Highlights
-
•
Development of a new Real-Time RT-PCR for detection of IDV.
-
•
The new Real-Time RT-PCR showed high sensitivity and specificity.
-
•
The new Real-Time RT-PCR can be used for diagnostics and as a confirmatory test.
Keywords: Influenzavirus D, IDV, Real-Time RT-PCR, Swine, Cattle
Abstract
The occurrence of virus belonging to the putative genus Influenzavirus D, has been demonstrated all-around the world arousing interest within the scientific community. Most of the published virological surveys are based on the first described Real-Time PCR method, designed on the PB1 gene of the first isolate. The necessity of extending investigation to different animal species and geographic areas, requires a continuous update of molecular tests, considering newly sequenced strains. Moreover, the availability of an alternative assay, is essential either to confirm data, or for ensuring the detection of the widest number of strains. A new Real-Time PCR, specific for influenza D virus (IDV), was developed and evaluated. The target sequences of primers and probe are highly conserved among IDV strains currently known. The specificity of the method was demonstrated in silico by BLAST, and in vitro with a huge panel of common swine and bovine respiratory pathogens. The analytical sensitivity of the Real-Time PCR was estimated through synthetic RNA molecules and the limit of detection was about 20 copies/μL. The assay was assessed in field and proved to be a valuable tool for the detection of IDV strains.
Orthomyxoviridae family includes three genera of Influenzavirus: Influenzavirus A (IAV), Influenzavirus B (IBV), Influenza virus C (ICV). In 2011, in Oklahoma, a viral strain belonging to a putative new genus, not yet acknowledged by official taxonomy, was isolated from pigs with influenza-like illness (Hause et al., 2013) and tentatively designated Influenzavirus D (IDV) (Hause et al., 2014). Despite its first isolation from swine, ensuing studies suggested cattle as reservoir hosts for this newly discovered virus (Hause et al., 2014, Collin et al., 2015, Ferguson et al., 2015, Ng et al., 2015, Ferguson et al., 2016). Indeed, IDV strains have been subsequently isolated from cattle in the United States (Hause et al., 2014, Collin et al., 2015, Ferguson et al., 2015, Ng et al., 2015), China (Jiang et al., 2014), France (Ducatez et al., 2015), Italy (Chiapponi et al., 2016), Mexico (Mitra et al., 2016) and Japan (Murakami et al., 2016). Besides, serological surveys indicated cattle as the species with the highest seroprevalence and the highest antibody titers for IDV (Hause et al., 2014). It has been experimentally demonstrated that IDV can also infect guinea pigs and ferrets, and replicates in their respiratory tract (Sreenivasan et al., 2015). Moreover, antibodies specific for IDV were detected in sera of small ruminants in North America (Quast et al., 2015). In humans, in spite of a low general seroprevalence (Hause et al., 2013), a recent study has shown that 94–97% of cattle-exposed workers have specific antibodies against IDV, raising concerns of a possible zoonotic risk (Ferguson et al., 2015, White et al., 2016). Further studies are necessary in order to better understand IDV epidemiology, hosts range, and pathogenic potential of this virus.
Few sequences of IDV strains are available in GenBank and only one Real-Time PCR method (Hause et al., 2013), based on the PB1 sequence of the first isolate (JQ922306) has been described (HM, Hause Method). Even though some research groups reported the usage of homemade alternative PCR methods available upon request (Ng et al., 2015, Jiang et al., 2014, Murakami et al., 2016), the majority of publications on IDV refers to this first method (Ferguson et al., 2015, Ducatez et al., 2015, Mitra et al., 2016, Chiapponi et al., 2016). Although PB1 is one of the most stable genes among the Influenzavirus genera, the necessity of extending virological investigation to other animal species and geographic areas, requires a continuous control of the fitness of molecular tests. Besides, an independent test targeting a different sequence is essential either to confirm data, when necessary, or for ensuring the detection of the widest number of strains.
The aim of this work was to develop and evaluate a new Real-Time Taqman assay (NM, New Method) to be applied as an alternative test to HM for the detection of IDV in diagnostic samples. Since, according to some phylogenetic analysis (Collin et al., 2015, Ducatez et al., 2015), PB1 is one of the most, if not the most, conserved region of IDV genome, we evaluated a new target in this region. Primers were designed by Primer 3 software (Rozen and Skaletsky, 2000) based on 14 PB1 sequences of IDV from GenBank (GenBank accession numbers: JQ922306, KM392469, KM392476, KM392483, KM392490, KM392497, KM392504, KF425653, KF425660, KF425667, KM015492, KM015499, KM015506, LN559121). Sequences were aligned using ClustalO (Li et al., 2015). A Taqman probe was designed in a conserved region between primers (Table 1 ). Viral RNA was extracted from clinical samples and cell cultures using One-For-All Vet Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Given the scarcity of sequence data, until the target will be better characterized, a two-step RT Real-Time PCR protocol was considered the most appropriate. A volume of 4 μL of RNA was reverse transcribed using 150 units of M-MLV RT RNase (H-) Point Mutant® (Promega, Madison, USA), 20 ng/μL Random hexamers, 1x M-MLV buffer, in a final volume of 15 μL. The reaction was carried out at 25 °C for 10 min, followed by 60 min at 50 °C and a final inactivation at 70 °C for 15 min. The subsequent Real-Time PCR was accomplished in 25 μL containing 5 μL of cDNA, 1 x GoTaq Probe qPCR Master Mix® (Promega, Madison, USA), 300 nM of each primer and 250 nM of probe. Two primers (70 nM each) and a probe (200 nM), for detection and amplification of Beta-actin mRNA, were included as an internal control (ß-actin F 5′ CTCCTTCYTGGGCATGGA 3′; ß-actin R 5′ GAGGCGCGATGATCTTGAT 3′; ß-actin Probe HEX 5′ AGGACCTCTACGCCAACACGGTGCT 3′ BHQ1). Cycling conditions were as follows: activation step at 95 °C for 2 min, followed by 45 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Fluorescence signal was detected after the annealing/extension step at each cycle. All PCR assays were performed on the CFX96 Real-Time PCR Systems (Bio-Rad, Milan, Italy). The HM was applied as previously described (Hause et al., 2013). In silico analysis of primers and probe by BLAST search was performed in order to assess specificity and inclusivity, taking account of newly published IDV sequences. The specificity of the Real-Time RT-PCR assay was also evaluated in vitro on the principal pathogens involved in respiratory diseases of cattle and swine: Bovine Diarrhea Virus (BVDv) Type 1, Type 2 and Type 3, Bovine Herpesvirus Type 1 (BHV1), Bovine Coronavirus, Bovine Parainfluenza 3 virus, Bovine Respiratory Syncytial virus, Mycoplasma bovis, Mannheimia haemolytica, Porcine Reproductive and Respiratory Syndrome virus (PRRSv) Type 1 and Type 2, Porcine Circovirus Type 2 (PCV2), Porcine Parvovirus, Porcine Encephalomyocarditis virus, Mycoplasma hyopneumoniae, Pasteurella multocida tossigena, Moraxella, Staphylococcus aureus. Among the Influenzavirus genera, the test was applied to cultures of IAV strains: A/swine/Finistere/2899/1982 (H1N1), A/swine/Italy/1523/1998 (H3N2), A/swine/Italy/284922/2009 (H1N2), A/swine/Italy/14820/2011 (H1N1), A/swine/Italy/224790/2012 (H1N1), A/swine/Italy/299116/2014 (H1N1), A/swine/Italy/331397-30/2015 (H1N1), A/swine/Italy/164892/2015 (H1N2), A/swine/Italy/323339/2015 (H1N2), A/swine/Italy/252641/2015 (H1N1pdm), A/swine/Italy/14535/2015 (H3N2), A/swine/Italy/20536/2016 (H3N2), A/turkey/Italy/90302/2005 (H5N2 LPAI), A/chicken/Italy/252465/2015 (H6N8), A/chicken/Italy/1227/1986 (H9N2), A/swine/Italy/170177/2010 (H1N2), A/Swine/Italy/173301/2013 (H3N2). Besides, the RNA from the IBV strain B/Bangladesh/3333/2007 and from two human clinical samples positive for ICV were tested.
Table 1.
Primers and probe of the new Real-Time PCR method (NM).
| Oligo | Sequence | Positiona |
|---|---|---|
| IDV F | TGGATGGAGAGTGCTGCTTC | 1215–1234 |
| IDV R | GCCAATGCTTCCTCCCTGTA | 1304–1323 |
| IDV Probe | FAM- CATGTTAAACATTCCCATCAGCATTCCT −BHQ1 | 1270–1243 |
Numbering is from the sequence of D/swine/Oklahoma/1334/2011 PB1 gene, accession number: JQ922306.
A recombinant plasmid, containing the synthetic sequence of the region from 1201 to 1600 of the PB1 sequence of the influenza D virus strain D/swine/Oklahoma/1334/2011 (accession number: JQ922306), was purchased (Eurofins Genomics, Ebersberg, Germany). For in vitro single strand RNA synthesis (ssRNA), the target fragment was sub-cloned into the plasmid vector pCR®2.1-TOPO® (Thermo Fisher Scientific, Waltham, USA), downstream of the T7 RNA polymerase promoter, according to the manufacturer’s instructions. The plasmid DNA was linearized by restriction enzyme (BamH1) digestion and then subjected to in vitro transcription using the RiboMaxTM Large Scale RNA Production System (Promega, Medison, USA), according to the instruction manuals. The ssRNA concentration was determined using the M-200 NanoQuant spectrophotometer (TECAN) and by droplet digital PCR (ddPCR, QX200™ Droplet Digital™ PCR System, Bio-Rad Laboratories, Milan, Italy). The ddPCR reaction was performed using the ddPCRTM Supermix for Probes (Bio-Rad Laboratories, Milan, Italy), with the same primers and probe concentrations used in the Real-time RT-PCR reaction. For a relative comparison of analytical sensitivity of the two methods, an IDV cell culture of the strain D/swine/Italy/199724-3/2015 was serially diluted. Three replicates of 10-fold serial dilution in nuclease-free water of both plasmid and ssRNA were tested to determine the Limit of Detection (LOD) of the Real-time RT-PCR assay. The last dilution showing 100% response was accepted as LOD. For an evaluation of concordance of the two assays in field, clinical samples from respiratory tract of swine and cattle were screened for IDV presence in parallel by Real-Time RT-PCR as previously described (Hause et al., 2013) and by the new method. Concordance between tests was evaluated by Cohen’s agreement test (kappa test) using resources available at VassarStats site (http://faculty.vassar.edu/lowry/VassarStats.html). In order to deeply assess field application, clinical samples from swine and cattle, conferred to our laboratories for diagnosis of respiratory diseases, were tested for IDV presence by the NM. For confirmation, positive samples were analyzed by HM. Moreover, they were submitted to viral isolation in cell cultures as previously described (Chiapponi et al., 2010, Collin et al., 2015). Besides, sequencing of 340 nucleotides of PB1 gene was attempted for at least one positive sample from each farm of origin. Sequencing was performed on both strands by BigDye Terminator Cycle Sequencing kit v1.1 on 3500xl genetic analyzer (Thermo Fisher Scientific, Waltham, USA) using the IDV F primer and the reverse primer of HM (Hause et al., 2013). Sequences were analyzed using Lasergene software (DNAStar, Madison, USA). BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to confirm the nucleotide identity of the amplicons.
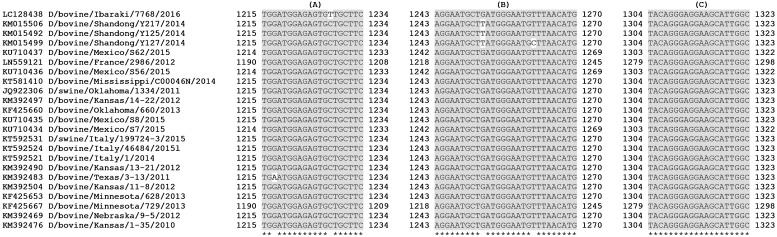
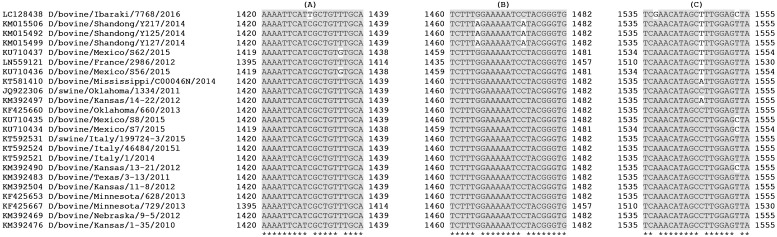
Primers and probe sequence analysis, including the new strains so far available on Genbank, has revealed that the target regions of the new assay are conserved (Fig. 1 ), even more than those of the HM (Fig. 2 ). In particular, BLAST search has demonstrated that the reverse primer is completely conserved among known strains, while the forward one presents only one internal mismatch with the sequence of the Japanese strain D/bovine/Ibaraki/7768/2016 (LC128438.1), and a further one with the strain D/bovine/Texas/3-13/2011 (KM392483.1). The probe, instead, differs by one nucleotide from strains D/bovine/Shandong/Y217/2014 (KM015506.1) and D/bovine/Shandong/Y125/2014 (KM015492.1), and by an additional one from D/bovine/Shandong/Y127/2014. Anyway, these bases are located in a central position, and the probe should be long enough to ensure the annealing at 60 °C also with these Chinese strains. Moreover, it is also remarkable that the region recognized by IDV probe is highly conserved among the Influenzavirus genera: minor substitutions are present in IAV, ICV and IBV strains. This represents an advantage for the assay, reducing the probability of mismatched sequences, difficult to detect. Indeed, the specificity of the test for IDV is assured by primers that are highly specific. Precisely, in silico analysis did not identify similar target sequences among IAV, IBV and ICV strains, included the one of swine origin: C/pig/Beijing/115/81 (Accession no. LC123406.1). Besides, the specificity of the test was extensively demonstrated in vitro: no cross reactivity was noticed with all the tested strains of IAV, IBV and ICV. Moreover, 38 of the analyzed field samples were positive for IAV and negative for IDV with the NM. Finally, none of the tested pathogens, involved in respiratory diseases of cattle and swine, gave positive results.
Fig. 1.
Nucleotide alignment of available IDV strains at primers and probe target regions of the NM. NM forward primer (A), NM Reverse primer (C) and NM Probe (B).
Fig. 2.
Nucleotide alignment of available IDV strains at primers and probe target regions of the HM. HM forward primer (A), HM reverse primer (C) and HM Probe (B).
In this study, the new method showed comparable sensitivity to the one previously described. Indeed, both were able to detect all replicates from dilution 10−1–10−7 of serially diluted viral culture, with R2 = 0.99 and slope = −3,78 for HM, R2 = 0.99 and slope −3.55 for NM. In particular NM detected 2 over 3 replicates of 10−8 dilution and HM only one, while both methods detected all replicates of dilution 10−7. Furthermore, the two tests gave similar results also with plasmid dilution series from 108copies/μL to 100 copies/μL. The limit of detection of the Real-Time PCR test was 101 plasmid copies/μL (R2 = 0.99 and slope = −3.17 for HM; R2 = 0.99 and slope = −3.11 for NM). Specifically, both assays were able to detect all replicates at 101 plasmid copies/μL and only some of those at 10 0copies/μL. Finally, the LOD of the Real-Time RT-PCR, estimated by the analysis of the quantified synthetic RNA, was 20 copies/μL for both methods (R2 = 0.99 and slope = −3.61 for HM; R2 = 0.99 and slope = −3.46 for NM). The mean Cq ± 2SD of replicates corresponding to LOD for NM was 38.11 ± 1.08. Based on these results, and considering that none of the plasmid or cell culture dilutions exceeded Cq 38, the analytical cutoff for positivity was provisionally established at Cq ≥ 39. The concordance of the two tests in field application was very high. A total of 157 field samples (19 lung tissues, 12 oral fluids, 21 nasal swabs of swine, and 27 lung tissues, and 78 nasal swabs of cattle) were analyzed in parallel with HM and NM. All results were concordant (Cohen's Kappa = 1): 18 positive samples (2 oral fluids and 7 nasal swabs from swine and 2 lung tissues and 7 nasal swabs from cattle) and 139 negative samples with both assays. To complete the field evaluation of the NM, more than 1 thousand clinical samples, 556 from cattle and 554 from swine, were tested. Among these 54 were found positive: 11 nasal swabs came from 2 pig farms, and 2 lung tissues and 41 nasal swabs were from 24 cattle farms. Sequencing was possible only when samples with a threshold cycle lower than 32 were available. Altogether 21 PB1 partial sequences were obtained. Sequence analysis confirmed all strains as belonging to IDV showing 99% of nucleotide identity towards PB1 sequence of the strain D/swine/Oklahoma/1334/2011 (JQ922306). A total of 12 isolates were achieved from positive samples detected by the new method. Moreover, all positives were confirmed by the HM.
These results demonstrate the new Real-Time RT-PCR assay is a valuable tool for diagnosis of IDV infection, suitable to detect currently known strains. It represents an advantageous alternative to the currently most diffuse method and can also be applied to confirm weakly positive results, that could not assessed by sequencing or isolation. The NM is extremely important mainly in this phase of research on IDV where is necessary to extend studies to other regions of the world and to different possible animal hosts. Due to the present scarcity of sequence data, the NM, as all PCR assays developed to date, will need to be assessed in the future, at least in silico, for the capability of recognizing all upcoming strains. When more information about sequence variability will be available the method could be implemented with a one-step RT Real-Time PCR procedure, more suitable for application in diagnostic routine.
Acknowledgements
We are grateful to Dr Paola Affanni of the Department of Biomedical, Biotechnological and Translational Sciences S.Bi.Bi.T. of the University of Parma, Italy for kindly providing IBV strain RNA. We are grateful to Prof Maria Angeles Marcos Maeso of the Section of Virology, Hospital Clínic, University of Barcelona, – Barcelona, Spain for providing RNA from ICV field samples. We thank Mrs Roberta Manfredi, Simonetta Sarzi, Giulia Donà and Verzeletti Sara for technical assistance.
References
- Chiapponi C., Zanni I., Garbarino C., Barigazzi G., Foni E. Comparison of the usefulness of the CACO-2 cell line with standard substrates for isolation of swine influenza A viruses. J. Virol. Methods. 2010;163:162–165. doi: 10.1016/j.jviromet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Chiapponi C., Faccini S., De Mattia A., Baioni L., Barbieri I., Rosignoli C., Nigrelli A., Foni E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016;22:352–354. doi: 10.3201/eid2202.151439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin E.A., Sheng Z., Lang Y., Ma W., Hause B.M., Li F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 2015;89:1036–1042. doi: 10.1128/JVI.02718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., Pelletier C., Meyer G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015;21:368–371. doi: 10.3201/eid2102.141449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L., Eckard L., Epperson W.B., Long L., Smith D., Huston C., Genova S., Webby R., Wan X. Influenza D virus infection in Mississippi beef cattle. Virology. 2015;486:28–34. doi: 10.1016/j.virol.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L., Olivier A.K., Genova S., Epperson W.B., Smith D.R., Schneider L., Barton K., McCuan K., Webby R.J., Wan X. Pathogenesis of Influenza D virus in Cattle. J. Virol. 2016 doi: 10.1128/JVI.03122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Ducatez M., Collin E.A., Ran Z., Liu R., Sheng Z., Armien A., Kaplan B., Chakravarty S., Hoppe A.D., Webby R.J., Simonson R.R., Li F. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W., Wang D., Nelson E.A., Li F. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5:e00031–14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.M., Wang S.C., Peng C., Yu J.M., Zhuang Q.Y., Hou G.Y., Liu S., Li J.P., Chen J.M. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes. 2014;49:493–496. doi: 10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–4. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N., Cernicchiaro N., Torres S., Li F., Hause B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016 doi: 10.1099/jgv.0.000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Endoh M., Kobayashi T., Takenaka-Uema A., Chambers J.K., Uchida K., Nishihara M., Hause B., Horimoto T. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg. Infect. Dis. 2016;22:1517–1519. doi: 10.3201/eid2208.160362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Kondov N.O., Deng X., Van Eenennaam A., Neibergs H.L., Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015;89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast M., Sreenivasan C., Sexton G., Nedland H., Singrey A., Fawcett L., Miller G., Lauer D., Voss S., Pollock S., Cunha C.W., Christopher-Hennings J., Nelson E., Li F. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015;180:281–285. doi: 10.1016/j.vetmic.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press Totowa; NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sreenivasan C., Thomas M., Sheng Z., Hause B.M., Collin E.A., Knudsen D.E., Pillatzki A., Nelson E., Wang D., Kaushik R.S., Li F. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J. Virol. 2015;89:11990–12001. doi: 10.1128/JVI.01630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.K., Ma W., McDaniel C.J., Gray G.C., Lednicky J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016;81:31–33. doi: 10.1016/j.jcv.2016.05.017. [DOI] [PubMed] [Google Scholar]