T vector (Takara, China).
The JEV replicon with a Renilla luciferase reporter was generated by cloning JEV SA14-14-2 strain cDNA into pBluescript-II KS (Agilent Technologies) under the control of a T7 promoter as the previous study (Chien et al., 2011). The Renilla luciferase gene was inserted after the first 102 bp of the JEV C gene, followed by the foot-and-mouth disease virus 2A self-cleaving protease (FMDV-2A), which enables the cleavage of the luciferase away from downstream nonstructural proteins (Lo et al., 2003, Varnavski and Khromykh, 1999). FMDV 2A was fused to the last 90 bp of the E gene which is necessary for the proper topology of the following viral proteins. To ensure RNA stability and processing, a hepatitis delta virus ribozyme was placed immediately adjacent to the 3′ end of the JEV cDNA followed by a bovine growth hormone (BGH) polyadenylation sequence. The control construct, a GAPDH 5′ and 3′ UTR-flanking luciferase reporter gene, was cloned into pMD-18T Vector (Takara, China).
2.3. Protein expression and purification
The expression plasmids of pCold-I-DDX5, pGEX-4T-3-core, pGEX-4T-3-NS3, pGEX-4T-3-NS5-MTase, pGEX-4T-3-NS5-RdRP, pGEX-4T-3-DDX5, pET-24a-core, pET-24a-NS3, pET-24a-NS5-MTase, and pET-24a-NS5-RdRP were transformed into Escherichia coli expression strain BL-21-Codon plus (DE3)-Rosetta 2 cells. A single colony was used to inoculate 10 ml of Luria-Bertani (LB) media containing ampicillin (100 μg/ml) or kanamycin (50 μg/ml), and the culture was grown at 37 °C overnight. The cultures were then diluted into the LB media containing ampicillin (100 μg/ml) and grown to an A600 of 0.6 at 37 °C, then protein expression was induced with 100 μM IPTG for 16–20 h at 21 °C. The cells were harvested by centrifugation at 5000g for 20 min at 4 °C and re-suspended in lysis buffer (100 mM NaCl, 20 mM Tris–HCl, pH 8.0, 0.1 mM PMSF). The cells were disrupted by sonication, and the supernatant was collected by centrifugation. The supernatant was then subjected to affinity purification using Bio-Rad Profinity™ IMAC or ProfinityTM GST column (Bio-Rad). The proteins were then eluted, pooled together and further purified using size exclusion chromatography (Superdex75, GE Healthcare). Concentration of the purified proteins was determined by Bradford assay.
2.4. Cytotoxicity assay
The cell viability of BHK-21 cells transfected with DDX5i, HA-DDX5 or DDX5 mutants plasmids was detected by CytoTox 96® NonRadioactive Cytotoxicity Assay (Promega) at 72 h after transfection. The Cytotoxicity of Leptomycin B (LMB) on BHK-21 cells was detected by CytoTox 96® NonRadioactive Cytotoxicity Assay at 48 h after added to the cells with different the concentrations (0–20 ng/ml).
2.5. Antibodies and reagents
The DDX5-specific polyclonal antibody was purchased from Santa Cruz Biotechnology (USA), anti-β-actin, anti-MYC, anti-GST and anti-HIS monoclonal antibodies were purchased from Abmart Company (Shanghai, China). Anti-MYC-HRP and Anti-HA–HRP monoclonal antibodies were purchased from cell signaling (USA). Leptomycin B (LMB) was purchased from Santa Cruz Biotechnology (USA). The monoclonal antibody against the JEV envelope glycoprotein was described previously (Ishag et al., 2012b). To produce anti-core, NS3 and NS5 polyclonal antibodies of mice source, the BALB/c mice free of pathogens was inoculated with 50 μg of NS5-RdRp protein emulsified with equal amount of freund’s complete adjuvant via subcutaneous injection. Booster doses were subsequently given at 1, 2 and 3 weeks later with 50 μg of core, NS3 and NS5-RdRp emulsified with incomplete adjuvant. Finally, the serum of the mice was collected. The serum of the mice without immunized any protein was collected as the negative serum. Both immunoblot and ELISA were performed to evaluate the immunoreactivity between JEV core, NS3 and NS5 proteins and their polyclonal antibodies. The animal used in this study and animal experiment was approved by Department of Science and Technology of Jiangsu Province and in compliance with the Nanjing Agricultural University Institutional Animal Care and Use Committee. The license number was SYXK (SU) 2010-0005.
2.6. Plaque assay
The viral culture supernatants with 10-fold dilutions (from 102 to 106) were added into 6-well plate with confluent monolayer of BHK-21 cells. The plate was then incubated at 37 °C for 2 h, with rocking at every 15-min interval. The excess virus inocula were removed by rinsing the wells with PBS for three times. Subsequently, overlay medium (2% low melting-point agarose with DMEM medium containing 2% FBS) was added to each well and further incubated at 37 °C with 5% CO2 for 3 days. The cells were stained with 0.5% crystal violet.
2.7. GST pull-down assay and competitive binding assay
50 μL of glutathione-Sparse 4B beads (GE Healthcare) were resuspended in binding buffer (20 mM Tris–HCl, pH 7.5, 500 mM NaCl, 0.5% NP-40). The purified GST tagged proteins were incubated with the beads for 2 h at 4 °C under gentle agitation. Unbounded proteins were washed away using washing buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40). Then the purified HIS-tagged proteins were then incubated with the beads for 3–4 h with gentle agitation at 4 °C respectively. The beads were washed four times with 1 ml of washing buffer. Finally the beads were resuspended in 200 μL of SDS–PAGE sample buffer, boiled and subjected to 12% or 15% SDS–PAGE gel, and then transferred to PVDF membrane. The presence of the targeted proteins was detected with the anti-DDX5 polyclonal antibody and anti-HIS monoclonal antibody, glutathione-Sparse 4B-beads with the GST proteins were used as control.
In competitive binding experiment, GST-DDX5 protein was first incubated with 50 μL of glutathione-Sparse 4B beads. After extensive washing by washing buffer, HIS-tagged core, NS3, NS5-Mtase or NS5-RdRp was incubated with the beads for 3–4 h with gentle agitation at 4 °C followed by washing 5 times with washing buffer. The different amount of competitive protein (core, NS3, NS5-Mtase or NS5-RdRp) (10 μg-50 μg) were mixed with the beads. The mixture was incubated for 3–4 h with gentle agitation at 4 °C. The mixture was centrifuged at 3000 rpm for 5 min, the supernatant and the beads were subjected to 12% or 15% SDS–PAGE gel and analyzed by Western blot using either anti-HIS monoclonal antibody, anti-NS3 or NS5 polyclonal antibody.
2.8. Co-immunoprecipitation assay
BHK-21 cells transfected with the plasmids as indicated or infected with JEV, 48 h later, The medium was removed and the cells were rinsed twice in cold phosphate-buffered saline, incubated for 3 h at 4 °C in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.2 mM PMSF, 1% NP-40) with gentle agitation and collected. Cell debris was removed by centrifugation at 10,000g for 10 min at 4 °C. Cell extracts were incubated with anti-MYC mAb coupled protein G-agarose beads or the beads (have been incubated with DDX5 polyclonal antibody) for 2 h at 4 °C. The beads were washed five times with 1 ml of washing buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40). The beads were resuspended in 200 μL of SDS–PAGE sample buffer and boiled and were separated by SDS–PAGE, and then transferred to PVDF membrane. The proteins were analyzed by anti-DDX5, core, NS3, NS5 polyclonal antibodies or anti-MYC-HRP monoclonal antibody.
2.9. Western blot and RT-PCR analysis
Cell lysates or proteins complexes for GST pulldown and Co-immunoprecipitation were separated by 12% or 15% SDS–PAGE and transferred to PVDF membrane (Millipore, USA) in a Trans-Blot SD semidry transfer cell (Bio-Rad, USA). The membrane was blocked with 5% non-fat milk powder in TBST buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20), then probed with anti-core, NS3 or NS5 polyclonal antibodies, anti-JEV E (Ishag et al., 2012a, Ishag et al., 2012b) or anti-β-actin monoclonal antibodies, anti-HA monoclonal antibody-HRP or anti-MYC monoclonal antibody-HRP (Cell Signaling, USA). For the anti-HA monoclonal antibody-HRP, anti-MYC monoclonal antibody-HRP detection, they were directly visualized using enhanced chemiluminescence (GE Healthcare). For the other antibodies, the bound antibody was detected using HRP-conjugated secondary antibodies, and visualized using enhanced chemiluminescence.
JEV-specific RNA copy number which was expressed as a ratio to the cellular β-actin cDNA copies was quantified using quantitative PCR (Q-PCR). The total RNA was extracted from JEV-infected BHK-21 cells with TRIzol reagent (Invitrogen) and purified according to the Manufacturer’s recommendations. Intracellular JEV genome levels were quantified with the SYBR Green Probe 3-step QRT-PCR kit (Takara, China) and fluorescent quantization meter (ABI PRISM 7300 sequence detection system, Applied Biosystems). For cDNA preparation, total RNA (1 μg) was reverse transcribed with first strand cDNA synthesis kit (Takara, China). Triplicate cDNA samples were amplified with the RT-PCR kit. The primer for the JEV NS1 and β-actin were the following: NS1 sense primer: 5′-acactcgtcagatcacaggttca-3′; antisense primer: 5′-gccagaaacatcaccagaagg-3′, β-actin sense primer: 5′-catccgtaaagacctctatgccaac-3′, antisense primer, 5′-atggagccaccgatccaca-3′ and cellular β-actin mRNA from the same RNA extract was used as an internal control in BHK-21 cells as the previous study(Ishag et al., 2012a; Ishag et al., 2012b).
2.10. In vitro RNA transcription and the interaction of JEV 5′ and 3′ UTR with DDX5
To synthesize JEV replicon, GAPDH-luciferase, JEV 5′, 3′ UTR and control RNAs in vitro, the purified plasmids DNA was first linearized with KpnI or BamHI enzyme. JEV 5′, 3′ UTR and control RNA were generated by in vitro transcription using Riboprobe® System-T7 Kit (Promega, China) following a standard method in a 20 μL reaction mixture containing 4 μL 5 × transcription buffer, 2 μL RNasin® RNA Inhibitor (40U/μL), 1 μL each NTP (10 mM), 1 μg of linearized DNA template and 1 μL T7 RNA polymerase (20 U/μL). The mixture was incubated at 37 °C for 2 h. 10 μL of RNase-free DNaseI (1 U/μL) was added to the mixture and incubated at 37 °C for 30 min. The RNA was quantified using spectrophotometry. 5 μg RNA was added to the matrix beads conjugated with tobramycin respectively (Hartmuth et al., 2004) and incubated by head-over-tail rotation for 1–1.5 h. The mixture was washed three times with PBS (prepared with DEPC-treated water), the supernatants of BHK-21 cell lysate or purified DDX5 mutants were added. The mixture was incubated by head-over-tail rotation for 2 h and washed three times with PBS and subjected to SDS–PAGE for Western blot using anti-DDX5 polyclonal antibody, The RNA of JEV 5′ and 3′ UTR and control RNA were detected by 1% agarose (stained by Ethidium bromide).
2.11. Confocal microscopy analysis
BHK-21 cells were plated onto cover glass in a 6-well plate. In the following day, cells were infected with JEV (MOI = 0.01). 24 h later, cells were fixed with ethanol for 30 min at 4 °C, the cells were then washed with 1% BSA/PBS, and endogenous proteins were directly stained with their antibodies respectively. The cells were then treated for 60 min at room temperature with Rhodamine-conjugated second antibodies (Dingguo, China). For the viral RNA staining, cells were first infected with JEV (MOI = 0.01). 3 h later, the cells were treated with actinomycin D (Sigma, China) for 4 h (15 μg/ml) (Xu et al., 2010) to block cellular RNA transcription, then 5-ethynyl uridine (EU) was added. The viral RNA was detected using the Click-iT® RNA Imaging Kits (Invitrogen, China) according to the manufacturer’s instructions. The nucleus was stained using 4′, 6-diamidino-2-phenylindole (DAPI). The cover slips were mounted onto slide glass using PBS containing 50% glycerol. Confocal images were obtained using a Zeiss LSM 710 scanning confocal microscope.
2.12. Luciferase reporter assay
For the replicon luciferase reporter assay, BHK-21 cells were seeded in 24-well plates and transfected with DDX5 shRNA plasmid. 24 h later, the cells were transfected with 0.6 μg in vitro-transcribed JEV replicons RNA plus 0.2 μg of a firefly luciferase RNA in vitro transcribed from a luciferase T7 control (Promega) as an internal control using TransFast™ (Promega). At the indicated times post transfection, cell lysates were collected for the dual-luciferase assay using Dual-Luciferase® Reporter Assay System Kit and GloMax® 20/20 Luminometer instrumentation (Promega).
2.13. Statistical analysis
All data were determined in triplicate and were representative of at least two separate experiments. The results represent the means ± standard deviations of triplicate determinations. The differences between means were considered significant at ∗ p < 0.05, very significant ∗∗ p < 0.01. All statistical analyses were performed by one-way ANOVA using a SPSS 16.0 software package (version 16.0, SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Endogenous DDX5 plays important role in JEV replication
It has been demonstrated that DDX5 is involved in the replication of several viruses (Chen et al., 2009, Goh et al., 2004). To determine whether DDX5 is required for JEV replication, we first used shRNA-mediated gene silence method (DDX5i) to decrease the expression of endogenous DDX5 in BHK-21 cells. Western blot analysis using anti-DDX5 antibody demonstrated 90% decrease in protein expression level compared with the control untargeted shRNA (Fig. 1 A). 24 h later, the cells previously transfected with DDX5i plasmid were infected with JEV (MOI = 0.01). 48 h later, the viral titers in the supernatants were detected. The results showed that a reduction of endogenous DDX5 expression accounted for more than 13-fold decrease in virus titers (p < 0.01) compared with the control shRNA (Fig. 1B, C). DDX5 knockdown also resulted in significant decrease of JEV genome RNA and viral proteins expression level as suggested by Q-PCR and Western blot analysis (Fig. 1D, E) compared with the control shRNA (p < 0.01). All the results showed that DDX5 is important for JEV replication.
Fig. 1.
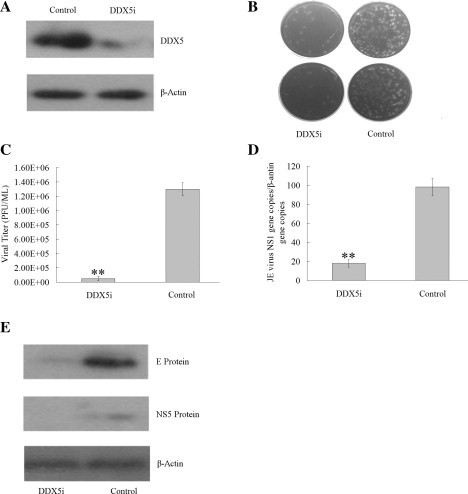
Requirement of DDX5 in JEV replication. (A). Western blot analysis of cellular lysates with anti-DDX5 or anti-β-actin antibody in BHK-21 cells after DDX5 knockdown (DDX5i), the untargeted shRNA was used as the control. (B). Viral titers determined by plaque assay after DDX5 knockdown. Representative plaque assay shows viral propagation in BHK-21 cells. (C). The statistical analysis on plaque assay. (D). The level of JEV genome RNA monitored by real-time PCR in JEV infected BHK-21 cells after DDX5 knockdown compared with the control shRNA treated cells. (E). Western blot analysis of the JEV proteins and DDX5 expression level in infected BHK-21 cells after DDX5 knockdown. Western blot of β-actin was served as the loading control. The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
We next determined whether cellular DDX5 knockdown decreases JEV infection by affecting virus assembly or release. The BHK-21 cells were transfected with DDX5i plasmid before infected with JEV (MOI = 0.01). JEV RNA copy numbers of the cell lysates and the supernatants were detected by Q-PCR, although the RNA copy numbers in cell lysates were nearly 12-fold higher than that in the supernatants, the ratios between them were almost the same between the cells treated with DDX5i and the control cells (p > 0.05) (Fig. 2 A), indicating that DDX5 knockdown did not affect the virus assembly. Meanwhile, the ratios of JEV infective titers between the supernatants and the cell lysates was also the same between DDX5i treated cells and the control cells (p > 0.05) (Fig. 2B), indicating that DDX5 knockdown did not affect the virus release. Therefore, DDX5 silencing did not affect JEV assembly or release.
Fig. 2.
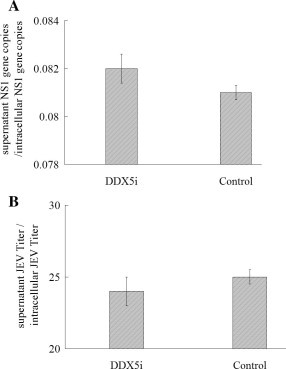
The influence of DDX5i on virus assembly or release. (A, B) The BHK-21 cells was infected with JEV (MOI = 0.01) after DDX5 silence, the ratio of JEV RNA copy numbers between the supernatants and the cell lysates was detected by Q-PCR, and the ratio of the virus titers in supernatants and cell lysates were determined by plaque assay, the untargeted shRNA was used as the control. The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
3.2. DDX5 helicase activity is important for JEV replication
To determine whether DDX5 helicase activity is important for JEV replication, DDX5 helicase-dead mutant lacking either ATPase activity (K144E) or RNA unwinding activity (S279L) (Garbelli et al., 2011, Tanner and Linder, 2001) was used. BHK-21 cells were first transfected with DDX5i plasmid to decrease the endogenous DDX5 expression level. 24 h later, the cells were then transfected with RNA interference-resistant plasmid (to exclude the influence of DDX5i plasmid): pCDNA3.1-DDX5r, pCDNA3.1-DDX5r-K144E or pCDNA3.1-DDX5r-S279L (Fig. 3 A) before the cells were infected (MOI = 0.01). The virus titers were detected 2 days later by plaque assay, the results showed that overexpression of DDX5r-K144E, DDX5r-S279L and the control plasmid pcDNA3.1 after DDX5 knock down resulted in the reduction of JEV replication for 10-fold (p < 0.01), 12-fold (p < 0.01) and 13-fold (p < 0.01). JEV replication was not significantly affected in DDX5r overexpressing cells and the control treated cells (Fig. 3B). Meanwhile, the viral RNA level was also reduced in DDX5 mutants overexpression cells which is consistent with the results of plaque assay (Fig. 3C) (p < 0.01). The expression of endogenous DDX5 or DDX5 mutants were confirmed by Western blot analysis using anti-HA monoclonal antibody and anti-DDX5 polyclonal antibody (Fig. 3D). Both of the results demonstrated that DDX5 helicase activity plays important roles in JEV replication.
Fig. 3.
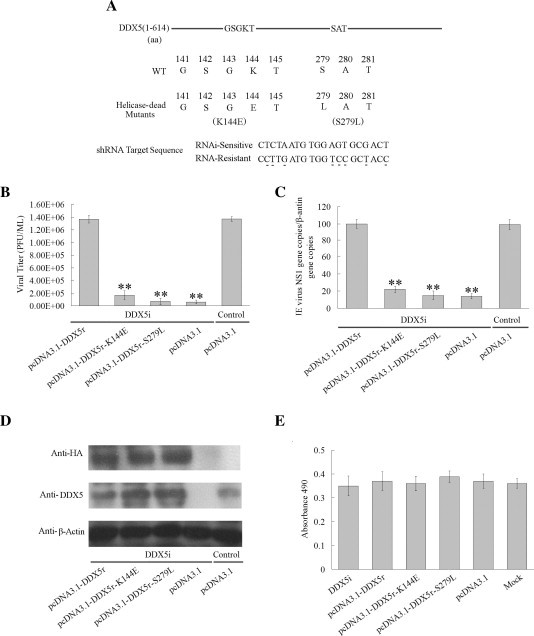
The effect of exogenous DDX5 and helicase-dead DDX5 on JEV replication. (A). The residues of DDX5 helicase-dead mutants (K144E, S279L) were shown. The sequences of DDX5 specific-shRNA and the shRNA-resistant targeting region were aligned. The different nucleotides are underlined. (B). BHK-21 cells were first transfected with DDX5i plasmid. 24 h later, the cells were transfected the pcDNA3.1-DDX5-HA, pcDNA3.1-DDX5r-K144E-HA, pcDNA3.1-DDX5r-S279L -HA or pcDNA3.1 plasmid, viral titers determined by plaque assay at 48hpi (MOI = 0.01). (C). The level of JEV genome RNA was detected by real-time PCR in the cells infected with JEV after first transfected with the DDX5i plasmid then with pcDNA3.1-DDX5r-HA, pcDNA3.1-DDX5r-K144E-HA or pcDNA3.1-DDX5r S279L-HA, the untargeted shRNA and pcDNA3.1 plasmid were used as the control. (D). The expression of transfected DDX5 mutants plasmids and the endogenous DDX5 were detected by Western blot using anti-HA monoclonal antibody and anti-DDX5 polyclonal antibody. The β-actin was used as loading control. (E). The cell viability of BHK-21 cells transfected with DDX5i, HA-DDX5 or DDX5 mutant plasmids was detected by CytoTox 96® NonRadioactive Cytotoxicity Assay (Promega) at 72 h after transfection. The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
The cell viability of all DDX5 constructs on BHK-21 cells was determined by measuring the cytotoxicity using CytoTox 96® NonRadioactive Cytotoxicity Assay Kit. Our result demonstrated that the viability of BHK-21 cells was not significantly affected (Fig. 3E).
3.3. The interactions between DDX5 and viral proteins and the interaction interface mapping with competition binding experiment
We next determined whether DDX5 is involved in JEV replication by interacting with JEV proteins related to viral RNA replication. It has been shown that JEV core, NS3 and NS5 are involved in the JEV replication (Chen et al., 1997, Uchil and Satchidanandam, 2003a). In flavivirus, NS5 can be separated into two domains: methyltransferase (MTase) domain and RNA-dependent RNA polymerase (RdRp) domain (Ackermann and Padmanabhan, 2001, Egloff et al., 2002, Guyatt et al., 2001, Koonin, 1993). During JEV infection, the NS5 protein is not cleaved into MTase and RdRp (Yu et al., 2007). In our study, we divided NS5 into two domains (MTase and RdRp domains) to detect their interactions with DDX5 in details. We used GST-pulldown assay to monitor the interactions between JEV core, NS3, NS5-MTase or NS5-RdRp and DDX5 or vice versa. The results showed GST-DDX5 can interact with His-tagged JEV core, NS3, NS5-Mtase and NS5-RdRp, while GST protein can not (Fig. 4 A). GST-core, GST-NS3, GST-NS5-MTase and GST-NS5-RdRp can interact with His-tagged DDX5 (Fig. 4B).
Fig. 4.
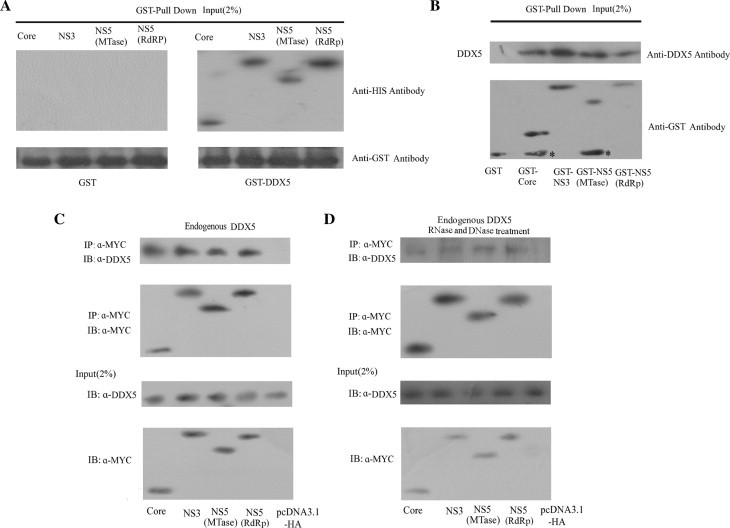
DDX5 interacts with JEV core, NS3, NS5-MTase and NS5-RdRp proteins. GST-pull down assay of the interaction between DDX5 and JEV core, NS3, NS5-MTase or NS5-RdRp proteins. (A). In vitro interaction analysis of DDX5 with JEV core, NS3, NS5-MTase and NS5-RdRp using GST-pull down experiment. GST fused DDX5, was incubated with GST beads, then with His-tagged JEV core, NS3, NS5-MTase and NS5-RdRp, and then detected by Western blot analysis using anti-His and GST antibodies. The GST protein was used as the control. JEV core, NS3, NS5-MTase and NS5-RdRp interact with DDX5. (B). In vitro interaction analysis of JEV core, NS3, NS5-MTase and NS5-RdRp with DDX5 using GST-pull down experiment. GST fused core, NS3, NS5-MTase and NS5-RdRp were incubated with GST beads, then with His-tagged DDX5, and then detected by western blot analysis using anti-His and GST antibodies. The GST protein was used as the control. (C). DDX5 interacts with JEV core, NS3, NS5-MTase and NS5-RdRp in BHK-21 cells. BHK-21 cells were transfected with the different viral protein constructs tagged with MYC epitope. MYC-tagged viral protein was used to immunoprecipitate the endogenous DDX5 protein from the cell lysates. The immunoprecipitated proteins were detected with anti-DDX5 polyclonal antibody and anti-MYC-HRP monoclonal antibody. (D). The interaction of DDX5 with JEV core, NS3, NS5-MTase and NS5-RdRp in BHK-21 cells is not mediated by RNA or DNA. BHK-21 cells were transfected with different MYC-tagged viral protein plasmid, MYC-tagged viral protein was used to immunoprecipitate the endogenous DDX5 protein from the cell lysates treated with RNase (100 μg/ml) and DNase (100 μg/ml). The cell lysates (input, 2%) were analyzed in parallel by Western blot. Asterisks represent the nonspecific recognition.
We next performed co-immunoprecipitation assay to confirm these interactions. The cells were transfected with either JEV core, NS3, NS5-MTase, or NS5-RdRp plasmids tagged with MYC epitope. The MYC-tagged proteins were immunoprecipitated with anti-MYC monoclonal antibody conjugated beads and the coimmunoprecipitated endogenous DDX5 were detected in Western blot analysis using anti-DDX5 polyclonal antibody. The results showed JEV core, NS3, NS5-MTase, or NS5-RdRp could interact with endogenous DDX5 (Fig. 4C). Since DDX5 is a nucleic acid-binding protein, it is possible that these interactions were mediated by RNA or DNA. To address this issue, we next examined the influence of RNA or DNA on the interaction between JEV core, NS3, NS5-MTase, NS5-RdRp and DDX5. The co-immunoprecipitation was performed following RNase and DNase treatment. The results showed JEV core, NS3, NS5-MTase, NS5-RdRp and DDX5 interaction was also detected after RNase (100 μg/ml) and DNase (100 μg/ml) treatment, suggesting that the interaction between JEV proteins and DDX5 were not mediated by RNA or DNA(Fig. 4D).
Because DDX5 can interact with JEV core, NS3, NS5-MTase, or NS5-RdRp proteins, we next tried to determine the interaction interface between JEV core, NS3, and NS5 (MTase and RdRp domains) proteins and DDX5 using competition binding experiment. The glutathione-Sparse 4B beads were first incubated with the GST-DDX5. After unbound GST-DDX5 protein was washed away, JEV core, NS3 or NS5 (MTase or RdRp domains) was mixed with the beads. The various amount of other viral protein was further incubated with the beads to compete the bound viral protein from DDX5. The results showed the JEV core protein bound with DDX5 decreased gradually when the mixture was incubated with increasing amount of NS3. Meanwhile, the amount of core protein in the supernatant increased (Fig. 5 A). The similar results were observed when use NS5-RdRp to compete the core protein (Fig. 5B), NS5-MTase could not compete with core protein (Fig. 5C). Furthermore, NS3 could be pulled down from DDX5 by NS5-MTase protein (Fig. 5D), but not by NS5-RdRp (Fig. 5E). NS5-MTase could not be pulled down from DDX5 by NS5-RdRp (Fig. 5F). The analysis of the proteins in the supernatant was consistent with the results on the beads. Therefore, the results suggested that interaction site of core/NS3, core/NS5-RdRp or NS3/NS5-MTase on DDX5 might be close to each other, while the interaction site of NS5-MTase/core, NS3/NS5-RdRp or NS5-MTase/NS5-RdRp on DDX5 is not. Meanwhile, all the purified recombinant proteins were analyzed by SDS–PAGE (Fig. S1).
Fig. 5.
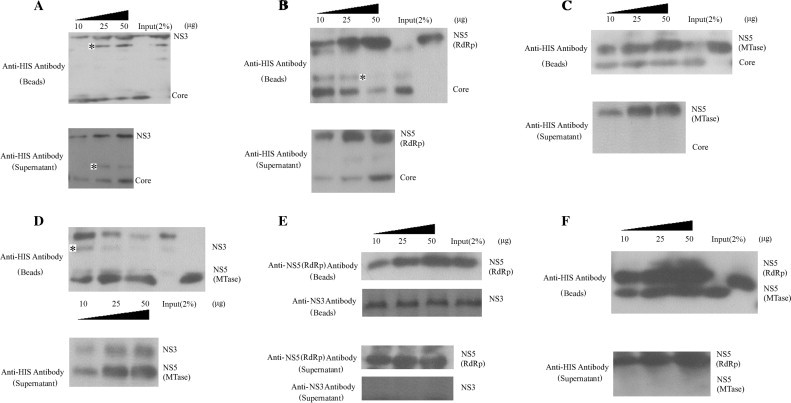
The interaction interface mapping between JEV core, NS3, and NS5 (MTase and RdRp domains) and DDX5 proteins. GST–DDX5 protein was first incubated with 50 μL of glutathione-Sparse 4B beads. After extensive washing, HIS-tagged core, NS3, NS5-Mtase or NS5-RdRp was incubated with the beads for 3–4 h with gentle agitation followed by washing 5 times with washing buffer. The different amount of competition protein (core, NS3, NS5-Mtase or NS5-RdRp) (10 μg-50 μg) was mixed with the beads to determine the interaction interface of NS3/core (A), NS5-RdRp/core (B), NS5-MTase/core (C), NS5-MTase/NS3 (D), NS5-RdRp/NS3 (E) and NS5-RdRp/NS5-MTase (F). The mixture was incubated for 3–4 h with gentle agitation. The mixture was centrifuged at 3000 rpm for 5 min, the supernatant and the beads were subjected to 12% or 15% SDS–PAGE gel and analyzed by western blot using either anti-HIS monoclonal antibody, anti-NS3 or NS5 polyclonal antibody. Asterisks represent the nonspecific recognition.
3.4. DDX5 was recruited from the nucleus to the cytoplasm during virus infection
As previously reported, DDX5 is primarily localized in the nucleus (Choi and Lee, 2012). We detected the cellular localization of endogenous DDX5 in BHK-21 cells infected with JEV. Confocal microscopy analysis demonstrated endogenous DDX5 primarily localized in the nucleus in the uninfected cells, while some of DDX5 was recruited to the cytoplasm in JEV infected cells (MOI = 0.01), suggesting DDX5 could be recruited to the cytoplasm to participate in JEV replication (Fig. 6 A). To investigate if the recruitment of DDX5 to the cytoplasm is necessary for JEV replication, we used leptomycin B (LMB, 5 ng/ml) to inhibit nuclear export of DDX5 (Wang et al., 2009a) at 0 h post-infection (MOI = 0.01), 24 h later, Confocal microscopy analysis showed that DDX5 was not recruited to the cytoplasm during JEV infection after LMB treatment (Fig. 6A). In the meantime, we determined the effect of LMB treatment on JEV infection, 5 ng/ml and 10 ng/ml LMB was added to the cell at 0 h post-infection, the virus titers was detected 2 days later by plaque assay. The results showed that LMB can significant inhibit JEV infection compared with the untreated cells (Fig. 6B), confirming that it is need to recruit DDX5 to the cytoplasm to participate in JEV replication. Cytotoxicity assay showed that LMB can not induce significantly cytotoxicity on BHK-21 cells even at the concentration of 20 ng/ml (Fig. 6C).
Fig. 6.
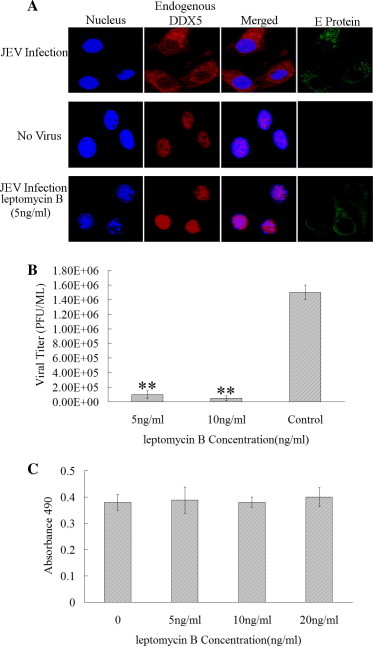
The localization of DDX5 in JEV infected BHK-21 cells. (A). Localization of endogenous DDX5 in BHK-21 cell infected, mock infected with JEV or LMB treated was detected using anti-DDX5 polyclonal antibody, the virus enveloped (E) protein was detected using anti-E monoclonal antibody. (B). LMB can inhibit JEV infection. BHK-21 cell was infected with JEV (MOI = 0.01), then LMB (5 ng/ml) was added, the viral titers were detected by plaque assay at 48 hpi, the untreated cells were used as the control. (C). The Cytotoxicity of Leptomycin B (LMB) on BHK-21 cells was detected by CytoTox 96® NonRadioactive Cytotoxicity Assay at 48 h after added to the cells with different the concentrations (0–20 ng/ml). The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
3.5. DDX5 interacts with the JEV core, NS3, NS5 proteins and colocalizes with them during virus infection
NS5 and NS3 have been hypothesized to seed the formation of virus replication complexes, and they may form replication complex together with 3′ noncoding region of JEV genomic RNA (Uchil and Satchidanandam, 2003a) and the core protein can bind to the viral RNA to participate in virus replication (Khromykh and Westaway, 1996). Since GST-pulldown and co-immunoprecipitation experiments showed that DDX5 can interact with the JEV core, NS3, NS5-MTase and NS5-RdRp proteins. We further determined if DDX5 can interact with these proteins during virus infection. The BHK-21 cells were infected with JEV (MOI = 0.01). 24 h later, the anti-DDX5 polyclonal antibody was used to co-immunoprecipitate proteins from the whole cell lysate which was also treated with RNase (100 μg/ml) and DNase (100 μg/ml). The results showed JEV core, NS3, and NS5 proteins can be co-immunoprecipitated by DDX5 compared with the control IgG (Fig. 7 A), confirming that DDX5 interacts with viral proteins that are important for virus replication and the interaction between them was not mediated by RNA or DNA. Furthermore, confocal microscopy analysis demonstrated that DDX5 colocalized with JEV core, NS3 and NS5 proteins mainly in the cytoplasm during virus infection. In uninfected cells, most of DDX5 still localized in the nucleus, the serum of the unimmunized mouse was used as the control (Fig. 7B).
Fig. 7.
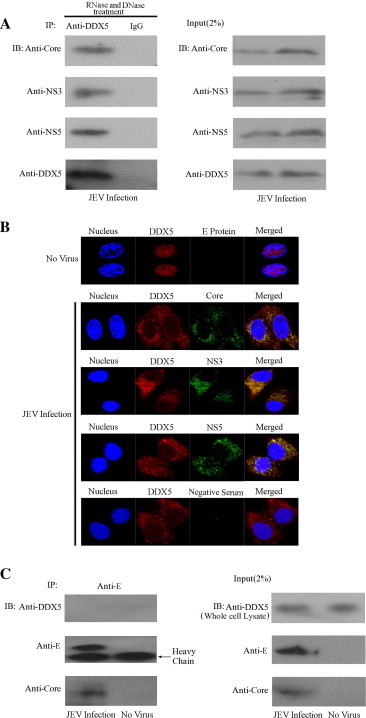
DDX5 interacts with the JEV core, NS3, NS5 proteins and colocalizes with them during virus infection. (A). DDX5 can interact with JEV core, NS3, NS5 proteins. BHK-21 cells was infected with JEV (MOI = 0.01). 48 h later, the anti-DDX5 polyclonal antibody was used to co-immunoprecipitate the proteins from the whole cell lysate treated with RNase (100 μg/ml) and DNase (100 μg/ml). The JEV core, NS3 and NS5 proteins were detected using their polyclonal antibodies respectively. The cell lysates (input, 2%) were analyzed in parallel by western blot. (B). DDX5 colocalizes with the JEV core, NS3, NS5 proteins. We infected the BHK-21 cells with JEV (MOI = 0.01) first. 48 h later, DDX5, JEV core, NS3 and NS5 proteins were detected using their polyclonal antibodies and the colocalization study was performed under confocal microscopy, the negative serum (the serum of unimmunized mouse) was used as the control. (C). DDX5 is not incorporated into the virions. BHK-21 cells were infected with JEV (MOI = 0.01) for 48 h, the anti-E monoclonal antibody was used to immunoprecipitate the mature virions in the medium, the proteins in the immunoprecipitated virions (IP) were detected with anti-DDX5, E or core antibodies. All cell lysates (input, 2%) were analyzed in parallel by western blot. The heavy chain of anti-E monoclonal antibody was labeled.
It has been shown DDX3, another cellular helicase, can be incorporation into the virons of HBV, Herpes simplex virus type 1(HSV-1) and Human cytomegalovirus (HCMV) (Stegen et al., 2013, Varnum et al., 2004, Wang et al., 2009b). We also determined whether DDX5 is also incorporated into JEV virions. We infected the BHK-21 cells with JEV (MOI = 0.01) for 48 h, the anti-EDIII monoclonal antibody was used to co-immunoprecipitate the mature virions in the medium, and the Western blot analysis showed that DDX5 is not incorporated into the JEV virions (Fig. 7C).
3.6. DDX5 can bind to JEV 3′ UTR and colocalize with the viral RNA
It has been shown that NS3 and NS5 of JEV can bind to 3′-UTR of viral RNA and engage in virus replication (Chen et al., 1997). We also determined whether DDX5 can bind to JEV 5′ or/and 3′-UTR. We synthesized JEV-5′, 3′ UTR and control RNA in vitro, and incubated RNAs with the Tobramycin Affinity Tag beads, the cell lysate was then incubated with the beads. The Western blot results showed only JEV 3′ UTR can pull down endogenous DDX5, while the control RNA can not (Fig. 8 A).
Fig. 8.
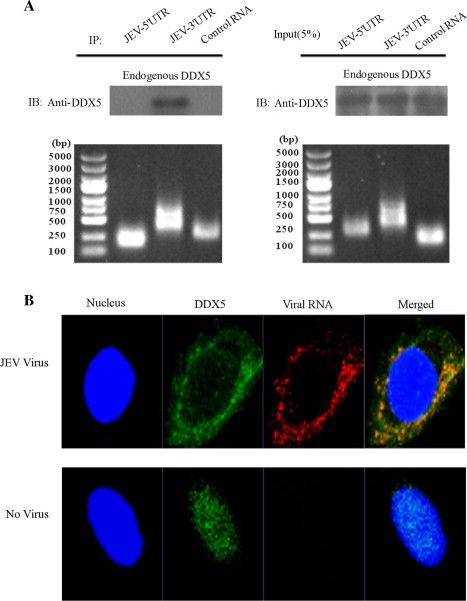
DDX5 can bind to JEV 5′, 3′UTR and colocalizes with viral RNA. (A). DDX5 can bind to JEV 3′ UTR. JEV 5′, 3′ UTR and control RNA with Tobramycin Affinity Tag were first transcripted in vitro, and then incubated with tobramycin conjugated matrix beads. The mixture was then incubated with BHK-21 cell lysate, the protein bounded on beads was detected by Western blot using anti-DDX5 polyclonal antibody. All the RNAs were also analyzed by 1% agarose, 5% input were also analyzed in parallel by western blot. (B). DDX5 colocalizes with viral RNA. BHK-21 cells was infected with JEV (MOI = 0.01), endogenous DDX5 was detected by anti-DDX5 polyclonal antibody and the viral RNA was detected (red) using the Click-iT® RNA Imaging Kit at 12hpi. The mock-infected cells were also included as a control.
Meanwhile, we also found that DDX5 colocalized with the viral RNA by confocal microscopy analysis at 12hpi (Fig. 8B), demonstrating DDX5 may be involved in JEV RNA synthesis.
3.7. DDX5 is a positive regulator for viral RNA synthesis but not for viral protein translation
Since 5′ and 3′ UTR of JEV are both involved in the first round of viral protein translation and virus RNA replication, we used a JEV replicon reporter system to determine whether DDX5 modulated viral translation and/or RNA replication. We constructed a luciferase-containing JEV replicon according to the previous study (Chien et al., 2011). BHK-21 cells were first transfected with DDX5i shRNA plasmid to knockdown the expression of DDX5, the non-targeting shRNA was used as the control. The cells were then transfected with in vitro-transcribed luciferase-containing JEV-replicon RNA and firefly luciferase RNA as the internal control. The cells were harvested for dual luciferase assays at 3, 6, 12, 24, 48 and 72 h post transfection. The results showed that there were two peaks of replicon luciferase activity corresponding to the first translation and the second replication peak (Fig. 9 A). The luciferase activities were nearly the same in the DDX5i and control treated BHK-21 cells in the first peak, while luciferase activities were lower for all of the time points in the DDX5i treated BHK-21 cells in the second peak. The data showed that viral RNA replication, but not the viral protein translation, was hampered by DDX5 knockdown. Meanwhile, the luciferase activity derived from a control reporter, Renilla luciferase flanked by cellular GAPDH 5′ and 3′ UTRs did not significantly changed between the shDDX5 and control treated cells (Fig. 9B), which means that the translation of cellular protein was not hampered by DDX5 knockdown. Collectively, our results strongly suggested that DDX5 is involved in viral RNA replication by binding to JEV 3′ UTR.
Fig. 9.
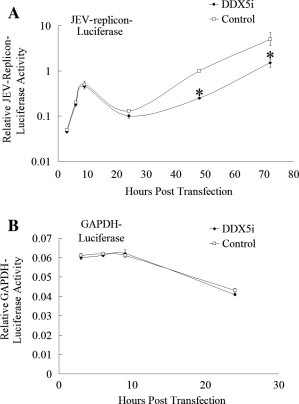
DDX5 positively regulates JEV RNA replication. BHK-21 cells were co-transfected with either JEV replicon. (A) or a Renilla luciferase reporter gene flanked by GAPDH 5′ and 3′ UTRs (B). The firefly luciferase reporter was used as the internal control. At various times post transfection, cell lysates were collected for dual-luciferase assays. Renilla luciferase activity was normalized to that of firefly luciferase activity. The differences between means were considered significant at ∗p < 0.05, very significant ∗∗p < 0.01.
4. Discussion
The high prevalence of disease caused by JEV and the limited efficacy of therapies stimulate the research for safer and more effective antiviral agents to treat JEV infection. However, viral resistance to antiviral agents has been a problem. One possible solution to overcome this problem is to target host cellular factors important for the viral life cycle. DDX5 plays important roles in many virus, such as SARS-CoV, influenza virus and HCV (Chen et al., 2009, Goh et al., 2004), and other cellular helicases such as DDX3, DDX1, DDX56 are also involved in many virus replication and the assembly of virions (Xu et al., 2011, Xu and Hobman, 2012). Two studies reported novel DDX3 inhibitors against HIV infection (Maga et al., 2011, Radi et al., 2012). Therefore, the cellular helicases have been proposed as a promising target to design novel agents to control viral infection that poses major global health threats (Geiss et al., 2009, Kwong et al., 2005, Stankiewicz-Drogoń et al., 2010).
In our studies, we demonstrated that a reduction of endogenous DDX5 and DDX5 helicase-dead mutants all significantly reduced the viral load and synthesis of viral RNA, suggesting DDX5 and its helicase activity are required for JEV replication. Our data also showed that DDX5 does not affect virus assembly and release or incorporate into the virions, implying its minimal influence on virus maturation.
It has been shown JEV core, NS3 and NS5 proteins play important roles in viral genome replication (Chen et al., 1997, Uchil and Satchidanandam, 2003a). By using GST-pulldown and co-immunoprecipitation, we demonstrated that DDX5 can interact with JEV core, NS3, NS5-MTase and NS5-RdRp, suggesting that DDX5 might act as a cofactor for JEV proteins in virus replication. It has been suggested that cellular proteins can bind to viral 5′ and/or 3′ UTR to modulate viral translation and/or replication (Chen et al., 1997). Our studies showed that DDX5 can only interact with viral 3′ UTR and colocalize with viral RNA. We further confirmed that DDX5 plays an important role in virus RNA replication but not in viral protein translation, which is consistent with its ability to bind to viral 3′ UTR. We performed the competition binding studies to map the interaction interfaces between DDX5 and viral proteins. The viral RNA replication is a quite dynamic process, and other host factors are also recruited by the viral proteins to facilitate this process (Lindenbach et al., 2005, Uchil and Satchidanandam, 2003a). We suspected that the interaction interfaces shown in our competition binding studies might not always exist during viral RNA replication.
Some negative-stranded RNA viruses use the host cytoplasm as the primary site of replication, they can also recruit nuclear factors and alter their nuclear-cytoplasm trafficking to enhance viral replication (Hiscox, 2003). The host nucleus proteins could be recruited to cytoplasm to facilitate flaviviruses replication (Uchil and Satchidanandam, 2003b, Zebovitz et al., 1974). It has been reported that DDX5 is redistributed into the cytoplasm during HCV infection (Goh et al., 2004). Consistent with this, our study showed that DDX5 might be recruited to the cytoplasm and colocalize with the JEV core, NS3 and NS5 and viral RNA during the infection to facilitate the unwinding the intermediate RNA duplexes. Since LMB can inhibit all the CRM1-mediated export of proteins from the nucleus to the cytoplasm (Wang et al., 2009a), we cannot rule out the trafficking of other cellular proteins inhibited by LMB which might also be important for virus replication. Our result showed that virus can still replicate at low titers after LMB treatment. Considering that DDX5 shuttles between nucleus and cytoplasma, we speculated the small amount of DDX5 resides in cytoplasma after LMB treatment still exerts its role in virus replication. Meanwhile, it is also possible that DDX5 is only used to increase the efficiency of RNA separation so that the viral RdRp can transcribe viral RNA more efficiently during virus replication.
In summary, our studies identified that DDX5 as a positive regulator for JEV replication, which will be useful for further understanding of cellular proteins involvement during JEV replication. Our results also suggest that DDX5 could be served as a novel targets to design small inhibitors against Japanese encephalitis virus infection.
Acknowledgments
We thank Prof. Ralf Janknecht for plasmid HA-DDX5. This project was funded by the priority academic program development of Jiangsu Higher Education Institutions.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.antiviral.2013.09.002.
Appendix A. Supplementary data
Supplementary Figure S1.
Reference
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Bortz E., Westera L., Maamary J., Steel J., Albrecht R.A., Manicassamy B., Chase G., Martínez-Sobrido L., Schwemmle M., García-Sastre A. Host-and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteinsmBio. 2. 2011:e00151–00111. doi: 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahar H.S., Chen S., Manjunath N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology. 2013;436:1–7. doi: 10.1016/j.virol.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Kuo M.D., Chien L.J., Hsu S.L., Wang Y.M., Lin J.H. RNA-protein interactions: involvement of NS3, NS5, and 3′noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.Y., Chen W.N., Poon K.M.V., Zheng B.J., Lin X., Wang Y.X., Wen Y.M. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch. Virol. 2009;154:507–512. doi: 10.1007/s00705-009-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien H.L., Liao C.L., Lin Y.L. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J. Virol. 2011;85:4698–4706. doi: 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.J., Lee S.G. The DEAD-box RNA helicase DDX3 interacts with DDX5, co-localizes with it in the cytoplasm during the G2/M phase of the cycle, and affects its shuttling during mRNP export. J. Cell. Biochem. 2012;113:985–996. doi: 10.1002/jcb.23428. [DOI] [PubMed] [Google Scholar]
- Clark E.L., Coulson A., Dalgliesh C., Rajan P., Nicol S.M., Fleming S., Heer R., Gaughan L., Leung H.Y., Elliott D.J. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′–O–)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh H., Maruyama K., Masuhiro Y., Kobayashi Y., Goto M., Tai H., Yanagisawa J., Metzger D., Hashimoto S., Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor α. Mol. Cell. Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fang J., Acheampong E., Dave R., Wang F., Mukhtar M., Pomerantz R.J. The RNA helicase DDX1 is involved in restricted HIV-1 Rev function in human astrocytes. Virology. 2005;336:299–307. doi: 10.1016/j.virol.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Fang J., Kubota S., Yang B., Zhou N., Zhang H., Godbout R., Pomerantz R.J. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Fuller-pace F.V., Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem. Soc. Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- Garbelli A., Beermann S., Di Cicco G., Dietrich U., Maga G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS ONE. 2011;6:e19810. doi: 10.1371/journal.pone.0019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss B.J., Stahla H., Hannah A.M., Gari H.H., Keenan S.M. Focus on flaviviruses: current and future drug targets. Future. 2009;1:327–344. doi: 10.4155/fmc.09.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.Y., Tan Y.J., Lim S.P., Tan Y., Lim S.G., Fuller-Pace F., Hong W. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 2004;78:5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer A.E., Hearing P., Ketner G. The adenovirus E4 11 k protein binds and relocalizes the cytoplasmic P-body component Ddx6 to aggresomes. Virology. 2011;417:161–168. doi: 10.1016/j.virol.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson E.A., Wessel G.M. DEAD-box helicases: posttranslational regulation and function. Biochem. Biophys. Res. Commun. 2010;395:1–6. doi: 10.1016/j.bbrc.2010.02.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt K.J., Westaway E.G., Khromykh A.A. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus kunjin. J. Virol. Methods. 2001;92:37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Hanna J.N., Ritchie S.A., Phillips D.A., Shield J., Bailey M.C., Mackenzie J.S., Poidinger M., McCall B.J., Mills P.J. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med. J. Aust. 1996;165:256–261. doi: 10.5694/j.1326-5377.1996.tb124960.x. [DOI] [PubMed] [Google Scholar]
- Harris, E., Holden, K.L., Edgil, D., Polacek, C., Clyde, K., 2006. Molecular biology of flaviviruses. Novartis Found Symp 277, 23–39; discussion 40, 71–23, 251–253. [PubMed]
- Hartmuth K., Vornlocher H.P., Luhrmann R. Tobramycin affinity tag purification of spliceosomes. Methods Mol. Biol. 2004;257:47–64. doi: 10.1385/1-59259-750-5:047. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 2003;95:13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishag H.Z.A., Li C., Huang L., Sun M., Ni B., Guo C., Mao X. Inhibition of Japanese encephalitis virus infection in vitro and in vivo by pokeweed antiviral protein. Virus Res. 2012;171:89–96. doi: 10.1016/j.virusres.2012.10.032. [DOI] [PubMed] [Google Scholar]
- Ishag H.Z.A., Li C., Huang L., Sun M., Wang F., Ni B., Malik T., Chen P., Mao X. Griffithsin inhibits Japanese encephalitis virus infection in vitro and in vivo. Arch. Virol. 2012;158:349–358. doi: 10.1007/s00705-012-1489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra R.K., Yi M., Lemon S.M. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J. Virol. 2010;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A., Westaway E. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 1996;141:685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Krishnan M.N., Ng A., Sukumaran B., Gilfoy F.D., Uchil P.D., Sultana H., Brass A.L., Adametz R., Tsui M., Qian F. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong A.D., Rao B.G., Jeang K.T. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Evans M.J., Syder A.J., Wölk B., Tellinghuisen T.L., Liu C.C., Maruyama T., Hynes R.O., Burton D.R., McKeating J.A. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Gubler D.J., Petersen L.R. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Maga G., Falchi F., Radi M., Botta L., Casaluce G., Bernardini M., Irannejad H., Manetti F., Garbelli A., Samuele A., Zanoli S., Este J.A., Gonzalez E., Zucca E., Paolucci S., Baldanti F., De Rijck J., Debyser Z., Botta M. Toward the discovery of novel anti-HIV drugs. Second-generation inhibitors of the cellular ATPase DDX3 with improved anti-HIV activity: synthesis, structure-activity relationship analysis, cytotoxicity studies, and target validation. ChemMedChem. 2011;6:1371–1389. doi: 10.1002/cmdc.201100166. [DOI] [PubMed] [Google Scholar]
- Radi M., Falchi F., Garbelli A., Samuele A., Bernardo V., Paolucci S., Baldanti F., Schenone S., Manetti F., Maga G., Botta M. Discovery of the first small molecule inhibitor of human DDX3 specifically designed to target the RNA binding site: towards the next generation HIV-1 inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2094–2098. doi: 10.1016/j.bmcl.2011.12.135. [DOI] [PubMed] [Google Scholar]
- Reed J.C., Molter B., Geary C.D., McNevin J., McElrath J., Giri S., Klein K.C., Lingappa J.R. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T. Recent advances in Japanese encephalitis. J. Neurovirol. 2003;9:274–283. doi: 10.1080/13550280390194037. [DOI] [PubMed] [Google Scholar]
- Stankiewicz-Drogoń A., Dörner B., Erker T., Boguszewska-Chachulska A.M. Synthesis of new acridone derivatives, inhibitors of NS3 helicase, which efficiently and specifically inhibit subgenomic HCV replication. J. Med. Chem. 2010;53:3117–3126. doi: 10.1021/jm901741p. [DOI] [PubMed] [Google Scholar]
- Stegen C., Yakova Y., Henaff D., Nadjar J., Duron J., Lippe R. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS ONE. 2013;8:e53276. doi: 10.1371/journal.pone.0053276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer L., Klostermeier D. RNA helicases in infection and disease. RNA Biol. 2012;9:751–771. doi: 10.4161/rna.20090. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. DExD/H Box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tsai T.F. New initiatives for the control of Japanese encephalitis by vaccination: Minutes of a WHO/CVI meeting, Bangkok, Thailand, 13–15 October 1998. Vaccine. 2000;18:1–25. doi: 10.1016/s0264-410x(00)00037-2. [DOI] [PubMed] [Google Scholar]
- Uchil P.D., Satchidanandam V. Architecture of the flaviviral replication complex protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Uchil P.D., Satchidanandam V. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology. 2003;307:358–371. doi: 10.1016/s0042-6822(02)00130-7. [DOI] [PubMed] [Google Scholar]
- Varnavski A.N., Khromykh A.A. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology. 1999;255:366–375. doi: 10.1006/viro.1998.9564. [DOI] [PubMed] [Google Scholar]
- Varnum S.M., Streblow D.N., Monroe M.E., Smith P., Auberry K.J., Pasa-Tolic L., Wang D., Camp D.G., 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R.D., Nelson J.A. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Urena L., Chaudhry Y., Goodfellow I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J. Virol. 2012;86:11977–11990. doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Gao X., Huang Y., Yang J., Liu Z.R. P68 RNA helicase is a nucleocytoplasmic shuttling protein. Cell Res. 2009;19:1388–1400. doi: 10.1038/cr.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Kim S., Ryu W.S. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 2009;83:5815–5824. doi: 10.1128/JVI.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H., Fu S.H., Wang H.Y., Liang X.F., Cheng J.X., Jing H.M., Cai G.L., Li X.W., Ze W.Y., Lv X.J. Japanese encephalitis outbreak, Yuncheng, China, 2006. Emerg. Infect. Dis. 2007;13:1123. doi: 10.3201/eid1307.070010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.M., Bidet K., Yinglin A., Ler S.G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M.A. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Khadijah S., Fang S., Wang L., Tay F.P.L., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Anderson R., Hobman T.C. The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J. Virol. 2011;85:5571–5580. doi: 10.1128/JVI.01933-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Hobman T.C. The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology. 2012;433:226–235. doi: 10.1016/j.virol.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Hasebe F., Inoue S., Mathenge E., Morita K. Identification and characterization of RNA-dependent RNA polymerase activity in recombinant Japanese encephalitis virus NS5 protein. Arch. Virol. 2007;152:1859–1869. doi: 10.1007/s00705-007-1007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.F., Lujan P., Jackson D.L., Emerman M., Linial M.L. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011;7:e1002303. doi: 10.1371/journal.ppat.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebovitz E., Leong J., Doughty S. Involvement of the host cell nuclear envelope membranes in the replication of Japanese encephalitis virus. Infect. Immun. 1974;10:204–211. doi: 10.1128/iai.10.1.204-211.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Luo J., Mills L., Wu S., Pan T., Geng G., Zhang J., Luo H., Liu C., Zhang H. DDX5 Facilitates HIV-1 Replication as a Cellular Co-Factor of Rev. PLoS ONE. 2013;8:e65040. doi: 10.1371/journal.pone.0065040. [DOI] [PMC free article] [PubMed] [Google Scholar]


