Abstract
In contrast to type I interferons that target various types of cells and organs, interferon lambda (IFN-L) primarily acts on mucosal epithelial cells and exhibits robust antiviral activity within the mucosal surface. Porcine epidemic diarrhea virus (PEDV), which causes high morbidity and mortality in piglets, is an enteropathogenic coronavirus with economic importance. Here, we demonstrated that both recombinant porcine IFN-L1 (rpIFN-L1) and rpIFN-L3 have powerful antiviral activity against PEDV infection of both Vero E6 cells and the intestinal porcine epithelial cell line J2 (IPEC-J2). Both forms of rpIFN-L inhibited two genotypes of PEDV (strain CV777 of genotype 1 and strain LNCT2 of genotype 2). rpIFN-L1 primarily controlled viral infection in the early stage and had less antiviral activity in IPEC-J2 than in rpIFN-L3 cells infected with PEDV. In addition, rpIFN-L1 exhibited greater antiviral activity against PEDV infection of IPEC-J2 cells than that of porcine IFN-alpha. Consistent with this finding, rpIFN-L1 triggered higher levels of certain antiviral IFN-stimulated genes (ISGs) (ISG15, OASL, and MxA) in IPEC-J2 cells than porcine IFN-alpha. Although IPEC-J2 cells responded to both IFN-alpha and lambda, transcriptional profiling of ISGs (specifically ISG15, OASL, MxA, and IFITMs) differed when induced by either IFN-alpha or rpIFN-L. Therefore, our data provide the experimental evidence that porcine IFN-L suppresses PEDV infection of IPEC-J2 cells, which may offer a promising therapeutic for combating PED in piglets.
Keywords: Interferon lambda, Interferon alpha, PEDV, Intestinal epithelial cell, IFN-stimulated genes
Highlights
-
•
Porcine IFN-lambda robustly inhibited PEDV in both Vero E6 and IPEC-J2.
-
•
IFN-lambda exhibited more anti-PEDV activity and induced a better antiviral response in IPEC-J2 than IFN-alpha.
-
•
Porcine IFN-lambda might represent a novel therapeutic agent for PEDV infection in the future.
1. Introduction
Interferons (IFNs) are the key components of innate immunity in response to viral infection. Among the three type IFNs (types I, II, and III), type III IFN-lambda (IFN-L) was recently discovered as a member of the IL-10 superfamily of cytokines (Dellgren et al., 2009, Kotenko et al., 2003, Lazear et al., 2015; review; Mordstein et al., 2010). Similar to type I IFN, IFN-L is a multigene family of closely related cytokines, consisting of four members in humans (IFN-L1, IFN-L2, IFN-L3, and IFN-L4) and two in mice (IFN-L2 and -L3) (Dellgren et al., 2009, Lazear et al., 2015; review). So far, only two members of porcine IFN-Ls (IFN-L1 and IFN-L3) have been cloned and expressed (Sang et al., 2010, Wang et al., 2011). Recently, IFN-L was shown to predominantly act in mucosal organs, including epithelial surfaces of the liver, respiratory, and gastrointestinal systems, which are major entry points for many pathogens (Lazear et al., 2015; review; Mordstein et al., 2010, Pott et al., 2011). Similar to type I IFNs, IFN-L is rapidly produced after infection and, following engagement with its receptor, induces IFN-stimulated gene (ISG) expression to mediate antiviral activity (Hernandez et al., 2015, Kotenko et al., 2003). Although type I IFN and IFN-L induce similar proximal signaling events and downstream transcriptional responses, the IFN-L receptor is structurally and genetically distinct from the type I IFN (alpha) receptor (IFNAR) (Kotenko et al., 2003, Lazear et al., 2015; review; Sommereyns et al., 2008). The IFN-L receptor is a single heterodimeric receptor complex consisting of the IFNLR1 signaling chain and the IL-10Rβ accessory chain (Kotenko et al., 2003, Sommereyns et al., 2008). IFNLR1 is preferentially expressed on epithelial cells unlike IFNAR and IL-10Rβ, each of which is expressed broadly on many types of cells and tissues (Sommereyns et al., 2008). Given that IFNLR1 is predominantly expressed on mucosal epithelia, IFN-L may provide a more focused antiviral response at mucosal barrier sites to reduce the side effects associated with a systemic pro-inflammatory immune response, such as that of type I IFN. Therefore, IFN-L may be a potential antiviral therapeutic for targeting mucosal infections.
Porcine epidemic diarrhea virus (PEDV) is an enteropathogenic alpha coronavirus and causes swine disease with huge economic relevance (Madson et al., 2014, Wang et al., 2014, Zhang and Yoo, 2016; review). PEDV primarily infects small intestinal epithelial cells in vivo, and infection causes pronounced villous atrophy that results in an acute malabsorption syndrome with symptoms of watery diarrhea, vomiting and anorexia in pigs of all ages (Li et al., 2012, Wang et al., 2014). PEDV infection causes high morbidity and mortality, particularly in newborn piglets, which results in a significant economic loss in the pig industry (Jung et al., 2015, Li et al., 2012). Although there are several commercial PEDV vaccines available in China, the PEDV endemic has persisted in China since 2011 (Chen et al., 2012, Li et al., 2012). Therefore, the development of anti-PEDV therapeutics is urgently needed. IFN-L has been shown to inhibit replication of many viruses in vitro and in vivo (Baldridge et al., 2015, Hernandez et al., 2015, Sommereyns et al., 2008). However, there are no reports regarding whether porcine IFN-L inhibits the coronavirus PEDV.
2. Materials and methods
2.1. Cells and viruses
African green monkey epithelial Vero E6 cells were used to amplify PEDV. Vero E6 cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with antibiotics (100 units/ml of penicillin and 100 μg/ml of streptomycin) and 10% heat-inactivated fetal bovine serum (FBS) (Gibco). The intestinal porcine epithelial cell line J2 (IPEC-J2) (kindly provided by Dr. Anthony Blikslager, North Carolina State University, Raleigh, NC, USA) was maintained in Dulbecco's Modified Eagle's Medium Nutrient Mixture F-12 (DMEM/F12) supplemented with antibiotics (100 units/ml of penicillin, 100 μg/ml of streptomycin, and 0.25 μg/ml of Fungizone®), 0.1 mM HEPES (Gibco), and 10% heat-inactivated fetal bovine serum (FBS) (Gibco). PEDV strain CV777 of genotype 1 (GenBank accession No KT323979) and PEDV strain LNCT2 of genotype 2 (GenBank accession No KT323980) were maintained at the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences, Harbin.
2.2. Determination of antiviral units (AUs) of rpIFN-L1 and L3
The biological antiviral activity of E. coli-derived recombinant porcine IFN-lambda (rpIFN-L)1 and 3 were prepared in our laboratory and evaluated in MDBK cells using a recombinant vesicular stomatitis virus (VSV) expressing a GFP reporter as described previously (Chen et al., 2011). The weight-activity unit (U/ml) of samples was calculated using porcine IFN-α as a reference.
2.3. Antiviral assay
To determine the anti-PEDV activity of E. coli-derived rpIFN-L1 and 3 prepared in our laboratory, Vero E6 or IPEC-J2 cells were seeded in 24-well plates and either left untreated or pre-treated with the indicated concentrations of rpIFN-L for 24 h. The cells were then infected with PEDV strain CV777 or strain LNCT2 at an MOI of 0.1 for 2 h and then cultured with medium in the absence of rpIFN-L for 36 h before harvesting the cell supernatant. For measuring the inhibition kinetics of rpIFN-L1 against PEDV infection, Vero E6 cells were stimulated or unstimulated for 24 h with 100 ng/ml of rpIFN-L1 and then infected with PEDV for 2 h followed by culturing in the absence of rpIFN-L for the indicated hours before quantifying PEDV infection. To examine the level of ISG expression in IPEC-J2 cells following IFN stimulation, the cells were stimulated with the indicated concentrations of IFNs in 24-well plates for 24 h. Cells were then lysed, and total RNA was extracted for subsequent qPCR analysis.
2.4. Real-time quantitative PCR (qPCR)
Total RNA was extracted from the cellular supernatant or cell lysates using the Simply P Total RNA Extraction Kit (BioFlux, China) according to the manufacturer's instructions. Reverse transcription was performed using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TAKARA), and qPCR was performed in a LightCycler480 Ⅱ (Roche, Switzerland) with power SYBR Green PCR Master Mix (Applied Biosystems). The thermal cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, and 60 °C for 1 min. All acquired data were obtained using LightCycler 480 real time PCR machines (Roche) and analyzed with LightCycler 480 software 1.5 based on the cycle threshold (ΔΔCT) method (Schmittgen and Livak, 2008). Primers were designed using Oligo 6 software and are shown in Table 1 .
Table 1.
qPCR primers used in the present study.
| Gene name | Primer sequences (5′—3′) | |
|---|---|---|
| qPCR primers: | ||
| PEDV S | Forward | GCAGTAATTCCTCAGATCCTC |
| Reverse | GTAGTGTCAGATGCAATGAGG | |
| MxA | Forward | CACTGCTTTGATACAAGGAGAGG |
| Reverse | GCACTCCATCTGCAGAACTCAT | |
| ISG15 | Forward | AGCATGGTCCTGTTGATGGTG |
| Reverse | CAGAAATGGTCAGCTTGCACG | |
| OASL | Forward | TCCCTGGGAAGAATGTGCAG |
| Reverse | CCCTGGCAAGAGCATAGTGT | |
| IFITM1 | Forward | TGCCTCCACCGCCAAGT |
| Reverse | GTGGCTCCGATGGTCAGAAT | |
| IFITM3 | Forward | GTCGTCTGGTCCCTGTTCAAC |
| Reverse | GAGTAGGCGAAAGCCACGAA | |
| Primers for IFITMs cloning: | ||
| pIFITM1-F | TTTGAATTCATGATCAAGAGCCAGCACGAGATGG | |
| pIFITM1-R | TTTGGTACCTAGTAGCCTCTGTTACTCTTTGCG | |
| pIFITM3-F | TTTGAATTCATGAACTGCGCTTCCCAGCCCTTCT | |
| pIFITM3-R | TTTGGTACCTAGTAGCCTCTGTAATCCTTTATGAG | |
Quantification of PEDV RNA was calculated based on a standard curve with known amounts of in vitro-transcribed PEDV RNA.
2.5. Immunofluorescence assay
Vero E6 or IPEC-J2 cells were seeded in 96-well plates, and each confluent monolayer of cells was stimulated with indicated concentrations of either type of rpIFN-L for 24 h before PEDV infection. The cells were then infected with PEDV strain CV777 at an MOI of 0.1. PEDV infection was analyzed using an immunofluorescence assay (IFA) at 36 h post-infection. Briefly, the cells were fixed with 4% paraformaldehyde at 4 °C for 30 min and washed with 1 × PBS. Fixed cells were permeabilized with 0.2% Triton X-100 for 15 min at room temperature and blocked with blocking buffer (PBS with 10% FBS) for 1 h. The preparations were labeled with the mouse anti-PEDV nucleocapsid monoclonal antibody (mAb) 2G3 stocked in our laboratory (1:100 dilution) at 37 °C for 2 h followed by labeling with the Alexa Fluor 546 goat anti-mouse IgG antibody (1:200 dilution) (ThermoFisher Scientific) for 1 h at 37 °C. DAPI (1:100 dilution) was used to stain cell nuclei. The stained cells were visualized using an AMG EVOS F1 florescence microscope.
2.6. Cloning and expression of porcine IFITM1 and IFITM3
Primers for swine IFITM1 and 3 were designed according to available sequences from the NCBI database (Genbank accession No. XM_003124230.2 for IFITM1 and NM_001201382 for IFITM3). Swine IFITM1 and 3 were amplified by PCR from cDNA obtained from IFN-stimulated IPEC-J2 cells using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TAKARA) according to the manufacturer's instructions. The PCR products of porcine IFITM1 and IFITM3 were amplified with the primers listed in Table 1, and then cloned into a pCAGGS-HA vector (Clontech) to obtain pIFITM1-HA and pIFITM3-HA using EcoRI and KpnI restriction enzyme sites. The recombinant plasmids were verified by sequencing. Expression of porcine IFITM1 and IFITM3 from the recombinant plasmids was verified by western blotting using the mouse anti-HA mAb HA-7 (Sigma).
2.7. Cell viability measurement
Cell viability was determined using the cell counting kit-8 (CCK-8) assay kit (Beyotime, Hangzhou, China) according to the manufacturer's instructions. Vero E6 or IPEC-J2 cells were seeded in 96-well cell culture plates at a density of 2 × 104 cells/well and grown at 37 °C for 24 h. Indicated concentrations of rpIFN-L1 or rpIFN-L3 were added to each well, and blank control wells (without cells and IFN) and cells only control wells (with cells but without IFN) were included. After 60 h, 10 μl of the CCK8 reagent was added to each well, and the plate was incubated for 4 h at 37 °C. The absorbance was measured at 450 nm using a microplate reader, and 50% cellular cytotoxicity (CC50) was calculated using GraphPad Prism (GraphPad Software, Inc).
2.8. Statistical analyses
All statistical analyses were analyzed in GraphPad Prism (GraphPad Software, Inc). The difference between two treatments was determined using an Unpaired T-test. Differences were considered significant if the P value was <0.05. P values are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
3. Results
3.1. rpIFN-L1 inhibits PEDV infection in Vero E6 cells and small intestinal epithelial cells
We prokaryotically expressed recombinant porcine IFN-L1 (rpIFN-L1) and rpIFN-L3 and purified each by Ni + affinity chromatography in our laboratory (unpublished results). rpIFN-L1 and rpIFN-L3 were determined to be biologically active and exhibit anti-VSV activity in MDBK cells using a VSV-GFP reporter assay. Previous studies have demonstrated that the antiviral unit (AU) of IFN is closely related to the virus and target cells used in the reporter assay; thus, we normalized the input of IFN by protein concentration instead of by AU throughout the study (Dellgren et al., 2009).
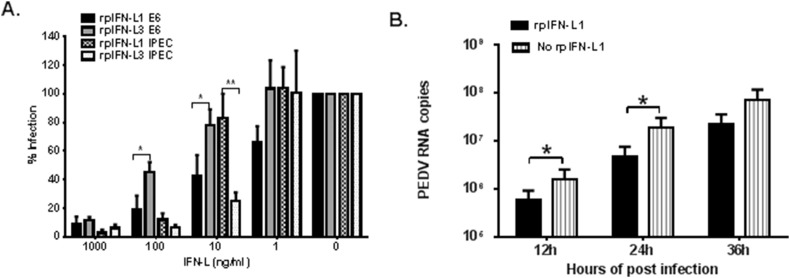
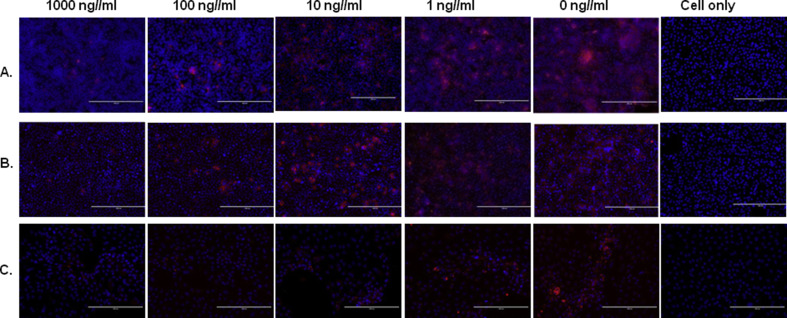
PEDV is generally grown and amplified in vitro in Vero E6 cells that have lost the capacity to produce type I IFNs due to a chromosomal deletion (Diaz et al., 1988). A previous study has demonstrated that Vero E6 cells can respond to recombinant human IFN-L1 and induce antiviral responses following human IFN-L1 stimulation (Stoltz and Klingstrom, 2010). First, we evaluated the antiviral activity of rpIFN-L1 against infection with the PEDV classical strain CV777 in Vero E6 cells. Vero E6 cells were pre-treated with increasing doses of rpIFN-L1 for 24 h prior to infection with PEDV strain CV777. rpIFN-L1 treatment potently inhibited PEDV replication in Vero E6 cells in a dose-dependent manner. By measuring PEDV viral RNA, it was found that rpIFN-L1 at concentrations of 1000 ng/ml and 100 ng/ml significantly inhibited PEDV infection. Compared with the control (1.77 × 108 copies), 1000 ng/ml and 100 ng/ml rpIFN-L1 reduced the level of PEDV viral RNA to 1.72 × 107 and 3.57 × 107 copies, respectively, resulting in more than 90% and 80% inhibition of PEDV infection, respectively. Additionally, 10 ng/ml rpIFN-L1 resulted in a 57.76% inhibition of infection in Vero E6 (Fig. 1 A). The dose-dependent inhibition of PEDV by rpIFN-L1 was further confirmed by measuring PEDV-infected cells using IFA detection of PEDV nucleocapsid (N) protein (Fig. 2 A). Next, we determined the stages of infection at which inhibition by rpIFN-L1 occurred by measuring virus production in the culture supernatant at different hours post-infection (hpi) (Fig. 1B). Pre-treatment with rpIFN-L1 at a concentration of 100 ng/ml reduced viral replication in Vero E6 cells at 12, 24, and 36 hpi (Fig. 1B). These results indicate that rpIFN-L1 exhibits antiviral activity against PEDV strain CV777 in Vero E6 cells independently of the responses induced by type I IFNs.
Fig. 1.
rpIFN-L1 and L3 inhibited PEDV (strain CV777) infection in Vero E6 and IPEC-J2 cells. Vero E6 or IPEC-J2 cells were stimulated with rpIFN-L1 or rpIFN-L3 at indicated concentrations for 24 h in 24-well plates and then were infected with PEDV at an MOI of 0.1. Vero E6 or IPEC-J2 cells were further cultured for 36 h prior to PEDV RNA quantification by RT-qPCR; B. Vero E6 cells were treated or untreated with 100 ng/ml of rpIFN-L1 for 24 h prior to infection with PEDV. The cell supernatant containing PEDV RNA was measured by RT-qPCR at 12, 24, 36, 48, 60 h post-infection. The results are presented as the mean ± SEM (N = 3). *P < 0.05; **P < 0.01 by Unpaired T test.
Fig. 2.
rpIFN-L1 and rpIFN-L3 inhibited PEDV infection both in Vero E6 and in IPEC-J2 cells by IFA. Vero E6 (A, B) or IPEC-J2 (C) cells were exposed to the indicated concentrations of rpIFN-L1 (A, C) or rpIFN-L3 (B) for 24 h prior to infection with PEDV, and then cells were infected with PEDV (strain CV777) for 2 h, maintained for 36 hpi without the presence of rpIFN-L, washed, and fixed with 4% paraformaldehyde. The virus was detected with PEDV N protein IFA. Nuclei were visualized by staining with DAPI. Scale bars represent 200 μm.
Intestinal epithelial cells of pig small intestine are the primary target cells of PEDV in vivo. We next investigated the antiviral effect of rpIFN-L1 against PEDV infection in intestinal epithelial cells in vitro using the IPEC-J2 cell line, a non-transformed, non-tumorigenic intestinal epithelial cell line isolated from the jejunal epithelium of a neonatal unsuckled piglet (Geens and Niewold, 2011, Zakrzewski et al., 2013). This cell line has been reported as an ideal in vitro model of porcine intestinal infections (Liu et al., 2010). As in the Vero E6 cell experiments, IPEC-J2 cells were infected with PEDV strain CV777 after stimulating with rpIFN-L1. At concentrations of 1000 ng/ml and 100 ng/ml, rpIFN-L1 significantly reduced infection to 2.89 ± 2.19% and 12.0 ± 4.37% of the untreated control, respectively. As observed in Vero E6 cells, pre-treatment with rpIFN-L1 reduced PEDV infection in a dose-dependent manner (Fig. 1A). These results demonstrate that rpIFN-L1 is capable of inhibiting PEDV infection in both IPEC-J2 and Vero E6 cells.
3.2. rpIFN-L3 inhibits PEDV infection in Vero E6 cells and small intestinal epithelial cells
Two members of porcine IFN-L have been reported (Sang et al., 2010); therefore, we next investigated the antiviral activity of rpIFN-L3 against PEDV infection. Analogous to treatment with rpIFN-L1, we measured the inhibitory effect of rpIFN-L3 in Vero E6 cells and IPEC-J2 infected with PEDV strain CV777 (Fig. 1A). As with rpIFN-L1, treatment with rpIFN-L3 significantly inhibited PEDV infection in both cell types in a dose-dependent manner (Fig. 1A). The inhibition of PEDV infection by rpIFN-L3 was further assessed by IFA (Fig. 2B). Compared with rpIFN-L1, rpIFN-L3 exhibited a reduced (2-fold) viral inhibitory effect in Vero E6 cells when treated with lower doses of rpIFN-Ls (18.73 ± 10.16% for rpIFN-L1 versus 45.06 ± 7.24% for rpIFN-L3 at 100 ng/ml; 42.24 ± 15.01% for rpIFN-L1 versus 77.84 ± 11.31% for rpIFN-L3 at 10 ng/ml) (Fig. 1A). Interestingly, unlike the infection of Vero E6 cells, rpIFN-L3 exhibited a more potent activity against PEDV infection of IPEC-J2 cells at lower doses than that of rpIFN-L1 (Fig. 1A). These results indicate that rpIFN-L3 substantially inhibits PEDV infection in both Vero E6 and IPEC-J2 cells in a similar manner to rpIFN-L1.
3.3. rpIFN-L inhibits genotype 2 PEDV (strain) LNCT2
There are two circulating PEDV genotypes, G1 and G2, which are based on the amino acid sequence of the PEDV spike protein (Wang et al., 2016). The classical strain CV777 is of the G1 genotype and is a Vero E6-adapted strain. Strain LNCT2, a wild isolate obtained by our laboratory, is of the G2 genotype and replicates less efficiently in both Vero E6 and IPEC-J2 cells in vitro compared with the cell-adapted CV777 strain (Wang et al., 2016). To determine whether rpIFN-L1 and rpIFN-L3 are capable of inhibiting infection with strain LNCT2, IPEC-J2 cells were infected with LNCT2 after pre-treatment with rpIFN-L1 or rpIFN-L3. Both rpIFN-L1 and rpIFN-L3 significantly suppressed infection of IPEC-J2 cells with strain LNCT2 in a dose-dependent manner, as determined by measuring viral RNA in the cellular supernatant (Fig. 3 ). Although rpIFN-L1 and rpIFN-L3 exhibited a similar level of viral inhibition at 1000 ng/ml, rpIFN-L3 more efficiently inhibited LNCT2 infection in IPEC-J2 when using concentrations of 100 ng/ml to 1 ng/ml than rpIFN-L1. These results demonstrate that rpIFN-L inhibits infection of PEDV G2 genotype in porcine intestinal epithelial cells.
Fig. 3.
rpIFN-L1 and rpIFN-L3 inhibited G2 PEDV LNCT2 infection in IPEC-J2 cells. IPEC-J2 cells were pre-treated with rpIFN-L1 or rpIFN-L3 at indicated concentrations for 24 h in 24-well plates and then were infected with PEDV (LNCT2). Cells were further cultured for another 36 h prior to PEDV RNA quantification by RT-qPCR. The results are presented as the mean ± SEM (N = 3). *P < 0.05; **P < 0.01.
3.4. rpIFN-L has greater antiviral activity against PEDV in IPEC-J2 than IFN-α
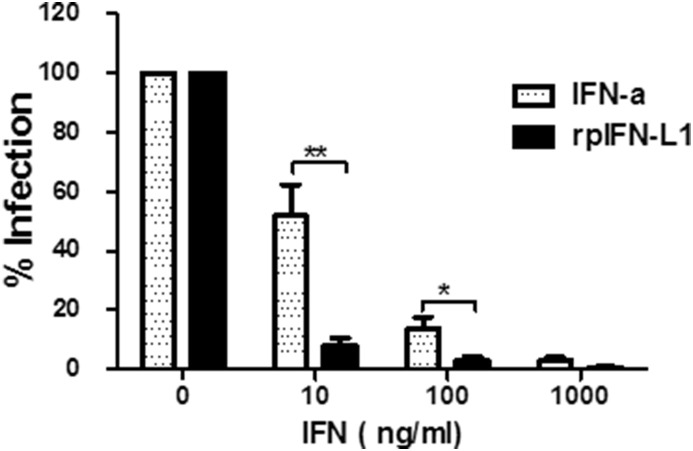
Growing evidences have demonstrated that the primary type of IFN acting in the gastrointestinal tract is type III rather than type I (Baldridge et al., 2015, Pott et al., 2011, Sommereyns et al., 2008). We compared the antiviral activity of rpIFN-L1 and porcine IFN-α against PEDV infection in IPEC-J2 cells. Although both IFN-α and rpIFN-L1 displayed antiviral effects against PEDV in IPEC-J2 cells in a dose-dependent manner, copies of viral RNA in the cellular supernatant were consistently higher in IFN-α-treated cells at concentrations of 1000 ng/ml to 10 ng/ml than in cells treated with equal concentrations of rpIFN-L1 (74,964 ± 58,768 copies for IFN-α versus 7845 ± 1196 for rpIFN-L1 at 1000 ng/ml; 1,095,148 ± 701,538 copies for IFN-α versus 140,109 ± 56,171 copies for rpIFN-L1 at 10 ng/ml), and rpIFN-L1 more significantly inhibited PEDV infection than IFN-α at 100 ng/ml and 10 ng/ml, indicating that IFN-α is less effective at inhibiting PEDV infection than rpIFN-L1 (Fig. 4 ). These results clearly demonstrate that IFN-L1 preferentially inhibits enteropathogenic PEDV in IPEC-J2 compared with IFN-α.
Fig. 4.
rpIFN-L1 displayed more efficiency to inhibit PEDV in IPEC-J2 cells than IFN-α. IPEC-J2 were treated with rpIFN-L1 or porcine IFN-a at indicated concentrations for 24 h in 24-well plates and then were infected with PEDV (CV777). PEDV infection was quantified by measuring RNA with RT-qPCR. The results are presented as the mean ± SEM (N = 3–4).
3.5. rpIFN-L inhibits PEDV infection by activating multiple mechanisms
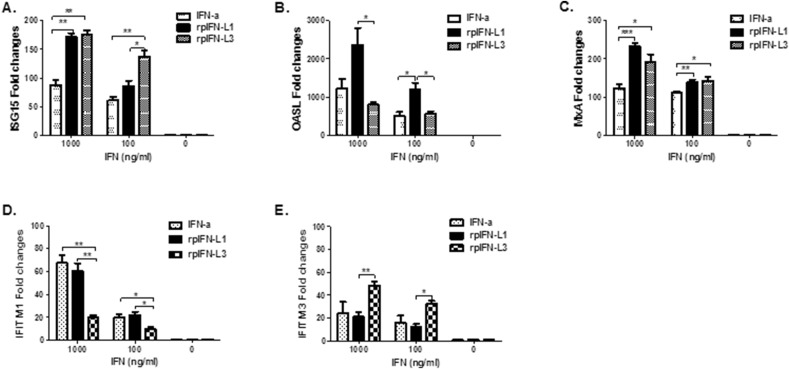
ISG15 (Lenschow et al., 2007), MxA (Kochs and Haller, 1999), and the 2′,5′-oligoadenylate synthetase (OAS)-directed RNaseL (OASL) (Melchjorsen et al., 2009) are three of the most important antiviral proteins induced by IFN. We next sought to determine whether an ISG antiviral response in IPEC-J2 was induced following rpIFN-L1 and rpIFN-L3 stimulation. The relative quantities of mRNA of 3 representative ISGs (i.e., ISG15, OASL, MxA) relative to their expression in control cells were determined by RT-qPCR. Transcriptional expression was up-regulated by approximately 171-fold for ISG15, 232-fold for MxA, and more than 2000-fold for OASL when cells were treated with 1000 ng/ml rpIFN-L1 (Fig. 5 ). The expression of the three antiviral genes exhibited a dose-dependent response to rpIFN-L1 stimulation in IPEC-J2 cells and was correlated with the antiviral activity against PEDV infection of IPEC-J2 cells. Similar to rpIFN-L1, both rpIFN-L3 and IFN-α up-regulated the expression of all three antiviral genes after 24 h stimulation of IPEC-J2 cells. Interestingly, rpIFN-L3 elicited significantly lower OASL than rpIFN-L1 (P < 0.05), even though rpIFN-L1 and rpIFN-L3 induced similar levels of MxA, and rpIFN-L3 induced significantly higher levels of ISG15 at 100 ng/ml than rpIFN-L1 (Fig. 5B and C). Consistent with a greater antiviral efficacy in IPEC-J2 cells by rpIFN-L1 compared with IFN-α, rpIFN-L1 induced greater mRNA expression of antiviral genes ISG15, OASL, and MxA than IFN-α, especially when using a high dose of 1000 ng/ml (Fig. 5). These data suggest that IPEC-J2 cells preferentially respond to rpIFN-L, although both IFN-α and rpIFN-L have antiviral activity in these cells.
Fig. 5.
rpIFN-L and IFN-α induced the expression of ISG genes in IPEC-J2 cells. IPEC-J2 were stimulated with IFN at indicated concentrations for 24 h in 24-well plates, and the mRNA of ISG15, OASL, MxA, IFITM1 or IFITM3 was measured by relative RT-qPCR. The results are presented as the mean ± SEM (N = 3–4).
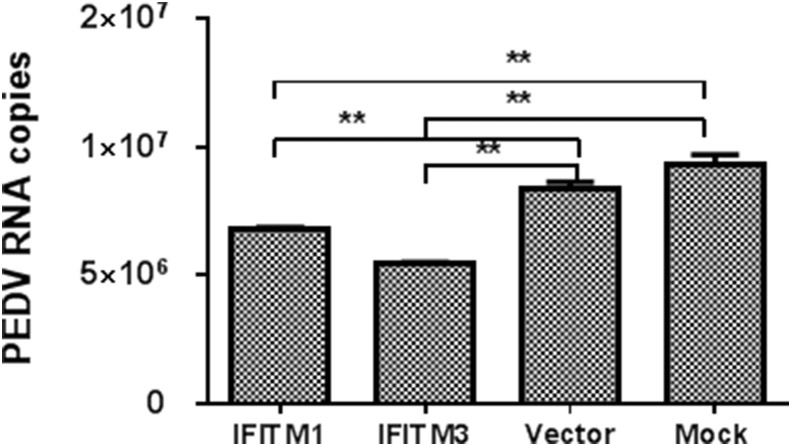
In addition, interferon-induced transmembrane proteins (IFITMs) have been recently identified as a related family of broadly potent antiviral cellular inhibitors, especially IFITM3, which exhibits robust antiviral activity against multiple enveloped RNA viruses include influenza virus, HIV-1, SARS coronavirus, and Degue virus (Brass et al., 2009, Diamond and Farzan, 2013; review). IFITMs primarily restrict the early life cycle of enveloped viruses and inhibit viral entry by blocking viral fusion. As expected, the expression of porcine IFITM1 and IFITM3 were induced in a dose-dependent manner in IPEC-J2 cells following stimulation with either rpIFN-L or IFN-α (Fig. 5D and E). Unexpectedly, unlike the expression profiling of ISG15, OASL, and MxA, IFN-α up-regulated similar levels of IFITM1 and IFITM3 expression to those of rpIFN-L1. By contrast, rpIFN-L3 induced the highest levels of IFITM3 expression among the three IFNs at both 1000 ng/ml and 100 ng/ml. These results indicate that both IFITM1 and IFITM3 are induced by IFNs but exhibit various sensitivity to the stimulation of different IFNs and that rpIFN-L1 and rpIFN-L3 induce a different profile of antiviral genes even though they use the same receptor. We further cloned porcine IFITM1 and IFITM3 into a pCAGGS-HA vector. The transient expression of IFITM1 and IFITM3 was verified by western blotting (data not shown). The transient over-expression of IFITM1 and IFITM3 in Vero E6 cells inhibited PEDV infection, especially for IFITM3, which resulted in a 40% decrease of PEDV infection (Fig. 6 ). All these results indicate that rpIFN-L induces the expression of multiple host antiviral genes to inhibit viral infection.
Fig. 6.
The transient expression of IFITMs inhibited PEDV replication. Porcine IFITM1 and IFITM3 were cloned and expressed in the eukaryotic expression vector pCAGGS-HA. Vero E6 cells were infected with PEDV (CV777) following transfection with pIFITM1-HA, pIFITM3-HA, or Vector for 24 h. The viral RNA was quantified at 36 hpi. The results are presented as the mean ± SEM (N = 3).
3.6. Cytotoxicity
To exclude the potential of cytotoxicity by rpIFN-L treatment of cells, we measured cell viability using the CCK-8 assay. No cytotoxicity was observed in the cells following IFN treatment for 60 h from 5000 ng/ml to 1 ng/ml. The percentages of CCK-8 OD values for rpIFN-L-treated Vero E6 or IPEC-J2 cells ranged from 90.69 ± 5.00% to 114.12 ± 4.36% of the controls, indicating no cytotoxicity. The CC50 of rpIFN-L1 and rpIFN-L3 was over 1.7 mg/ml, which is 1000-fold more than the concentration of rpIFN-L used in this study.
4. Discussion
Interferons are one of the most important molecules of innate immunity in response to viral infection. IFN-L is increasingly being shown to play a pivotal role in inhibiting viral infections at mucosal surfaces. However, there is no information available about whether porcine IFN-L inhibits swine enteric pathogenic coronaviruses. PEDV is an alpha coronavirus that causes economically significant swine diseases. In this study, we investigated the relative contribution of porcine IFN-L toward controlling the infection of PEDV in vitro.
IFN-L preferentially acts on epithelial cells because IFNLR1 is expressed primarily on the epithelial cells of the gastrointestinal and respiratory tracts (Mordstein et al., 2010, Pott et al., 2011, Sommereyns et al., 2008). Here we report that both rpIFN-L1 and rpIFN-L3 have robust antiviral activity against PEDV infection in both IPEC-J2 and Vero E6 cells. Recent studies have demonstrated that IFN-L plays a vital role in restricting enteric viral infections relative to type I IFN (Mordstein et al., 2010, Pott et al., 2011, Sommereyns et al., 2008). Our results of the comparison of rpIFN-L1 and IFN-α against PEDV infection in IPEC-J2 cells also demonstrate that rpIFN-L more efficiently inhibits PEDV infection in IPEC-J2 cells than IFN-α, although both inhibit PEDV infection. Consistent with these results, rpIFN-L1 induced higher levels of antiviral gene expression (ISG15, OASL, and MxA) in IPEC-J2 cells compared with IFN-α, indicating that the ISGs induced by IFN-L confer an antiviral state in IPEC-J2. Timothy J. Nice et al. demonstrated that IFN-L alone can prevent and cure the enteric persistent infection of murine norovirus (MNoV) without the help of adaptive immunity (Nice et al., 2015). The study of another virus targeting intestinal epithelia, rotavirus, confirms that IFN-L but not type I IFN inhibits rotavirus infection in vivo (Pott et al., 2011). All these imply that IFN-L preferably provides the critical antiviral defenses of the intestinal epithelium compared with type I IFN.
The IFN-L family consists of two members in mice and four members in humans (Lazear et al., 2015; review). Two members of porcine IFN-L have been reported so far (Sang et al., 2010). Although porcine IFN-L1 and IFN-L3 both belong to the type III interferon family, porcine IFN-L1 (GenBank accession no. ACZ63406.1) and IFN-L3 (GenBank accession no. NP_001159962.1) have only 52.4% amino acid identity. Among the four members of human IFN-L, IFN-L1 and IFN-L3 are studied most and show some differences in their antiviral activities (Dellgren et al., 2009, Lazear et al., 2015; review). Previous studies have shown that human IFN-L3 has the highest antiviral activity in an in vitro model of encephalomyocarditis viral infection (Palma-Ocampo et al., 2015). This finding is consistent with our results comparing the anti-PEDV activity of rpIFN-L1 and L3, as we found that rpIFN-L3 exhibited more anti-PEDV activity at lower concentrations (from 100 ng/ml to 10 ng/ml) than rpIFN-L1 in IPEC-J2 cells (Fig. 1, Fig. 3). In addition, rpIFN-L3 exhibited approximately 30-fold more anti-VSV activity in the bovine kidney MDBK cell line than rpIFN-L1 (data not shown). However, the higher anti-PEDV activity of rpIFN-L3 in IPEC-J2, compared to rpIFN-L1 is not observed in the Vero E6 infection model of PEDV. The difference in antiviral activity between Vero E6 and IPEC-J2 cells may be attributed to a difference in protein affinity of rpIFN-L1 or rpIFN-L3 to IFNLR1, given that both recombinant proteins only have a 52.4% amino acid identity. Our results indicate the importance of the specific cell type used when evaluating the antiviral activity of IFN. Therefore, pIFN-L3 exhibits a more potent, or at least similar, antiviral activity than pIFN-L1.
Type I IFN-α and Type III IFN-L induce overlapping proximal signaling events and downstream antiviral responses, despite engaging different heterodimeric receptors (Kotenko et al., 2003, Lazear et al., 2015; review; Zhang and Yoo, 2016; review). Consistent with this result, all three IFNs (IFN-α, rpIFN-L1, and rpIFN-L3) provoked the expression of the antiviral genes ISG15, OASL, MxA, and IFITM1, 3 in IPEC-J2 cells. However, the magnitude of these antiviral responses triggered by IFN-α and rpIFN-L were dissimilar, as 1000 ng/ml IFN-α stimulation resulted in an approximately 2-fold reduced expression of ISG15 and MxA compared with rpIFN-L 1 and rpIFN-L3, but IFN-α more efficiently up-regulated IFITM1 expression than rpIFN-L1 (Fig. 5). Although the different expression of IFNAR and IFNLR1 in epithelial cells accounts for the variation of the profiles of the induced antiviral genes, distinct signaling events of IFN-α and IFN-L might contribute to the different levels of antiviral genes observed in this study. Recent studies have demonstrated that IFN-L specifically activates JAK2, although both IFN-L and IFN-α/β lead to the activation of the receptor-associated tyrosine kinases JAK1 and TYK2 (Odendall et al., 2014, Odendall and Kagan, 2015; review). In addition, interestingly, the transcriptional profiles of antiviral genes elicited by rpIFN-L1 and rpIFN-L3 are not the same, even though rpIFN-L1 and rpIFN-L3 engage the same receptor (Fig. 5). Further studies are needed to identify the different transcriptional profiles of antiviral genes by different IFNs, which is vital to clarify the various antiviral effects of different IFN-L members.
Collectively, we demonstrated that rpIFN-L1 and rpIFN-L3 displayed robust antiviral activity against PEDV infection in both IPEC-J2 and Vero E6 cells. rpIFN-L up-regulated the expression of several antiviral proteins, including OASL, ISG15, MxA, and IFITMs. The transient expression of IFITM1 and 3 inhibited PEDV infection in vitro. Moreover, rpIFN-L1 provided better viral inhibition against PEDV infection of IPEC-J2 cells than porcine IFN-α. Our findings indicate that porcine IFN-L might represent a promising therapeutic agent for PED in the future.
Acknowledgments
This work was supported by grants from the Foundation of the Chinese Academy of Agricultural Sciences (302014005) and (2015ZL061).
Contributor Information
Li Feng, Email: fengli_h@163.com.
Pinghuang Liu, Email: liupinghuang@caas.cn.
References
- Baldridge M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., Diamond M.S., Ivanova Y., Artyomov M., Virgin H.W. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Cao W., Zhao H., Hu Q., Qu L., Hu S., Ge J., Wen Z., Wang X., Li H., Huang K., Bu Z. Establishment of a stable CHO cell line with high level expression of recombinant porcine IFN-beta. Cytokine. 2011;54:324–329. doi: 10.1016/j.cyto.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang J., Yu F., Ge J., Lin T., Song T. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) samples from field cases in Fujian, China. Virus Genes. 2012;45:499–507. doi: 10.1007/s11262-012-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellgren C., Gad H.H., Hamming O.J., Melchjorsen J., Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M.O., Ziemin S., Le Beau M.M., Pitha P., Smith S.D., Chilcote R.R., Rowley J.D. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geens M.M., Niewold T.A. Optimizing culture conditions of a porcine epithelial cell line IPEC-J2 through a histological and physiological characterization. Cytotechnology. 2011;63:415–423. doi: 10.1007/s10616-011-9362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P.P., Mahlakoiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Holscher C., Dumoutier L., Renauld J.C., Suerbaum S., Staeheli P., Diefenbach A. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G., Haller O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2082–2086. doi: 10.1073/pnas.96.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- Lazear H.M., Nice T.J., Diamond M.S. Interferon-lambda: immune functions at barrier surfaces and beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T., Osiak A., Levine B., Schmidt R.E., Garcia-Sastre A., Leib D.A., Pekosz A., Knobeloch K.P., Horak I., Virgin H.W.t. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Li G., Wen K., Bui T., Cao D., Zhang Y., Yuan L. Porcine small intestinal epithelial cell line (IPEC-J2) of rotavirus infection as a new model for the study of innate immune responses to rotaviruses and probiotics. Viral Immunol. 2010;23:135–149. doi: 10.1089/vim.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Magstadt D.R., Arruda P.H., Hoang H., Sun D., Bower L.P., Bhandari M., Burrough E.R., Gauger P.C., Pillatzki A.E., Stevenson G.W., Wilberts B.L., Brodie J., Harmon K.M., Wang C., Main R.G., Zhang J., Yoon K.J. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet. Microbiol. 2014;174:60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J., Kristiansen H., Christiansen R., Rintahaka J., Matikainen S., Paludan S.R., Hartmann R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., Schwemmle M., Gunther S., Drosten C., Michiels T., Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., Virgin H.W. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., Kagan J.C. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall C., Kagan J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015;12:47–52. doi: 10.1016/j.coviro.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Ocampo H.K., Flores-Alonso J.C., Vallejo-Ruiz V., Reyes-Leyva J., Flores-Mendoza L., Herrera-Camacho I., Rosas-Murrieta N.H., Santos-Lopez G. Interferon lambda inhibits dengue virus replication in epithelial cells. Virol. J. 2015;12:150. doi: 10.1186/s12985-015-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Rowland R.R., Blecha F. Molecular characterization and antiviral analyses of porcine type III interferons. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2010;30:801–807. doi: 10.1089/jir.2010.0016. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sommereyns C., Paul S., Staeheli P., Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz M., Klingstrom J. Alpha/beta interferon (IFN-alpha/beta)-independent induction of IFN-lambda1 (interleukin-29) in response to Hantaan virus infection. J. Virol. 2010;84:9140–9148. doi: 10.1128/JVI.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Zhao F., Luo R., Chen H., Xiao S. Molecular cloning, expression and antiviral activity of porcine interleukin-29 (poIL-29) Dev. Comp. Immunol. 2011;35:378–384. doi: 10.1016/j.dci.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Shi D., Shi H., Zhang X., Yuan J., Jiang S., Feng L. Immunogenicity and antigenic relationships among spike proteins of porcine epidemic diarrhea virus subtypes G1 and G2. Arch. Virol. 2016;161:537–547. doi: 10.1007/s00705-015-2694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewski S.S., Richter J.F., Krug S.M., Jebautzke B., Lee I.F., Rieger J., Sachtleben M., Bondzio A., Schulzke J.D., Fromm M., Gunzel D. Improved cell line IPEC-J2, characterized as a model for porcine jejunal epithelium. PLoS One. 2013;8:e79643. doi: 10.1371/journal.pone.0079643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Yoo D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016;226:128–141. doi: 10.1016/j.virusres.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]