Abstract
There is currently no approved antiviral therapy for treatment of Ebola virus disease. To discover readily available approved drugs that can be rapidly repurposed for treatment of Ebola virus infections, we screened 1280 FDA-approved drugs and identified glycopeptide antibiotic teicoplanin inhibiting Ebola pseudovirus infection by blocking virus entry in the low micromolar range. Teicoplanin could be evaluated further and incorporated into ongoing clinical studies.
Keywords: Ebola, Pseudovirus, Teicoplanin, High-throughput screening, Antiviral drug discovery
Highlights
-
•
We developed a high-throughput cell-based screening assay using Ebola pseudovirus.
-
•
We identified teicoplanin inhibiting Ebola pseudovirus infection.
-
•
Teicoplanin blocked virus entry but did not inhibit replication.
-
•
Teicoplanin inhibits enveloped viruses but not nonenveloped viruses.
Ebola virus (EBOV), a member of the Filoviridae, is an enveloped, filamentous, non-segmented negative-sense RNA virus that can cause deadly Ebola virus disease (EVD) (Feldmann and Geisbert, 2011). In February 2014, the largest known EVD outbreak started in Guinea, and virologic investigation identified Zaire ebola virus (ZEBOV) as the causative agent (Baize et al., 2014). As of 26 July 2015, a total of 27,784 cases and 11,294 deaths were reported (http://www.who.int/csr/disease/ebola/situation-reports/en/), resulting in a fatality rate of 40.6%. To date, no antiviral or therapeutic has been approved for treating patients with EVD, and treatment remains limited to supportive care. Therefore there is an urgent need for the discovery and development of antiviral agents against EBOV infection.
EBOV is a biosafety level 4 (BSL-4) pathogen and work with infectious EBOV is restricted to only a few BSL-4 laboratories. Hence the biology of EBOV infection remains relatively poorly understood hampering vaccine and drug development. In order to overcome this limitation, surrogate systems which allow modeling of the virus life cycle under BSL-2 conditions have been developed (Hoenen and Feldmann, 2014). Pseudoparticles expressing EBOV glycoprotein (GP) is the most commonly used tool for the study of EBOV entry and identification of EBOV entry inhibitors. Virus attachment and entry offer numerous targets for antiviral therapy and T20 (enfuvirtide), a peptide inhibitor of gp41-mediated virus entry has been successfully used in the treatment of HIV-1 infection (Altmeyer, 2004). We set out to screen approved drugs for identification of potential therapeutic options for EVD. Drug repurposing is a valid approach, and several existing drugs have been proven to be effective in the new indications (Ashburn and Thor, 2004, Chong and Sullivan, 2007).
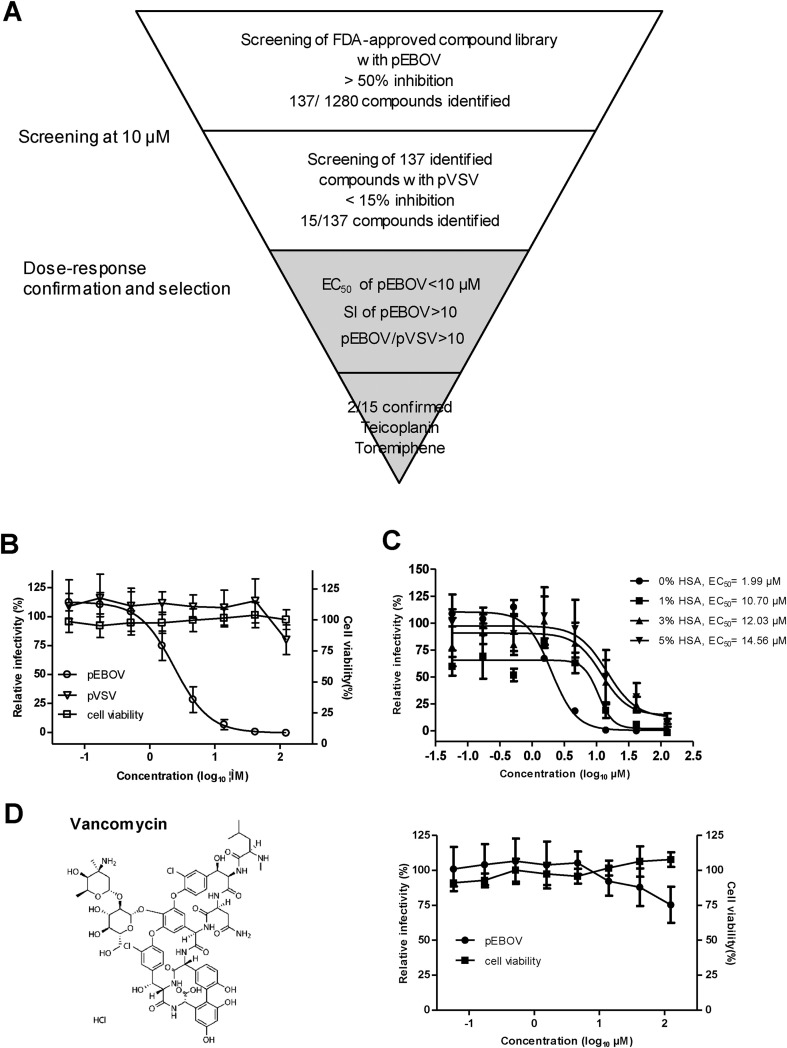
We performed a screen of 1280 FDA-approved drugs using EBOV (Zaire strain) GP/HIV core pseudovirus containing firefly luciferase (FLuc) reporter gene (designated as pEBOV, kindly provided by Prof. Paul Zhou from Institut Pasteur of Shanghai) to identify new inhibitors. Fig. 1 A shows the scheme of primary screening and hits selection process. Briefly, Vero cells (10,000 cells in 50 μl of DMEM) were seeded into each well of a white 96-well plate (Corning Costar) and incubated at 37 °C with 5% CO2 for 24 h prior to infection. Five microliters of each test compound at a final concentration of 10 μM (diluted in assay media with a final DMSO concentration of 0.25%) were added to the plates (one compound per well). In cell control and pEBOV infection control wells, 0.25% DMSO alone was added. Within 10 min of compound addition, 45 μl of 1:20-diluted pEBOV was added to each well. In cell control wells, 45 μl of assay medium was added. The final assay volume was 100 μl/well. Plates were then incubated at 37 °C with 5% CO2 for 72 h and allowed to equilibrate to room temperature for 30 min. Afterward, 50 μl of Bright-Glo (Promega) reagent was added to each plate well, and the plates were incubated at room temperature for 5 min before being read by Veritas Microplate Luminometer (Turner BioSystems). The Z factors of screening plates were in the range of 0.67–0.98 with an average of 0.83, indicative of robust assay performance (Zhang et al., 1999). The average signal-to-background (S/B) ratio and signal-to-noise (S/N) ratio was 239.1 and 29.9, respectively. A total of 137 compounds with greater than 50% inhibition of pEBOV infection were identified for secondary screening against VSV G/HIV core pseudovirus containing FLuc reporter gene (designated as pVSV, also provided by Prof. Paul Zhou) with the same assay condition. 15 out of 137 compounds with less than 15% inhibition of pVSV infection were considered as primary hits, yielding a hit rate of 1.17%. These compounds include several previously reported inhibitors such as estrogen receptor modulators tamoxifen (Johansen et al., 2013, Kouznetsova et al., 2014), clomiphene (Johansen et al., 2013), and toremiphene (Johansen et al., 2013, Kouznetsova et al., 2014, Madrid et al., 2013), histamine antagonist clemastine (Kouznetsova et al., 2014) (Johansen et al., 2015), anti-depressant paroxetine (Johansen et al., 2015, Madrid et al., 2013), ion channel inhibitor amiodarone (Gehring et al., 2014, Salata et al., 2015), and glycopeptide antibiotic teicoplanin (Johansen et al., 2015).
Fig. 1.
Identification of teicoplanin as an inhibitor of EBOV pseudovirus (pEBOV). (A) Flowchart of screening procedure. 1280 compounds from the FDA-approved compound library were screened in single dose at 10 μM for activity against pEBOV. 137 compounds that had activity (>50% inhibition) against pEBOV were subsequently screened against VSV pseudovirus (pVSV), leading to 15 compounds that were selectively active against pEBOV. Dose-response analysis confirmed that two compounds (teicoplanin and toremiphene) met the selection criteria of EC50 < 10 μM, SI > 10 against pEBOV, and pEBOV/pVSV > 10. (B) Activity of teicoplanin against pEBOV and pVSV. Three-fold serial dilutions of teicoplanin were added to Vero cells, after 72 h of incubation, the relative infectivities were analyzed by measuring the luciferase and presented as a percentage of luciferase derived from the compound-treated cells compared with that from the mock-treated cells. Cytotoxicity was also examined by incubation of Vero cells with the indicated concentrations of teicoplanin and was presented as a percentage of luminescence derived from the compound-treated cells compared with that from the mock-treated cells (with medium). (C) Effect of human serum albumin (HSA) on the anti-pEBOV activity of teicoplanin. Activity of teicoplanin against pEBOV was evaluated in the presence of indicated concentrations of HSA and EC50s were calculated using Prism's nonlinear regression (GraphPadPrism5). (D) Activity of vancomycin against pEBOV. Left, chemical structure of vancomycin; right, activity of vancomycin against pEBOV and cytotoxicity of vancomycin. For (B), (C), and (D), average results from three experiments are shown. Error bars represent the standard deviations (B and D) or standard error (C) of means of three independent measurements.
To validate the activity of 15 primary hits, all the compounds were repurchased as dry powders and dissolved in DMSO except teicoplanin which was dissolved in medium, and then eight-point dose-response analysis was performed in duplicate in three assays, including the same assay for primary screening against pEBOV and pVSV and a cytotoxicity assay. The cytotoxicity of each compound was assessed in parallel without the addition of pseudovirus and measured by CellTiter-Glo (Promega) reagent to determine the concentration that resulted in 50% inhibition of cell viability (CC50). For each compound, selectivity indexes (SI) were calculated as SI = CC50/EC50, and the ratio of EC50 of pVSV to EC50 of pEBOV (pEBOV/pVSV) was used to rule out compounds that inhibited FLuc expression or HIV replication. The chemical structure, CC50, EC50, SI values, and pEBOV/pVSV for each compound are listed in Table 1 . Our selection criteria for further compound characterization required that each compound has an EC50 < 10 μM, SI > 10 against pEBOV, and pEBOV/pVSV > 10. Two compounds (teicoplanin and toremiphene) met our criteria, and teicoplanin was selected for additional characterization because previous studies have shown that toremiphene interferes with a step late in viral entry and likely affects the triggering of fusion (Johansen et al., 2013).
Table 1.
Compounds with activity against EBOV pseudovirus.
| Name | Structure | CC50 (μM) | EC50 (μM) |
SI |
pEBOV/pVSVa | ||
|---|---|---|---|---|---|---|---|
| pEBOV | pVSV | pEBOV | pVSV | ||||
| Teicoplanin | 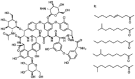 |
>125 | 2.38 | >125 | >52.52 | N.D | >52.52 |
| Tamoxifen | 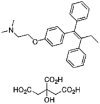 |
10.09 | 0.75 | 4.94 | 13.47 | 2.04 | 6.59 |
| Clemastine | 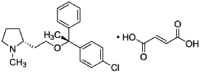 |
15.80 | 3.01 | 10.31 | 5.25 | 1.53 | 3.43 |
| Toremiphene | 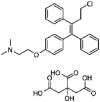 |
10.16 | 0.38 | 7.53 | 26.73 | 1.35 | 19.82 |
| Paroxetine | 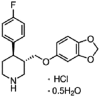 |
18.10 | 2.40 | 7.39 | 7.54 | 2.45 | 3.08 |
| Amiodarone | 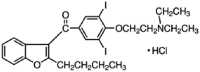 |
54.18 | 4.03 | 8.18 | 13.44 | 6.62 | 2.03 |
| Clomiphene | 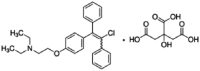 |
15.30 | 1.83 | 8.17 | 8.36 | 1.87 | 4.46 |
| Securinine | 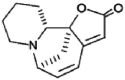 |
113.58 | 27.11 | 26.64 | 4.19 | 4.26 | 0.98 |
| Glafenine | 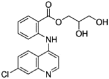 |
>100 | 11.13 | 10.89 | >8.98 | >9.18 | 0.98 |
| Oxeladin | 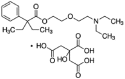 |
>100 | 8.06 | 38.08 | >12.41 | >2.63 | 4.72 |
| Pimethixene maleate | 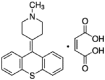 |
19.76 | 10.66 | 14.83 | 1.85 | 1.33 | 1.39 |
| Artemisinin | 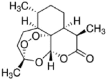 |
>500 | 74.14 | 161.60 | >6.74 | >3.09 | 2.18 |
| Indoprofen | 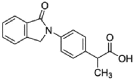 |
91.85 | 9.37 | 8.51 | 9.80 | 10.79 | 0.91 |
| Iodoquinol | 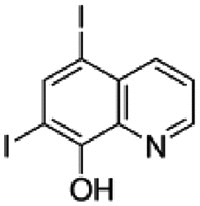 |
16.64 | 3.50 | 3.45 | 4.75 | 4.82 | 0.99 |
| Levonordefrin | 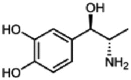 |
>50 | >50 | >50 | N.D | N.D | N.D |
pEBOV/pVSV = EC50 of pVSV/EC50 of pEBOV, N.D, not determined.
Teicoplanin, a glycopeptide antibiotic used for the treatment of gram-positive bacterial infections (Pea et al., 2003, Sancar et al., 2008), effectively inhibited pEBOV in a dose-dependent manner with an EC50 of 2.38 μM, EC90 of 9.36 μM, and SI of greater than 52.5 (Fig. 1B). At 125 μM, teicoplanin completely inhibited pEBOV infection without cytotoxicity but only slightly (less than 20%) inhibited pVSV infection suggesting it is a selective inhibitor of pEBOV. Since teicoplanin is highly (90%) bound to plasma protein (Wilson, 2000, Yagasaki et al., 2003), we evaluated the effect of human serum albumin (HSA) on the anti-pEBOV activity of teicoplanin. In the presence of HSA, the dose-response curves shifted toward higher EC50 values as the concentration increased (Fig. 1C). The fold shift in EC50 was 5.4, 6.0, and 7.3 in the presence of 1%, 3%, and 5% HSA, respectively. As albumin is present in human serum at concentrations in the range of 35–45 mg/ml, the EC50 of teicoplanin against pEBOV in humans should be in the range of 12.03–14.56 μM (20.56–24.89 mg/L). Recently, Johansen and coworkers (Johansen et al., 2015) also showed that teicoplanin is active against eGFP-EBOV in vitro, however, no protection was observed in EBOV-infected C57BL/6 mice after treatment with teicoplanin at a dose of 14 mg/kg of body weight once daily for 10 days. We speculate that the failure was due to the therapeutic concentration of teicoplanin not being achieved. Perhaps a higher dose e.g. daily dose of 40 mg/kg that has shown to be safe and effectively in decreasing bacteremia (Domenech et al., 2004) may lead to significant survival benefits. To maintain serum concentration of teicoplanin above the EC50 in patients infected with EBOV is the key to successful outcome in clinical settings. Given that a trough plasma concentration (Cmin) of 20–60 mg/L (>the estimated EC50 against pEBOV in human) is achieved in 30–70% samples and toxicity is not seen until Cmin reach 60 mg/L (Tobin et al., 2010), teicoplanin may be of potential for treatment of EVD. Nevertheless, new dosage guidelines should be developed to ensure optimal drug exposure for the majority of patients who may have EVD. Moreover, vancomycin which is also used for the treatment of gram-positive bacterial infections and structurally related to teicoplanin only showed 24.7% inhibition at 125 μM (Fig. 1D), suggesting the structural difference between teicoplanin and vancomycin, e.g. a long fatty acid chain attached to teicoplanin that is absent in vancomycin and different structures of their aglycones is crucial for anti-pEBOV activity.
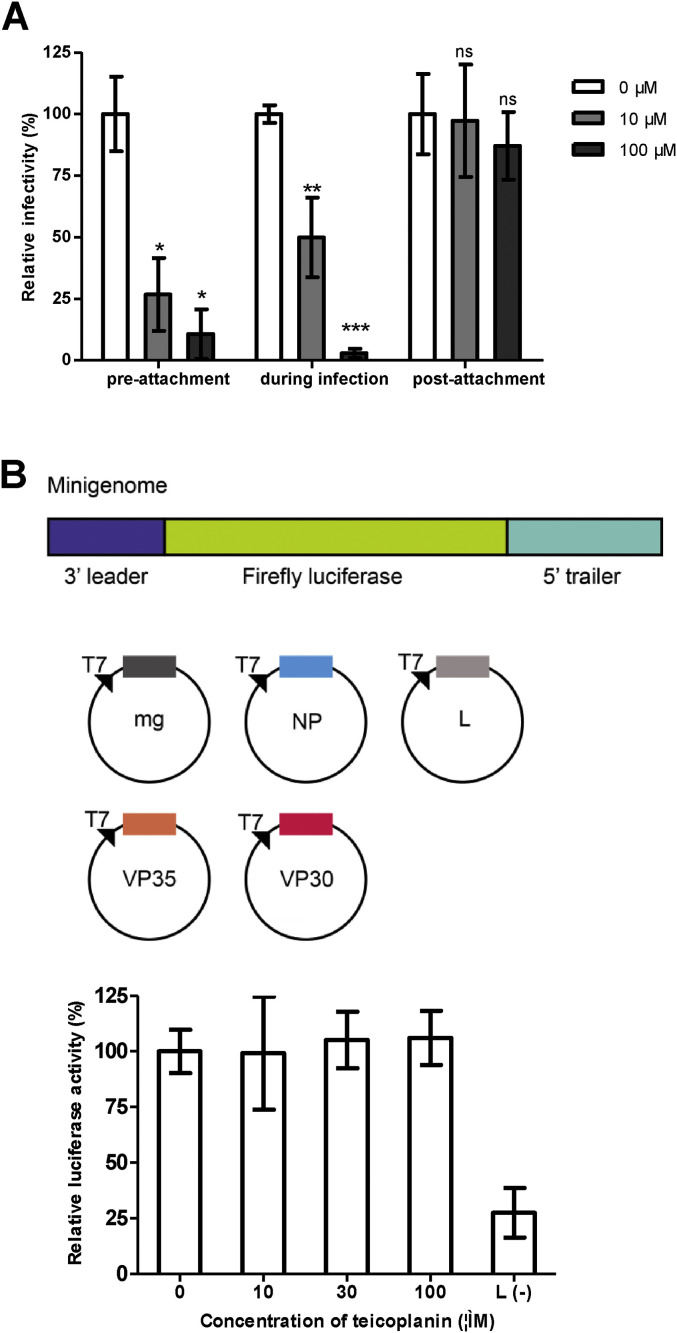
EBOV binds to target cells through interactions of either glycans on GP with C-type lectins (CLECs) or virion-associated phosphatidylserine with phosphatidylserine receptors to initiate entry (Moller-Tank and Maury, 2015), after which it is internalized via macropinocytosis and traffics to the endosome (Nanbo et al., 2010, Saeed et al., 2010), where GP is cleaved by host proteases such as cathepsins (Chandran et al., 2005). Binding of cleaved GP to the endosomal membrane protein Niemann-Pick C1 (NPC1) triggers fusion and facilitates the release of the viral nucleoprotein into the cytoplasm prior to the initiation of virus replication (Carette et al., 2011, Cote et al., 2011). To determine the stage of the viral entry pathway at which teicoplanin inhibits infection, we performed pre- and post-attachment assays. Teicoplanin was incubated with pEBOV for 1 h before or after pEBOV binding to a monolayer of Vero cells, and infection was measured by firefly luciferase assay. As shown in Fig. 2 A, teicoplanin efficiently inhibited infection when premixed with pEBOV or added during infection. However, no inhibition was detected when added after virus adsorption to the cell surface, indicating that teicoplanin blocks viral entry. Because teicoplanin aglycone derivatives were reported to inhibit subgenomic HCV replication (Obeid et al., 2011), we took advantage of an EBOV minigenome system (schemed in Fig. 2B, and all constructs were synthesized by Generay Biotech Ltd, Shanghai, China based on the sequences of GenBank accession number AY354458) to test whether teicoplanin inhibits EBOV replication. BSR T7/5 cells stably expressing the T7 RNA polymerase (kindly provided by Prof. Dr. Karl-Klaus Conzelmann from Max-von-Pettenkofer Institut, Germany) were seeded at 2 × 105 cells/well in a 24-well plate (Corning Costar) 24 h in advance. Cells were transfected with plasmids encoding for EBOV NP (100 ng), VP35 (100 ng), VP30 (60 ng), L (600 ng) proteins, and minigenome (50 ng) containing the FLuc reporter gene using Lipofectamine 2000 (Invitrogen). Plasmid pRL-SV40 (5 ng) encoding renilla luciferase was co-transfected as an internal control to normalize transfection efficiency, and transfection without plasmid expressing L protein served as negative control. Teicoplanin was added after transfection, and left to incubate with the cells. After 24 h of transfection, cells were lysed in luciferase lysis buffer and firefly luciferase as well as renilla luciferase signals were measured for each well with dual-luciferase reporter assay system (Promega). In contrast to inhibitory effects on subgenomic HCV replication, teicoplanin did not inhibit replication of the EBOV minigenome at all tested concentrations (Fig. 2B). This observation together with the results of pre- and post-attachment assays suggest that the mechanism of inhibition of pEBOV by teicoplanin is by blocking virus entry.
Fig. 2.
Teicoplanin blocks pEBOV entry but does not inhibit viral RNA replication. (A) Time-of-addition studies with teicoplanin. Vero cells were infected with pEBOV at 37 °C for 1 h in the presence (during infection) of 10 or 100 μM of teicoplanin and washed to remove unbound pEBOV and compounds, and then fresh medium was added and incubated at 37 °C for 72 h. For the pre-attachment assay, pEBOV was incubated with 10 or 100 μM of teicoplanin at 4 °C for 1 h prior to addition to cells. The virus-compound mixture was incubated with cells at 37 °C for 1 h, after which the cells were washed with medium and incubated at 37 °C for 72 h. For the post-attachment assay, Vero cells were prechilled to 4 °C, and pEBOV was added to cells, and virus adsorption was allowed for 1 h at 4 °C. Cells were washed three times with cold medium to remove unbound virus, and 10 or 100 μM of teicoplanin were added for 1 h at 4 °C, and cells then were washed and incubated at 37 °C for 72 h. The data are the averages for quadruplicate wells for two independent experiments, and the error bars represent the standard deviations of the means. Statistical significance between treated and control group was analyzed by t-test (ns, not significant, *, p < 0.05, **, p < 0.01 ***, p < 0.001). (B) Minigenome assay with teicoplanin. Upper, schematic diagram of the Ebola virus (Zaire strain) minigenome used in this study, in which the viral genes are removed and replaced with a firefly luciferase (FLuc) reporter gene, but the nontranscribed leader and trailer regions as well as the noncoding regions upstream and downstream of NP gene are retained. The minigenome (mg) was flanked by the T7 RNA polymerase promoter (T7) and a ribozyme (HDVr). The minigenome is replicated and transcribed by NP, VP35, VP30, and L provided in trans from expression plasmids. Lower, BSR T7/5 cells were transfected with minigenome and expression plasmids for the RNP proteins NP, VP35, VP30, and L, and after 2 h of incubation with transfection complex, cells were treated with 10 μM, 30 μM, or 100 μM of teicoplanin. Firefly and renilla luciferase activities were measured at 24 h post-transfection. Cells transfected without L plasmid were set as background. The effects of teicoplanin were presented as a percentage of firefly luciferase (normalized against renilla luciferase) derived from the compound-treated cells compared with that from the mock-treated cells. The data presented were obtained from two independent experiments. Error bars represent the standard deviations from two independent experiments.
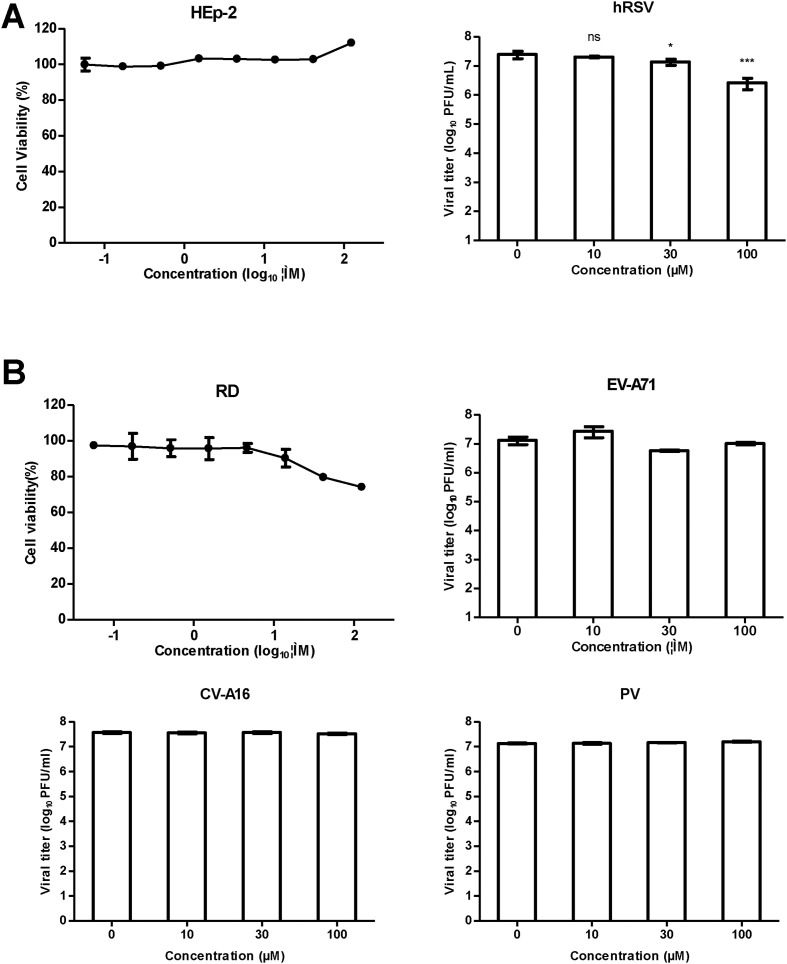
Teicoplanin and its derivatives have been reported to inhibit several viruses such as HIV (Balzarini et al., 2003, Preobrazhenskaya and Olsufyeva, 2006), influenza (Bereczki et al., 2014), HCV (Maieron and Kerschner, 2012, Obeid et al., 2011), dengue virus and other flaviviruses (De Burghgraeve et al., 2012), and coronaviruses including SARS-CoV and FIPV (Balzarini et al., 2006). Because all of these viruses are enveloped viruses, we questioned whether teicoplanin inhibits a wide variety of enveloped viruses but not nonenveloped viruses. We then evaluated the activity of teicoplanin against another enveloped virus human respiratory syncytial virus (hRSV, strain Long) and three nonenveloped viruses including enterovirus 71 (EV-A71, strain G082), Coxsackievirus A16 (CV-A16, strain SHZH05-1), and poliovirus 1 (PV1, strain Sabin). Interestingly, teicoplanin reduced viral titers of hRSV by 1.8- and 9.4-fold at 30 and 100 μM, respectively (Fig. 3 A), however, no inhibition was observed against EV-A71, CV-A16, or PV1 at all tested concentrations (Fig. 3B), suggesting teicoplanin probably targets a common component among those enveloped viruses or target cells.
Fig. 3.
Effect of teicoplanin on hRSV, EV-A71, CV-A16, and PV1. (A) Left, cytotoxicity of teicoplanin on HEp-2 cells was assessed in eight-point dose-response. Right, HEp-2 cells were infected with hRSV (subtype A, strain Long) at an MOI of 0.1 and treated with teicoplanin at the indicated concentrations. Supernatants were collected at 48 h post-infection, and viral titers were determined by immunostaining assay. (B) Cytotoxicity of teicoplanin on RD cells was assessed in eight-point dose-response. Cells were infected with EV-A71 (MOI = 0.1), CV-A16, and PV1 (MOI = 0.01) and treated with teicoplanin at the indicated concentrations. Supernatants were collected at 42 h post-infection, and viral titers were determined by plaque assay. For (A) and (B), the data presented were obtained from two independent experiments. Error bars represent the standard deviations from two independent experiments. Statistical significance between treated and control group was analyzed by t-test (ns, not significant, *, p < 0.05, ***, p < 0.001).
In conclusion, we identified glycopeptide antibiotic teicoplanin as an inhibitor of EBOV pseudovirus in cell culture by blocking entry. Based on its long history of clinical use and achievable high enough human exposure, teicoplanin has the potential to be rapidly advanced to clinical settings. Moreover, teicoplanin also provides a good tool to gain novel insights into the entry process of EBOV.
Acknowledgments
We thank Dr. Ke Xu for helpful discussions during the study. This research was supported by funding from Key Laboratory of Molecular Virology & Immunology, Institute Pasteur of Shanghai, Chinese Academy of Sciences (KLMVI-Ebola-2014004) and National Natural Science Foundation of China (31400148) to G.Z. and Special Presidential Foundation of the Chinese Academy of Sciences, China to R.A.. G.Z. gratefully acknowledges the support of SA-SIBS scholarship program.
References
- Altmeyer R. Virus attachment and entry offer numerous targets for antiviral therapy. Curr. Pharm. Des. 2004;10:3701–3712. doi: 10.2174/1381612043382729. [DOI] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N., Soropogui B., Sow M.S., Keita S., De Clerck H., Tiffany A., Dominguez G., Loua M., Traore A., Kolie M., Malano E.R., Heleze E., Bocquin A., Mely S., Raoul H., Caro V., Cadar D., Gabriel M., Pahlmann M., Tappe D., Schmidt-Chanasit J., Impouma B., Diallo A.K., Formenty P., Van Herp M., Gunther S. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- Balzarini J., Keyaerts E., Vijgen L., Egberink H., De Clercq E., Van Ranst M., Printsevskaya S.S., Olsufyeva E.N., Solovieva S.E., Preobrazhenskaya M.N. Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antivir. Res. 2006;72:20–33. doi: 10.1016/j.antiviral.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini J., Pannecouque C., De Clercq E., Pavlov A.Y., Printsevskaya S.S., Miroshnikova O.V., Reznikova M.I., Preobrazhenskaya M.N. Antiretroviral activity of semisynthetic derivatives of glycopeptide antibiotics. J. Med. Chem. 2003;46:2755–2764. doi: 10.1021/jm0300882. [DOI] [PubMed] [Google Scholar]
- Bereczki I., Kicsak M., Dobray L., Borbas A., Batta G., Keki S., Nikodem E.N., Ostorhazi E., Rozgonyi F., Vanderlinden E., Naesens L., Herczegh P. Semisynthetic teicoplanin derivatives as new influenza virus binding inhibitors: synthesis and antiviral studies. Bioorg. Med. Chem. Lett. 2014;24:3251–3254. doi: 10.1016/j.bmcl.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong C.R., Sullivan D.J., Jr. New uses for old drugs. Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Burghgraeve T., Kaptein S.J., Ayala-Nunez N.V., Mondotte J.A., Pastorino B., Printsevskaya S.S., de Lamballerie X., Jacobs M., Preobrazhenskaya M., Gamarnik A.V., Smit J.M., Neyts J. An analogue of the antibiotic teicoplanin prevents flavivirus entry in vitro. PloS One. 2012;7:e37244. doi: 10.1371/journal.pone.0037244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech A., Ribes S., Cabellos C., Dominguez M.A., Montero A., Linares J., Ariza J., Gudiol F. A mouse peritonitis model for the study of glycopeptide efficacy in GISA infections. Microb. Drug Resist. 2004;10:346–353. doi: 10.1089/mdr.2004.10.346. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring G., Rohrmann K., Atenchong N., Mittler E., Becker S., Dahlmann F., Pohlmann S., Vondran F.W., David S., Manns M.P., Ciesek S., von Hahn T. The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J. Antimicrob. Chemother. 2014;69:2123–2131. doi: 10.1093/jac/dku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T., Feldmann H. Reverse genetics systems as tools for the development of novel therapies against filoviruses. Expert Rev. Anti-infective Ther. 2014;12:1253–1263. doi: 10.1586/14787210.2014.948848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.M., Brannan J.M., Delos S.E., Shoemaker C.J., Stossel A., Lear C., Hoffstrom B.G., Dewald L.E., Schornberg K.L., Scully C., Lehar J., Hensley L.E., White J.M., Olinger G.G. FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 2013;5:190ra179. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen L.M., DeWald L.E., Shoemaker C.J., Hoffstrom B.G., Lear-Rooney C.M., Stossel A., Nelson E., Delos S.E., Simmons J.A., Grenier J.M., Pierce L.T., Pajouhesh H., Lehar J., Hensley L.E., Glass P.J., White J.M., Olinger G.G. A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci. Transl. Med. 2015;7:290ra289. doi: 10.1126/scitranslmed.aaa5597. [DOI] [PubMed] [Google Scholar]
- Kouznetsova J., Sun W., Martinez-Romero C., Tawa G., Shinn P., Chen C.Z., Schimmer A., Sanderson P., McKew J.C., Zheng W., Garcia-Sastre A. Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg. Microbes Infect. 2014;3:e84. doi: 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid P.B., Chopra S., Manger I.D., Gilfillan L., Keepers T.R., Shurtleff A.C., Green C.E., Iyer L.V., Dilks H.H., Davey R.A., Kolokoltsov A.A., Carrion R., Jr., Patterson J.L., Bavari S., Panchal R.G., Warren T.K., Wells J.B., Moos W.H., Burke R.L., Tanga M.J. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PloS One. 2013;8:e60579. doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maieron A., Kerschner H. Teicoplanin therapy leading to a significant decrease in viral load in a patient with chronic hepatitis C. J. Antimicrob. Chemother. 2012;67:2537–2538. doi: 10.1093/jac/dks217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Tank S., Maury W. Ebola virus entry: a curious and complex series of events. PLoS Pathog. 2015;11:e1004731. doi: 10.1371/journal.ppat.1004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid S., Printsevskaya S.S., Olsufyeva E.N., Dallmeier K., Durantel D., Zoulim F., Preobrazhenskaya M.N., Neyts J., Paeshuyse J. Inhibition of hepatitis C virus replication by semi-synthetic derivatives of glycopeptide antibiotics. J. Antimicrob. Chemother. 2011;66:1287–1294. doi: 10.1093/jac/dkr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pea F., Brollo L., Viale P., Pavan F., Furlanut M. Teicoplanin therapeutic drug monitoring in critically ill patients: a retrospective study emphasizing the importance of a loading dose. J. Antimicrob. Chemother. 2003;51:971–975. doi: 10.1093/jac/dkg147. [DOI] [PubMed] [Google Scholar]
- Preobrazhenskaya M.N., Olsufyeva E.N. Polycyclic peptide and glycopeptide antibiotics and their derivatives as inhibitors of HIV entry. Antivir. Res. 2006;71:227–236. doi: 10.1016/j.antiviral.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M.F., Kolokoltsov A.A., Albrecht T., Davey R.A. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salata C., Baritussio A., Munegato D., Calistri A., Ha H.R., Bigler L., Fabris F., Parolin C., Palu G., Mirazimi A. Amiodarone and metabolite MDEA inhibit Ebola virus infection by interfering with the viral entry process. Pathogens Dis. 2015;73 doi: 10.1093/femspd/ftv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A.A., Yegenoglu S., de Vries R., Postma M.J., Simsek N., Pechlivanoglou P., Unal S. Vancomycin vs teicoplanin in the treatment of gram-positive infections: a pharmacoeconomic analysis in a Turkish university hospital. Pharm. World & Sci. PWS. 2008;30:916–923. doi: 10.1007/s11096-008-9251-2. [DOI] [PubMed] [Google Scholar]
- Tobin C.M., Lovering A.M., Sweeney E., MacGowan A.P. Analyses of teicoplanin concentrations from 1994 to 2006 from a UK assay service. J. Antimicrob. Chemother. 2010;65:2155–2157. doi: 10.1093/jac/dkq266. [DOI] [PubMed] [Google Scholar]
- Wilson A.P. Clinical pharmacokinetics of teicoplanin. Clin. Pharmacokinet. 2000;39:167–183. doi: 10.2165/00003088-200039030-00001. [DOI] [PubMed] [Google Scholar]
- Yagasaki K., Gando S., Matsuda N., Kameue T., Ishitani T., Hirano T., Iseki K. Pharmacokinetics of teicoplanin in critically ill patients undergoing continuous hemodiafiltration. Intensive Care Med. 2003;29:2094–2095. doi: 10.1007/s00134-003-1914-9. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]