Short abstract
Objective
Malignant melanoma is a highly invasive cancer whose pathogenesis remains unclear. We analyzed the microarray dataset GDS1375 in the Gene Expression Omnibus database to search for key genes involved in the occurrence and development of melanoma.
Methods
The dataset included 52 samples (7 normal skin and 45 melanoma samples). We identified differentially expressed genes (DEGs) between the two groups and used integrated discovery databases for Gene Ontology (GO) and Kyoto Gene and Genome Encyclopedia (KEGG) pathway analyses. In addition, we used the STRING and MCODE plugins of Cytoscape to visualize the protein-protein interactions (PPI) for these DEGs.
Results
A total of 509 upregulated and 618 downregulated DEGs were identified, which were enriched in GO terms including integrin binding, protein binding, and structural constituent of cytoskeleton, and in KEGG pathways such as melanogenesis, prostate cancer, focal adhesion, and renin secretion. Three major modules from the PPI networks and 10 hub genes were identified, including CDC20, GNB2, PPP2R1A, AURKB, POLR2E, and AGTR1. Overall survival was low when these six hub genes were highly expressed.
Conclusion
This bioinformatics analysis identified hub genes that may promote the development of melanoma and represent potential new biomarkers for diagnosis and treatment of melanoma.
Keywords: Melanoma, bioinformatics, biomarkers, genes, Gene Expression Omnibus, protein–protein interactions
Background
Melanoma is a common skin tumor, and epidemiological studies have shown that the incidence of melanoma has increased dramatically in the past 50 years. In 2010, the incidence of melanoma in the United States was 35.4 and 24.2 cases per 100,000 men and women, respectively. In contrast, in the 1960s, the respective incidences were only 9.4 and 8.2 cases per 100,000.1 The prognosis of advanced melanoma is poor and the median overall survival rate is only 23 months.2
Although great progress has been made in the diagnosis and treatment of melanoma (e.g., BRAF V600E inhibitors, early surgical resection), the long-term survival rate remains low in patients with advanced melanoma, mainly due to distant metastases.3,4 Thus, there is an urgent need to elucidate the molecular mechanisms underlying the development and metastasis of melanoma to develop new and better therapeutic strategies.
Many recent studies have found that biomarkers can be used to screen for and diagnose skin melanoma. For examples, Song et al.5 reported that CDKL1 (cyclin dependent kinase like 1) inhibits the growth and colony formation of melanoma cells by increasing apoptosis. Liu et al.6 showed that the metastasis of melanoma can be inhibited by microRNA (miR)-425, which represses the PI3K-Akt pathway by targeting insulin-like growth factor-1. Kubic et al.7 found that PAX3 (paired box 3) and FOXD3 (forkhead box D3) upregulate CXCR4 (C-X-C motif chemokine receptor 4) expression in melanoma. However, the discovery of these biomarkers still fails to fully explain the mechanisms underlying the growth and metastasis of melanoma.
To better understand the molecular mechanisms of melanoma, we analyzed the microarray dataset GDS1375 from Gene Expression Omnibus (GEO), which contains expression data from both melanoma and normal tissues to identify differentially expressed genes (DEGs) and subsequently construct a protein–protein interaction (PPI) network to identify highly connected central genes for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. In addition, we performed an overall survival (OS) analysis to further elucidate the biological significance of the identified genes. The findings could provide new insights into molecular mechanisms related to melanoma pathogenesis and clues to develop better biomarker and therapeutic approaches for the cancer.
Materials and methods
Ethics and consent
This study used bioinformatics analysis and did not involve humans or animals. Therefore, local ethics committee approval and informed consent were not needed.
DEG identification
The microarray dataset GDS1375 was obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). It included 52 samples—7 normal skin samples and 45 melanoma samples. DEGs in the samples were identified via the limma package of R (www.r-project.org). The Bonferroni and Hochberg method was used to correct the P-values. Thresholds of P < 0.02 and |log2 (fold change)| ≥2 were set. In addition, hierarchical clustering analysis of the DEGs was performed and volcano maps were plotted.
GO and KEGG pathway analysis
GO analysis is a common method for annotating genes and gene products as well as characterizing biological properties of high-throughput genome or transcriptome data. KEGG is a set of databases dealing with genomes, biological pathways, diseases, drugs, and chemicals.8 DAVID (https://david.ncifcrf.gov/) is a web-based online bioinformatics tool designed to interpret the functions of a large number of genes or proteins.9 We used these tools to analyze and visualize the core biological processes, molecular functions, cellular components, and pathways associated with these DEGs.
PPI network and module analysis
The search tool for retrieval of interacting genes (STRING; https://string-db.org/) is an online tool that assesses PPI.10 To detect potential relationships between DEGs, we used the STRING plug-in in Cytoscape (https://cytoscape.org/) to map the interactive networks of the DEGs. The confidence score and the maximum number of interactors were set at ≥0.9 and 0, respectively. In addition, the MPI application was used to screen the PPI network in Cytoscape with a cutoff of 2, a node score cut-off of 0.2, a k-core of 2, and a depth of 100. Gene pathway analysis in each module was done using DAVID. In addition, 10 hub genes were mapped using STRING with a confidence score of ≥0.4 and a maximum value of the interaction parameter of ≤5.
Survival analysis of hub genes and construction of miRNA-hub gene network
GEPIA (http://gepia.cancer-pku.cn/index.html) is a newly developed interactive web server for the analysis of gene expression from The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEx) projects; we used GEPIA to analyze the survival response of the hub genes as boxplots. In addition, we identified the microRNA (miRNA) targets of the hub genes using Targetscan (http://www.targetscan.org/vert_72/). The most-matched miRNAs were used to construct a miRNA-hub gene network using Cytoscape.
Results
Identification of DEGs
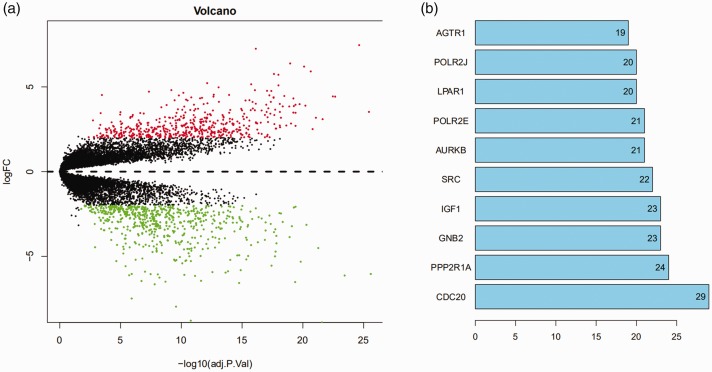
Seven normal skin samples and 45 melanoma samples were analyzed in this study; 509 upregulated and 618 downregulated genes were identified, as well as 10 hub genes (Figure 1a and b).
Figure 1.
Differential expression profiles of mRNAs in cutaneous melanoma and control tissues. (a) The horizontal dotted line delimits up- and downregulation. Red and green dots represent significant mRNAs with fold change >2 and P < 0.02. Red dots represent up-regulated genes, green dots represent down-regulated genes. (b) Ten hub genes with their fold changes. FC, fold change.
GO function and KEGG pathway enrichment analysis
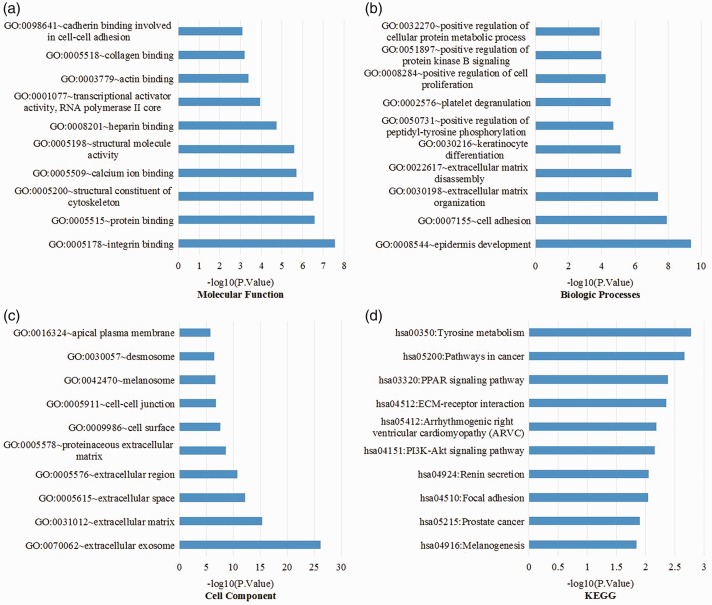
To gain a better understanding of the DEGs, we used DAVID for GO function and KEGG analysis. GO analysis showed that the DEGs were enriched in molecular function, integrin binding, protein binding, structural constituent of cytoskeleton, calcium ion binding, structural molecule activity, heparin binding, transcriptional activator activity, RNA polymerase II core promoter proximal region, actin binding, collagen binding, and cadherin binding involved in cell–cell adhesion (Figure 2a). In biological processes, enriched terms included epidermis development, cell adhesion, extracellular matrix (ECM) organization, ECM disassembly, keratinocyte differentiation, positive regulation of peptidyl-tyrosine phosphorylation, platelet degranulation, positive regulation of cell proliferation, positive regulation of protein kinase B signaling, and positive regulation of cellular protein metabolic process (Figure 2b). In addition, GO cell component analysis showed that the DEGs were significantly enriched in extracellular exosome, ECM, extracellular space, extracellular region, proteinaceous ECM, cell surface, cell–cell junction, melanosome, desmosome, and apical plasma membrane (Figure 2c). KEGG pathway analysis showed that the DEGs were enriched in melanogenesis, prostate cancer, focal adhesion, renin secretion, PI3K-Akt signaling pathway, arrhythmogenic right ventricular cardiomyopathy, ECM-receptor interaction, PPAR signaling pathway, and pathways in cancer and tyrosine metabolism (Figure 2d).
Figure 2.
GO analysis and KEGG pathway analysis of differentially expressed genes. (a) Molecular functions, (b) biological processes, (c) cell components, and (d) KEGG pathways. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Hub genes and PPI network modules
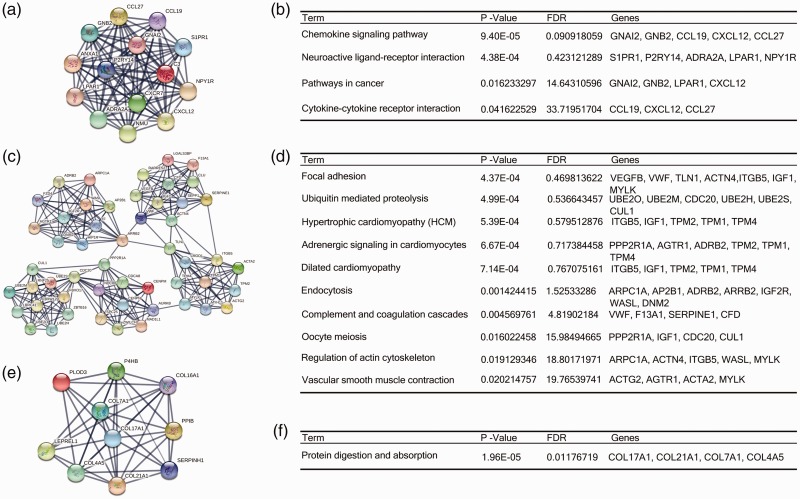
Based on the STRING protein query information from the public database, we created PPI networks of the top 10 hub genes with high connectivity (Figure 3). In addition, we used the MCODE plug-in to select the first three modules. KEGG pathway enrichment analysis showed that the modules were primarily associated with cell cycle (Figure 3).
Figure 3.
The protein–protein interaction networks of top three modules. Module 1 (a) and the enriched pathways of module 1 (b); module 2 (c) and the enriched pathways of module 2 (d); module 3 (e) and the enriched pathways of module 3 (f). FDR, false discovery rate.
miRNA-hub gene network and prognostic value of hub genes
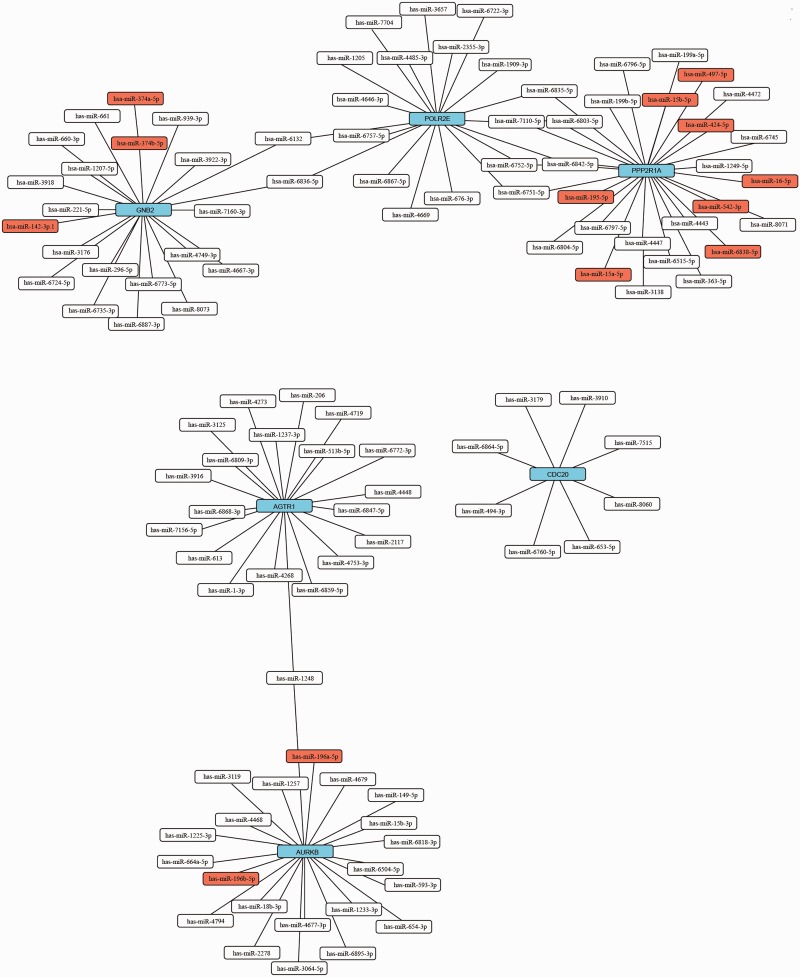
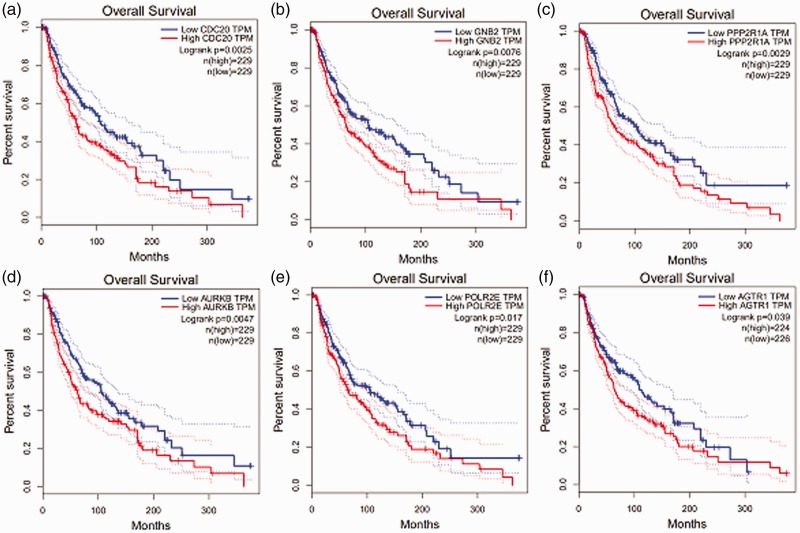
We screened the miRNA of the hub genes using Targetscan (http://www.targetscan.org/vert_72/). The most-matched miRNAs were used to make the miRNA-hub gene network (Figure 4). GEPIA (http://gepia.cancer-pku.cn/index.html) is a newly developed interactive web server for the analysis of RNA sequencing expression of 9736 tumors and 8587 normal samples from the TCGA and GTEx projects. Prognostic information associated with the 10 hub genes was available in GEPIA. We analyzed prognosis associated with the genes CDC20, GNB2, PPP2R1A, AURKB, POLR2E, and AGTR1; prognosis was poor when these genes were highly expressed (Figure 5).
Figure 4.
miRNA-hub gene network. Blue boxes represent hub genes, red boxes represent conserved miRNA, and white boxes represent poorly conserved miRNA. Straight lines indicate a regulatory relationship between miRNA and gene. miRNA, microRNA.
Figure 5.
Prognostic value (overall survival) of six genes: CDC20 (a), GNB2 (b), PPP2R1A (c), AURKB (d), POLR2E (e), and AGTR1 (f) in patients with cutaneous melanoma. HR, hazard ratio; CI, confidence interval; TPM, transcripts per million.
Discussion
To illustrate the mechanisms underlying the pathogenesis of melanoma, we analyzed microarray expression profiles of melanoma tissues. Compared with normal skin samples, 1127 DEGs were identified in melanoma samples, including 509 upregulated and 618 downregulated genes.
The GO analysis revealed that DEGs are involved in melanogenesis, epidermis development, keratinocyte differentiation, extracellular matrix, melanosome, and integrin binding. The KEGG pathways showed that DEGs are mainly involved in melanogenesis, prostate cancer, and other pathways in cancer. In addition, we used the MCODE plug-in to select the first three modules. KEGG pathway analysis showed that the three most important modules were related to pathways in focal adhesion, cancer, and protein degradation and absorption.
To identify the hub genes, we selected 10 genes with the highest connectivity in DEGs in the PPI network. Survival analysis showed that 6 out of the 10 genes (CDC20, GNB2, PPP2R1A, AURKB, POLR2E, and AGTR1) were associated with prognosis. It is likely, therefore, that these genes are involved in the pathogenesis of melanoma and would be candidates for further functional analysis.
An increasing number of studies show that CDC20 (cell division cycle 20) plays a role in tumor pathogenesis. In many types of tumors such as breast cancer, pancreatic cancer, and prostate cancer, CDC20 is highly expressed.11–13 Mainly through the activation of APC, CDC20 forms an E3 ubiquitin ligase complex called the APC complex (APCCdc20) to degrade its downstream substrates, regulate the mitogenesis cycle, and promote apoptosis.11–14 APCCdc20 regulates the activity of downstream pluripotency-related transcription factor SOX2, which promotes the invasion and renewal of glioma stem cells.13 Of note, the CDC20 short interfering (si)RNA that knocks down CDC20 expression inhibits the growth of solid melanoma tumor.14 Therefore, CDC20 is likely involved in the pathogenesis of melanoma. However, whether it activates the stem cells of melanoma is still unclear.
GNB2 (G protein subunit beta 2) is a member of the Gβ protein family. GNB2 and its related family member GNB1 confer cytokine-independent growth and activate the canonical G protein signaling.15 G proteins and their downstream signaling targets are involved in the initiation and progression of some cancers, resulting in aberrant cell growth and reduced survival, largely by activating the AKT/mTOR, MAPK, and Hippo signaling pathways.16 However, the role of GNB1 in the pathogenesis of melanoma is still not clear. Expression of GNB2 K78E in A375 melanoma cells that harbor the BRAF V600E mutation was shown to confer resistance to vemurafenib.15 This suggests that GNB2 may play an important role in the pathogenesis and drug resistance of melanoma.
PPP2R1A (protein phosphatase 2 scaffold subunit A alpha) is a scaffolding subunit of protein phosphatase 2A (PP2A), one of four major serine/threonine protein phosphatases.17 Mutations in PPP2R1A occur in breast cancer, lung cancer, and melanoma.17 PPP2A1A may mediate the survival and resistance to apoptosis of the type B malignant melanoma cell lines.18 PPP2R1A has also been reported to promote the metastasis of melanoma cells through the interaction of tumor cells and lymphatic endothelial cells.19 Therefore, PPP2R1A is a potential target associated with the melanoma process.
AURKB (Aurora B kinase) is a chromosomal passenger protein regulating early mitotic stage transition from prophase to metaphase.20 AURKB is overexpressed in a variety of tumors including melanoma.20 Studies have reported that AURKA controls proliferation, epithelial–mesenchymal transition (EMT), metastasis, and self-renewal capacity of cancer stem cell (CSC), and regulates the cell cycle and survival of cancer cells.20 In melanoma cells, studies have found that Aurora kinase promotes melanocyte proliferation and reduces apoptosis.21 Therefore, AURKB is a valuable therapeutic target for melanoma, but its role in melanoma needs further study.
The POLR2E gene encodes the fifth largest subunit of RNA polymerase II (RNA polymerase II subunit E), which is responsible for synthesis of messenger RNA in eukaryotes.22 Meta-analysis results have revealed that POLR2E is associated with the risk of esophageal cancer.22 The relationship between POLR2E and melanoma has yet to be established. Our study suggests that POLR2E is likely a valuable gene to be further studied in melanoma.
AGTR1 (angiotensin II receptor type 1) is a subtype of the renin-angiotensin system (RAS)23 and has tumor suppressive function in melanoma. Methylation of an AGTR1 CpG island is a biomarker of metastatic melanoma23 and could be used as a new target for gene therapy.
In conclusion, our analysis identified several DEGs associated with the development, progression, and prognosis of melanoma. A total of 1127 DEGs and 10 hub genes were found this study. Based on their high connectivity in the PPI network, CDC20, GNB2, PPP2R1A, AURKB, POLR2E, and AGTR1 might be the core genes of melanoma. Further studies are needed to further validate the functions of these genes, including in vitro and in vivo expression and functional analysis.
Acknowledgement
We thank Ronny Mason for language instructions and Wenhui Chen for software assistance.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81202699 and 81573980), the Natural Science Foundation of Hubei Province (grant no. 2018CFB289), and the Science Foundation of Health Commission of Hubei Province (grant no. WJ2019M074).
ORCID iD
References
- 1.Berwick M, Buller DB, Cust Aet al. Melanoma epidemiology and prevention. Cancer Treat Res 2016; 167: 17–49. [DOI] [PubMed] [Google Scholar]
- 2.Ribas A, Hamid O, Daud Aet al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 2016; 315: 1600–1609. [DOI] [PubMed] [Google Scholar]
- 3.Ascierto PA, Minor D, Ribas Aet al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 2013; 31: 3205–3211. [DOI] [PubMed] [Google Scholar]
- 4.Guo J, Qin S, Liang Jet al . Chinese guidelines on the diagnosis and treatment of melanoma (2015 edition). Chin Clin Oncol 2016; 5: 57. [DOI] [PubMed] [Google Scholar]
- 5.Song Z, Lin J, Sun Zet al. RNAi-mediated downregulation of CDKL1 inhibits growth and colony-formation ability, promotes apoptosis of human melanoma cells. J Dermatol Sci 2015; 79: 57–63. [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Hu Y, Ma Let al. miR-425 inhibits melanoma metastasis through repression of PI3K-Akt pathway by targeting IGF-1. Biomed Pharmacother 2015; 75: 51–57. [DOI] [PubMed] [Google Scholar]
- 7.Kubic JD, Lui JW, Little ECet al. PAX3 and FOXD3 promote CXCR4 expression in melanoma. J Biol Chem 2015; 290: 21901–21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Franceschini A, Wyder Set al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao Y, Li K, Lu Let al. Overexpression of Cdc20 in clinically localized prostate cancer: relation to high Gleason score and biochemical recurrence after laparoscopic radical prostatectomy. Cancer Biomark 2016; 16: 351–358. [DOI] [PubMed] [Google Scholar]
- 12.Chang DZ, Ma Y, Ji Bet al. Increased CDC20 expression is associated with pancreatic ductal adenocarcinoma differentiation and progression. J Hematol Oncol 2012; 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karra H, Repo H, Ahonen Iet al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer 2014; 110: 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee A, Bhattacharyya J, Sagar MVet al. Liposomally encapsulated CDC20 siRNA inhibits both solid melanoma tumor growth and spontaneous growth of intravenously injected melanoma cells on mouse lung. Drug Deliv Transl Res 2013; 3: 224–234. [DOI] [PubMed] [Google Scholar]
- 15.Yoda A, Adelmant G, Tamburini Jet al. Mutations in G protein beta subunits promote transformation and kinase inhibitor resistance. Nat Med 2015; 21: 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Hayre M, Degese MS, Gutkind JS. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol 2014; 27: 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin GA, di Iasio MG, Caprini Eet al. Low frequency of alterations of the alpha (PPP2R1A) and beta (PPP2R1B) isoforms of the subunit A of the serine-threonine phosphatase 2A in human neoplasms. Oncogene 2000; 19: 1191–1195. [DOI] [PubMed] [Google Scholar]
- 18.Su DM, Zhang Q, Wang Xet al. Two types of human malignant melanoma cell lines revealed by expression patterns of mitochondrial and survival-apoptosis genes: implications for malignant melanoma therapy. Mol Cancer Ther 2009; 8: 1292–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christianson DR, Dobroff AS, Proneth Bet al. Ligand-directed targeting of lymphatic vessels uncovers mechanistic insights in melanoma metastasis. Proc Natl Acad Sci U S A 2015; 112: 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang A, Gao K, Chu Let al. Aurora kinases: novel therapy targets in cancers. Oncotarget 2017; 8: 23937–23954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonet C, Giuliano S, Ohanna Met al. Aurora B is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem 2012; 287: 29887–29898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu H, Chen Y, Yuan Qet al. The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget 2017; 8: 43271–43283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renziehausen A, Wang H, Rao Bet al. The renin angiotensin system (RAS) mediates bifunctional growth regulation in melanoma and is a novel target for therapeutic intervention. Oncogene 2019; 38: 2320–2336. [DOI] [PubMed] [Google Scholar]