Abstract
The Global Virus Network (GVN) was established in 2011 in order to strengthen research and responses to current viral causes of human disease and to prepare against new viral pandemic threats. There are now 38 GVN Centers of Excellence and 6 Affiliate laboratories in 24 countries. GVN scientists meet annually to learn about each other's current research, address collaborative priorities and plan future programs. The 2016 meeting was held from October 23–25 in Hokkaido, Japan, in partnership with the Japanese Society for Virology, the National Institute of Infectious Diseases of Japan and the Research Center for Zoonosis Control of Hokkaido University. This report highlights the accomplishments of GVN researchers in many priority areas of medical virology, including the current Zika epidemic, infections by human papillomavirus, influenza, Ebola, Lassa, dengue, HIV, hepatitis C, and chikungunya viruses, and the development of improved diagnostics and new vaccines.
Keywords: Global Virus Network, Emerging viruses, International collaborations, Medical virology
Highlights
-
•
The GVN is an international research network comprised of 38 Centers of Excellence and 6 Affiliates in 24 countries.
-
•
The 2016 Global Virus Network (GVN) Meeting was held in Sapporo, Japan from October 23–25.
-
•
New data were presented on all various aspects of medical virology, therapy, and vaccines were presented.
-
•
International collaboration is needed to develop effective viral vaccines and therapeutics.
-
•
The 2017 international GVN meeting will be held on September 25–28 in Melbourne, Australia.
1. Introduction
The Global Virus Network (GVN) was established in 2011 in order to strengthen research in response to viral causes of human disease and to prepare for new viral pandemic threats (Mann, 2011). The GVN now has 38 Centers of Excellence and 6 Affiliates in 24 countries. Network scientists meet annually to address research and collaborative priorities, learn about each member's current work and plan future programs. These international conferences have become critical platforms for the exchange of ideas (for further information, go to http://www.gvn.org).
The 2016 international GVN meeting was held from October 23–25 in Hokkaido, Japan, in partnership with the Japanese Society for Virology (JSV), the National Institute of Infectious Diseases of Japan and the Research Center for Zoonosis Control of Hokkaido University. It brought together directors of GVN Centers and their colleagues for two days of scientific presentations and discussions of emerging and re-emerging viral threats. This meeting report highlights accomplishments of GVN researchers in many priority areas of human virology, including the current Zika epidemic, infections by human papillomavirus, influenza, Ebola, Lassa, dengue, HIV, hepatitis C and chikungunya viruses, and the development of improved diagnostics and new vaccines.
The Sapporo meeting was the first GVN event held jointly with a local national virology society. More than 1000 JSV members had the opportunity to attend scientific sessions, thereby expanding opportunities for collaborative dialogue, particularly with young Japanese virologists. The main objectives of the meeting were to present and discuss current findings in medical virology, including advances in research on HIV vaccines and other important retroviruses; provide a framework to encourage collaborations among world experts; and address the GVN's annual strategy for continued development.
2. The Global Virus Network
The GVN was founded because there was no research organization independent of national governments which had the depth of expertise needed to respond promptly to new pandemic viral outbreaks and to prepare for such outbreaks. The GVN was not intended to replace organizations such as the Centers for Disease Control and Prevention (CDC) or the World Health Organization (WHO), but to complement their activities with independent opinions and expertise. In parallel, a second major goal of the GVN was to advocate for the strengthening of virology as a core discipline in infectious diseases and to help develop the next generation of virologists (DiMaio, 2014).
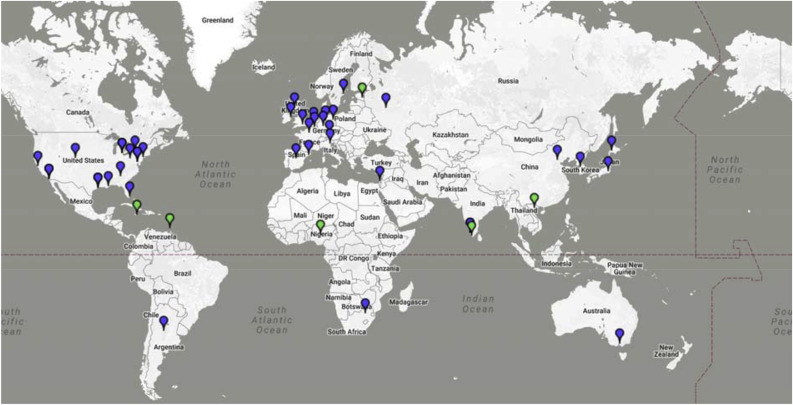
In March 2011, 30 of the world's leading virologists gathered in Washington, D.C. to pledge their support for an international coalition of virology institutions, ready to act in times of outbreaks and committed to advancing knowledge of pathogenic viruses. Today, the GVN is comprised of 38 Centers of Excellence and 6 Affiliates in 24 countries (Fig. 1 ). Centers are led by world-class virologists who have expertise in two or three areas of virology, strong publication records, a commitment to building technical and scientific capacities in resource-poor countries, and the readiness to support the GVN infrastructure.
Fig. 1.
The global reach of the GVN in 2017. Centers of Excellence are shown in blue and Affiliate sites in green. For information about Centers, Affiliates and participating scientists, go to http://www.gvn.org/.
3. The 2016 Robert C. Gallo Award
Albert Osterhaus, PhD, DVM, Director of the Research Center for Emerging Infections and Zoonoses (RIZ) at the University of Veterinary Medicine Hannover, Germany, a GVN Center of Excellence, was presented with the Robert C. Gallo Award for Scientific Excellence and Leadership for his pioneering contributions to influenza and coronavirus research as well as his contributions to advancing the GVN mission. Osterhaus was cited for his decades-long career, which has included the discovery of more than 50 new human and animal viruses and helping the WHO to combat outbreaks of MERS (Middle East Respiratory Syndrome), SARS (Severe Acute Respiratory Syndrome), and pandemic influenza. He was also praised as a leader within the GVN for his active participation in international conferences and virology training programs, for making important basic science contributions to medical virology and for translating those contributions into broad public health initiatives.
The award is named for Robert C. Gallo, MD, the Director of the Institute of Human Virology at the University of Maryland at Baltimore, and the Co-founder and Scientific Director of the GVN. Dr. Gallo and colleagues discovered the first human retroviruses, human T-cell leukemia virus (HTLV)-1 and -2; and human herpesvirus-6, which was subsequently demonstrated by others to cause roseola (Gallo, 2005). Gallo co-discovered the human immunodeficiency virus (HIV) as the cause of AIDS and together with colleagues discovered one of the first cytokines (IL-2). IL-2 later became critical to growing continuous in vitro cell-lines, and is now important in many immune therapy protocols. Dr. Gallo and colleagues also developed the first blood tests for HTLV-1 and HIV.
4. Joint GVN-JSV symposium on translational virology
Robert Gallo reviewed recent HIV vaccine trials that used four different methods. The Thai-U.S. Army trial (RV144) appeared to have given very modest protection in the early months after vaccination, while the others failed to prevent infection or in one case was associated with greater infection. Gallo explained that for the last several years, the emphasis has been on induction of antibodies to the HIV envelope component, gp120, rather than to a primary induction of cellular immunity to prevent infection. This approach was based on the logic of trying to prevent integration and establishment of a permanent infection. This effort has mainly evolved around two programs. The first was a detailed analysis of the RV144 results, particularly correlates of protection, which have mainly highlighted antibodies interacting with VIV2 gp120 epitopes and epitopes induced by gp120 interactions with CD4, referred to as CD4i Abs, which target conserved envelope regions needed for gp120 interaction with CCR5. Functional correlations were mainly with Fc effector functions rather than neutralizing antibodies (Nab). The second major approach is one that is geared toward inducing broad Nab, which is a reasonable goal, but one that is not being met, as it is proving far more difficult than supposed. Moreover, in studies in nonhuman primates (NHP), as well as the results with RV144, correlation of protection is rare with Nab, even when they are generated.
Gallo also described a candidate immunogen expressed as a chimeric protein of gp120 and CD4, which has shown some efficacy against heterologous challenges in primates, and correlates with CD4i antibodies that have Fc effector activity (Fouts et al., 2015). This vaccine is in Phase I trials (https://clinicaltrials.gov/show/NCT02756208). Like all anti-gp120 antibodies correlating with protection, these antibodies do not persist, and attempts to increase persistence have resulted in hyper-activation of T-cells, which then form more targets for HIV infection. Persistent boosting to maintain the antibodies leads to changes in the antibody isotype, which can reduce their efficacy. Based on the RV144 trial and recent NHP studies, Gallo concluded that two critical basic science problems remain to be solved for the development of a successful anti-envelope HIV vaccine: durability of the antibodies and a correct immune balance.
Yasushi Kawaguchi, at the University of Tokyo, described cell factors that are involved in a unique nuclear-pore-independent export system for macromolecular complexes in the nucleus, and which may be potential targets for novel anti-herpetic drugs. Vesicle-mediated nucleocytoplasmic transport is a unique mechanism for nuclear export of macromolecular complexes in which a complex in the nucleus buds through the inner nuclear membrane to form a vesicle in the perinuclear space (primary envelopment). The vesicle then fuses with the outer nuclear membrane to release the complex into the cytoplasm (de-envelopment). This type of transport is observed in herpesvirus-infected mammalian cells for export of viral nucleocapsids, but is not common in other types of cells, indicating that herpesviruses may expropriate this mechanism (Oda et al., 2016). However, the cellular mechanism for vesicle-mediated nucleocytoplasmic transport remains largely unknown. He went on to describe recently identified host cell proteins involved in this unique nuclear export system, and how these proteins could be targets for novel anti-herpetic drugs, since this transport system is essential for the life cycle of herpesviruses and is unique in biology.
Hideki Hasegawa, a GVN center director at the National Institute of Infectious Diseases in Tokyo, explained that secretory IgA antibodies on mucosal surfaces play an important role in protection against influenza virus infection. Secretory, polymeric IgA antibodies induced by intranasal, inactivated influenza vaccine have higher neutralizing and cross-neutralizing ability against homologous and heterologous influenza viruses compare to monomeric IgA antibodies in humans (Suzuki et al., 2015). A method of producing secretory multimeric IgA antibodies in vitro was established recently, and Hasegawa and colleagues examined the improvement of neutralizing effects by multimerization using monoclonal IgA antibodies. The method of in vitro production of multimeric IgA antibodies was established by introducing cDNA constructs of H, L, and J chains of IgA with secretory component (SC) into Expi293F cells. This method allowed production of large amounts of recombinant monoclonal polymeric IgA antibody. The antibody-coding cDNA was isolated and cloned from single plasma cell from the peripheral blood mononuclear cells of a subject previously vaccinated intranasally with inactivated influenza A (H5N1) vaccine. Monomeric and multimeric IgA antibody specific for influenza virus was prepared, and functional analysis—such as ELISA and neutralization tests—was performed. Similar to secretory IgA antibodies in human nasal washes, the recombinant IgA showed that polymerization of the antibody molecule enhanced the neutralizing activity. Mass spectrometry analysis found that the antibodies had a molecular weight of 720 kDa, suggesting they consisted mostly of the tetramer form. These tetrameric IgA antibodies may be clinically useful as mucosal agents for the prevention and treatment of influenza.
Erica Ollmann Saphire, a GVN center co-director at the Scripps Research Institute (USA), reported on recent outbreaks of Ebola virus disease and other viral diseases and the urgent need for novel therapeutics. Monoclonal antibodies offer a promising solution to the lack of currently available therapies, but it is often unclear how to select the most effective antibodies or combinations for optimal therapy (Saphire and Aman, 2016). Integrated analysis of a large array of antibodies provides a rational basis for the selection of optimized therapeutics, and can lead to rapid understanding of the mechanisms underlying protective efficacy. She described the results of a field-wide analysis of antibodies against Ebola virus, a particularly potent antibody against Marburg virus, and a novel panel of antibodies against Lassa virus. A newly formed international group, the Viral Hemorrhagic Fever Immunotherapeutic Consortium, aims to galvanize and focus efforts against Ebola and similar viruses, using multidisciplinary approaches (Saphire et al., 2017).
Sharon Lewin, a GVN center director at the Doherty Institute for Infection and Immunity (Australia), reported on HIV cure research (Lederman et al., 2016). HIV persists predominantly in long-lived resting CD4+ T cells that are found in blood and at greater frequency in tissue. HIV can also persist in specific T-cell subsets enriched in certain tissue sites, including T follicular helper cells in lymph nodes and Th17 (CCR6+) cells in the gastrointestinal tract. Long-lived infected macrophages, specifically microglia in the brain or Kupffer cells in the liver, may also play a role, though this remains controversial. On ART, HIV-specific T cells express multiple exhaustion markers, and these cells are often unable to recognize and kill infected cells due to multiple immune escape mutations that are archived in long lived latently infected cells. Furthermore, in some tissue sites such as B-cell follicles in lymph nodes, there is limited penetration of HIV-specific T-cells.
Lewin described several approaches that are being tested to achieve HIV remission, including reducing latently infected cells through early ART or latency reversal, boosting immune clearance, reducing immune activation and homeostatic proliferation, eliminating tissue reservoirs, and gene therapy to make cells resistant to HIV. Recently completed clinical trials of latency-reversing agents (LRA) in HIV-infected patients on ART have included histone deacetylase inhibitors (HDACi) and high-dose disulfiram (Delagrèverie et al., 2016). Other interventions, including activation of protein kinase C (PKC) with bryostatin and ingenols, are under evaluation. Studies of HDACi and disulfiram have demonstrated that HIV transcription can be activated in vivo with varying efficiency, but to date there is no evidence that these interventions alone can clear latently infected cells. In addition, recent in vitro studies have shown that activating transcription with HDACi, but not other LRAs, may lead to profound changes in HIV RNA splicing, therefore limiting potency. Other concerns regarding HDACi include short- and long-term toxicity, changes in host gene expression and adverse effects on immune function. More potent and specific LRAs are necessary and in development for use alone and in combination. Additional interventions to promote cell death may be needed.
Targeting BCL2 or inhibitors of anti-apoptotic proteins (IAP) is being evaluated in vitro, and immune-mediated clearance with bi-specific antibodies, broadly neutralizing antibodies and T-cell vaccines are in clinical development. However, even if the number of latently infected cells is reduced, long-term immune control will be required through therapeutic vaccination and/or immunomodulation. Immune checkpoint blockers, including antibodies to CTLA-4 and PD-1 that have recently been licensed for the treatment of melanoma, are potentially attractive tools to enhance HIV-specific T-cells and reverse latency. Clinical trials of these agents are now starting in HIV-infected individuals with malignancies.
5. Scientific presentations
5.1. Hepatitis C
Shyamasundran Kottilil, at the Institute of Human Virology (USA), described the challenges of current treatments of chronic hepatitis C. Eighty-five percent of HCV infections become chronic; 20–30% of those cases will develop cirrhosis, and 5–10% of these patients will progress to hepatocellular carcinoma and/or decompensation of the liver, leading either to death or liver transplantation.
In contrast to chronic hepatitis B or HIV infection, chronic HCV infection can be cured with direct-acting antiviral (DAA) treatment. “Cure” is defined in terms of sustained virologic response (SVR), which is the absence of detectable levels of HCV RNA in the plasma after 12 or 24 weeks of DAA treatment (Jazwinski and Muir, 2011). Current HCV inhibitors act against the NS3/4 protease (e.g., asunaprivir and grazoprevir), the NS5A protein (e.g., daclatasvir and velpatasvir) and the NS5B polymerase (e.g., sofosbuvir and beclabuvir). Treatment with a combination of the two inhibitors, sofosbuvir and velpatasvir, for 12 weeks, will result in a cure of more than 99% of treated patients. Still, clinical challenges remain, including treatment of patients with end-stage liver and renal disease owing to metabolism and/or toxicity of the drugs. Other challenges include the treatment of HIV/HCV co-infections, injection drug users, pediatric patients and maternal-fetal transmission and pregnancy.
Estaban Domingo, at the Centro de Biología Molecular Severo Ochoa in Spain, presented studies on the population dynamics of HCV after antiviral interventions in infected human hepatoma Huh-7.5 cells. Unexpectedly, serial passage of HCV in these cells in the absence of antiviral drugs resulted in increased resistance to inhibitors that target viral or cellular proteins, and was associated with a fitness gain of the virus.
If DAA-resistance mutations become widespread in human populations, alternative drug treatments will be needed. One such drug is favipiravir (Furuta et al., 2013). This is a broad-spectrum antiviral agent active against numerous RNA viruses, including influenza, norovirus, rabies, and Rift Valley fever virus. It acts as a lethal mutagen (de Ávila et al., 2016). Efficacy and safety trials of favipiravir for treating influenza, as well as Ebola virus disease, already have been done. In cell cultures], the efficacy of favipiravir against HCV was found to be comparable to its efficacy against other RNA viruses. In HCV-infected Huh-7.5 cells, favipiravir demonstrated features typical of lethal mutagenesis. It was concluded that favipiravir offers an alternative for patients chronically infected with HCV.
5.2. Special session
Research on biomarkers of progression in human papillomasvirus (HPV)-related cancers was highlighted in this session by Franco Buonaguro (National Cancer Institute, Italy). Buonaguro described persistent infections with high-risk HPVs that are often associated with progression to mucosal cancers in anogenital as well as oropharyngeal areas of the body. He also noted that for cervical cancers, for which screening programs are available, it is difficult to identify the lesions likely to progress to invasive cancers. Given that only 1 in 500 HPV16-positive cervical dysplasias will eventually progress further, biomarkers clearly are needed. Tumor progression is characterized by increased expression of the viral E6 gene and E6-dependent degradation of p53, and increased expression of E7, which is known to bind and inactivate pRb; and integration of viral DNA into the host genome with the consequent disruption of the E2 viral gene (Annunziata et al., 2012).
Molecular markers able to identify viral infections associated with progressing cervical neoplasia are strongly needed for screening and triage. In particular, predictive biomarkers are needed to detect lesions at high risk of recurrence or progression, in order to implement appropriate treatment and to avoid over-treatment of patients with a high probability of regression. To achieve such goals, expression profile analysis of p53-related genes in HPV-16-positive genital carcinomas, along with autologous non-tumor tissue, was performed, and significant differences in the expression levels of genes involved in regulation of apoptosis, cell cycle, proliferation and DNA repair pathways were identified. Moreover, oncogenic driver mutations in critical genes (such as PIK3CA and TP53), recurrent chromosomal gain/loss, and oncogene activation by HPV integration have all been recognized to cause deregulation of cell signaling pathways and cancer progression (Tornesello et al., 2014). More recently, analysis of the regulatory regions of human genes encoding for cell proliferation proteins has shown a high frequency of mutations in lesions associated with less oncogenic HPV genotypes, and even in HPV-negative lesions. Validation of these candidate biomarkers is currently under way on a larger number of cases, including different grades of HPV-related neoplastic lesions (CIN1-3 and invasive cervical cancer). Such studies will contribute to the development of new tools for the identification of premalignant lesions at high risk of progression to invasive cervical carcinoma.
Jonathan Gershoni, a GVN center director at Tel Aviv University (Israel), described an analysis of the vast repertoire of antibodies circulating in polyclonal serum (the “IgOme.”) He highlighted how the paratope region of an antibody mirrors the corresponding region of an antigen (e.g., a virus protein) and therefore favors an indirect approach of IgOme profiling by describing all peptides interacting with the entity of antibodies. The spectrum of antibody specificities is dynamic and varies with age, physiology, and exposure to pathological insults. The IgOme is an extraordinarily rich source of information–a molecular record of all previous encounters, as well as a status report of current immune activity (Weiss-Ottolenghi and Gershoni, 2014). The ability to profile antibody specificities of polyclonal serum at very high resolution has been an important and serious challenge, which can now be met by probing, recording, and qualifying the archive of antibodies present in the serum of an individual with a vast library of peptides.
Gershoni also described “Deep Panning”, a methodology that merges the flexibility of combinatorial phage-display peptide libraries with the power of next-generation sequencing to enable high-resolution/high-throughput interrogation of the IgOme (Ryvkin et al., 2012). This approach produces heat-maps that characterize the interaction between the immune system and antigens. This method was used to characterize and differentiate sera from HIV-infected individuals versus HCV-infected individuals. By analyzing the immune response to influenza antigens in birds, Jonathan showed that this technique could be used to monitor and characterize the efficiency of vaccines, suggesting that IgOme analysis may become a very useful tool for vaccine development.
5.3. Arboviruses
Since the appearance of chikungunya virus (CHIKV) in the Western Hemisphere in 2014, the GVN has had a special focus on arboviruses. In 2014, the Network sponsored a joint symposium with the American Society of Tropical Medicine and Hygiene on “The Global Spread of Chikungunya: Epidemiology, Evolution, Pathogenesis and Global Needs” and published a special report describing research goals and public health priorities for the outbreak (McSweegan et al., 2015). A task force of international experts also was formed to provide a source of expert information and a platform for collaboration.
Marc Lecuit at the GVN center at the Institut Pasteur in Paris described CHIKV, its geographical spread, the clinical presentation of infection and the development of a mouse model that has allowed pathophysiology studies and the evaluation of novel therapeutics. Murine fibroblasts of skeletal muscles, joint capsules and dermis were found to be primary viral targets, and type I IFN-sensing, non-hematopoietic cells were critical for innate immune control. A genome-wide siRNA screen identified 156 proviral and 41 antiviral host factors affecting viral replication, which allowed identification of potential drug targets. Twenty-one small molecule inhibitors that affect six host pathways have been identified; three showed prophylactic effects in the mouse model. A calmodulin inhibitor (pimozide) and a fatty acid synthesis inhibitor (TOFA) showed antiviral effects in mice when used together (Karlas et al., 2016). Other experimental data showed that CHIKV infection does not directly target the placental barrier, therefore vertical transmission from viremic mothers to neonates likely occurs via labor-induced placental breaches. CHIKV also was found to be a significant cause of CNS disease in larger human outbreaks, with the virus detected in affected tissues (Gérardin et al., 2016).
Scott Weaver, Chair of a GVN task force on Zika virus (ZIKV), reviewed the sudden emergence of ZIKV from relative obscurity to global menace. Serosurveys have shown its historical incidence in parts of Asia and Africa (Weaver et al., 2016). A widespread outbreak in the Micronesian island of Yap in 2007 later expanded to neighboring islands in South Pacific. Coincident with these outbreaks, a 20-fold higher incidence of Guillain-Barre syndrome (GBS) was noted. The explosive outbreak in Brazil that began in 2015 and has involved millions of people has since spread to 47 other countries and territories in the Western Hemisphere. The peridomestic mosquito, Aedes aegypti, and the invasive Aedes albopictus mosquito have been identified as the major insect vectors. Surprisingly, sexual transmission of ZIKV also was confirmed in travelers returning to non-endemic regions. A most disturbing aspect of the recent outbreaks is the high incidence of fetal microcephaly and other birth defects directly related to infection in pregnant women.
Weaver also discussed important knowledge gaps. When and how should infants be tested for possible congenital Zika infection? Is microcephaly among some neonates just the tip of the neurological iceberg? Why does the overall risk of microcephaly and other birth defects seem to differ in other parts of Latin America and the Caribbean, compared to Brazil? It appears that these new outbreaks in the Western Hemisphere have not been caused by novel viral strains. Instead, the introduction of ZIKV into naïve populations via rapid global travel, the growth of tropical cities, and changing vector populations seem to have allowed virus amplification and subsequent epidemic spread. Prospects for control are dependent on mosquito control, new therapeutics and development of an effective vaccine. Vaccine prospects appear good, with about 40 candidates now in development and animal models available for preclinical testing. Two DNA-based vaccines have entered Phase I trials, and an inactivated vaccine is nearing Phase I. Deploying a vaccine, however, remains dependent on developing an accurate test for ZIKV. Current tests are hampered by antigenic cross-reactivity between ZIKV and other closely related flaviviruses such as dengue, yellow fever, and St. Louis encephalitis, some of which are endemic in the same geographic areas.
Richard Scheuermann, GVN center co-director at the J. Craig Venter Institute (USA), discussed using the Influenza Research Database (IRD, fludb.org) and the Virus Pathogen Resource (ViPR, viprbrc.org) to identify diagnostic peptide regions in Zika virus (ZIKV) and other flaviviruses. These databases identified 76 and 82 diagnostic amino acid sites from the ZIKV NS1 and E proteins, respectively, that distinguish it from other flaviviruses, with sensitivity/specificity above 98% (Sun et al., 2017). Similar numbers of such diagnostic sites were also identified for other flaviviral species. Sliding window analysis revealed several contiguous peptide regions that contain multiple diagnostic sites specific for each of the different flaviviruses. These regions are now being used to develop peptide arrays for the detection of antibodies specific for each of the viruses including ZIKV. These peptide arrays could have a significant impact on diagnosis and epidemiological analysis of future disease outbreaks.
Diane Griffin at the GVN center at Johns Hopkins University (USA) noted that insect-borne viruses causing encephalitis have become a global health concern due to their recent expansion into new regions. While the fatality rates vary, many patients recovering from serious episodes of viral encephalitis are likely to have lifelong physical and mental disabilities. Viral persistence and immune-mediated clearance are important factors in these sequelae. Griffin described experiments with Sindbis virus—a prototype alphavirus—using a variety of mouse models (Griffin, 2016). Infection of neurons in adult mice resulted in non-fatal encephalomyelitis. Recovery was mediated through a coordinated effort between antibody to the E2 viral surface protein and IFN-γ. While IFN-γ signaling leads to clinical disease via pro-inflammatory cytokines, it synergistically helps clear the virus by increasing B-cell-attracting chemokine production for recruitment of antibody-producing cells to the central nervous system (CNS). However, the non-cytolytic immune phenomenon does not lead to elimination of intracellular viral RNA from long-lived neurons, so that continued suppression of virus and the prevention of reactivation and reemergence in the CNS require immune control after recovery. Evidence was presented that B and T cells that infiltrated during the acute phase of infection remain there after recovery and continue to exert their protective effect locally. Long-term residence of these immune cells in the CNS is likely facilitated by the continued presence of viral proteins.
Arthropods make up the largest phylum of the animal kingdom and are known to harbor and transmit viruses of agricultural, veterinary and medical importance. The true extent of the ‘virosphere’ of arthropods, however, is largely unknown and needs to be more completely explored. Yong-Zhen Zhang, an investigator at the National Institute for Communicable Disease Control and Prevention in Beijing, China, presented work on newly identified RNA viruses from arthropods collected in different regions of China. Among recent examples are Jingmen tick virus (JMTV), and viruses belonging to a new family, Chuviridae (Qin et al., 2014). JTMV has four genomic segments, and, sequence analysis has revealed an unexpected connection between segmented and non-segmented viruses. Chuviruses exhibit various genomic organizations, including non-segmented, bi-segmented, and circular genomes. These viruses provided evidence of an evolutionary link between segmented and non-segmented negative strand viruses. Overall, many novel viruses representing five new families, have been discovered in China and await further characterization.
5.4. Vaccines
Peter Palese, a GVN center director at the Icahn School of Medicine at Mount Sinai (USA), described ongoing efforts to design a universal influenza vaccine and to understand the immune responses to such constructs. He said that, despite the availability of FDA-approved vaccines and antivirals, seasonal and pandemic influenza remain a serious threat associated with substantial morbidity and mortality. While annual seasonal influenza virus vaccination is effective – albeit underutilized in most countries – a safe, universal vaccine providing broad and long-lasting immunity would represent a major breakthrough. Today's licensed vaccines focus on eliciting humoral immunity to the globular head (HA1) of the hemagglutinin (HA) glycoprotein, an antigen subject to antigenic drift, which requires continued surveillance of circulating influenza viruses. Since the 2009 H1N1 pandemic, research efforts on universal vaccines have refocused, looking to shift immunity to the more conserved domains of HA, which is expected to cover many more circulating influenza viruses.
Palese also has developed vaccine constructs that express chimeric hemagglutinins (cHA) that result in the redirection of the immune response away from the immunodominant (variant) HA1 head domain towards the much more conserved stalk (HA2) region, and which also express the highly conserved neuraminidase (NA) (Tran et al., 2016). This type of immunogen has been demonstrated to be efficacious following vaccination in mice, conferring hetero-subtypic protection against virus challenge. The mechanism by which this approach confers antiviral activity suggests that re-directing immunity to the stalk region preferentially supports induction of antibody-dependent cell-mediated cytotoxicity (ADCC). Data presented showed that broadly neutralizing HA stalk-specific antibodies require FcγR interactions for protection. Palese proposed a two-contact model for optimal induction of ADCC by influenza virus-specific mAbs targeting the stalk region—and not the globular head—and he noted the importance of the HA binding to effector cells via its sialic acid receptor was required for optimal ADCC induction. Passive transfer studies in mice further supported the role of H1-specific stalk antibodies and protection against weight loss following virus challenge.
This novel technology is being supported through government and industry support, and GMP-quality batches of two cHA immunogens (cH5/1N1 & cH8/1N1) that display heterogenous globular heads have been produced for evaluation in the clinic. Such studies will make it possible to assess the role of additional effector mechanism in protecting against influenza virus infection and to evaluate the importance of reducing the immunodominance of the variable HA head domain. This type of translational study will bring us a step closer to a universal influenza vaccine.
5.5. Zoonoses
Hiroshi Kida at Hokkaido University in Japan reported on the establishment of a library of 2900 avian influenza virus strains isolated from ducks, and reassortant viruses generated in the laboratory, with 144 combinations of the nine NA and sixteen HA subtypes (virusdb.czc.hokudai.ac.jp). Because influenza A viruses of all known subtypes can perpetuate among migratory ducks and contribute genes to the generation of reassortant viruses in pigs, none of the 144 combinations can be ruled out as possible future pandemic strains. This virus library is kept as a source for vaccine candidates to assure effective preparedness for future pandemics (Haredy et al., 2013). Vaccine constructs prepared from H1N1, H5N1, H6N2, H7N7, H7N9 and H9N2 viruses in the library elicited sufficient immune responses to protect chickens, mice, and macaques from challenge with isolates from poultry and humans, suggesting their utility against pandemic influenza.
In addition, he noted that current seasonal vaccines prepared by ether or detergent disruption are not sufficiently immunogenic, especially in children and the elderly, and need to be significantly improved. Furthermore, methods for controlling pandemic influenza should be based on measures that are used for the control of seasonal influenza. Hiroshi noted that inactivated whole virus-based vaccine candidates might provide solutions to the limitations of current vaccines. To accomplish the goal of a new and effective influenza vaccine, a collaborative research group, the All Japan Collaborating Study Group For the Development of Influenza Vaccines, was established in April 2015. Five Japanese manufacturers of influenza vaccines participate in the program. The goals of the study group are: to compare the immunogenicity and safety of whole virus particle vaccine and “split” vaccines; use the results of pre-clinical and clinical trials to revise the standard of influenza vaccines for human use; and determine the optimum route of inoculation and evaluate proprietary adjuvants.
Massimo Palmarini, a GVN center director at the MRC-University of Glasgow Center for Virus Research (Scotland), reported on studies of bluetongue, a major infectious disease of ruminants, caused by the double-stranded RNA virus known as bluetongue virus (BTV). Studies of BTV in sheep offer unique perspectives for understanding the pathogenesis of arboviral diseases, as observations made in the naturally occurring disease can be effectively reproduced in a convenient experimental setting, using the same animal species (Caporale et al., 2014). The clinical outcome of BTV infection in sheep is extremely variable, but similar to other arbovirus infections, in that a rapid onset of the antibody response correlates with a more favorable clinical outcome. BTV infection of its natural sheep host was used to examine a previously uncharacterized mechanism adopted by an arbovirus to manipulate host immunity at the early stages of infection. BTV is transported rapidly via the lymph to the peripheral lymph nodes, where it infects and disrupts follicular dendritic cells (FDC). These cells of stromal origin promote the formation and maintenance of the germinal centers in which B cells differentiate into memory cells and plasma cells, and are also responsible for supporting antibody class-switching and affinity maturation. This work showed that BTV hindered B-cell division in germinal centers, resulting in the delayed production of high-affinity and virus-neutralizing antibodies. Importantly, the humoral immune response to a second antigen is also hampered in infected sheep. Thus, an arbovirus can evade the host antiviral response by inducing an acute immunosuppression. Although transient, this immunosuppression occurs at the critical, early stage of infection, when a delayed host humoral immune response likely affects the systemic dissemination of the virus and the clinical outcome of disease (Melzi et al., 2016).
Ab Osterhaus, the GVN center director at the Research Center for Emergin, reviewed the complex relationships among human and animal species that have promoted cross-species transmission, emergence and the eventual evolution of a plethora of human pathogens. Changes affecting modern human populations worldwide and their dramatic impact on the global environment have taken domestication, agriculture, urbanization, and industrialization to unprecedented levels. This has created new and global multi-faceted human-animal interfaces, associated with major epidemiological transitions, accompanied by an unexpected surge of emerging and re-emerging human infectious diseases that have their origin in animal reservoirs.
Until the beginning of the last century, infectious diseases were the major cause of human mortality. Around 1900, infections caused about fifty percent of deaths in the western world, but in the following decades, this percentage decreased to less than a few percent. This was largely due to the implementation of public health measures such as the installation of sewers and the development of clean drinking water systems, but also to development of vaccines and antimicrobials. Major successes in this regard were the eradication of smallpox and rinderpest through well-orchestrated vaccination campaigns in humans and cattle, respectively. These successes prompted policymakers and scientists to predict that infectious diseases of humankind and of their domestic animals would eventually be brought under control in the industrialized world.
Paradoxically the following decades confronted the world with an ever-increasing number of emerging or re-emerging infectious diseases, some causing true human or animal pandemics. Pathogens spilling over from wildlife reservoirs, either directly or via intermediate hosts, were the basis of most of the outbreaks. Striking examples in humans were the emergence of AIDS from chimpanzees, avian flu from migratory birds, and SARS, MERS, and Ebola virus disease from bat reservoirs. A complex mix of predisposing factors in our globalizing world, linked to major changes in the societal environment and global ecology, collectively created opportunities for viruses and other pathogens to infect and adapt to new animal and human hosts. This paved the way for the unprecedented spread of infections, with dramatic consequences for public and animal health, animal welfare, food supplies, economies, and biodiversity. It is important to realize that, because of the complex and largely interactive nature of the predisposing factors, it is virtually impossible to predict the next pathogen threat, where it will come from and when it will strike. However, a better understanding of the underlying processes may eventually lead to enhanced predictive capacity, improving preparedness for outbreaks in humans and animals. Importantly, the increased emergence of viral infections has been largely paralleled by medical, veterinary, technological, and scientific progress. Investments to better understanding human-animal interfaces should offer a future head start in the struggle against infectious diseases of humans.
Ayato Takada at the Global Institution for Collaborative Research and Education at Hokkaido University in Sapporo, Japan, described an Ebola virus glycoprotein-specific monoclonal antibody (mab 6D6) that was broadly cross-reactive with all known Ebola virus species. This mab recognized the putative epitope in the highly conserved internal fusion loop (IFL) and neutralized infectivity by inhibiting membrane fusion. Mab 6D6 may, therefore, have potential as a therapeutic agent in animal models. He also described a novel antibody-dependent enhancement (ADE) mechanism in which Fc receptor-mediated intracellular signaling increased uptake of Ebola virus into cells in vitro (Furuyama et al., 2016). Activation of the Fc-receptor-mediated signaling pathway was essential for ADE of Ebola virus infection; this finding provides new insights into mechanisms of ADE and the development of treatments for ADE-associated diseases. Finally, he noted that neutralizing and ADE antibodies show balanced effects depending on total antibody concentration, raising theoretical concerns about the use of convalescent human sera or plasma with low antibody titers. Therapeutic treatment with convalescent sera with in vitro neutralizing activity was not sufficient to protect against Ebola virus infection in nonhuman primates (Mire et al., 2016). It may be that ADE antibodies counterbalanced the neutralizing capacity of other antibodies. These laboratory phenomena warrant further evaluation.
5.6. Retroviruses
Luc Willems, a GVN investigator at Gembloux Agro-Bio Tech in Belgium, described how bovine delta-retroviral ribonucleic acids induce cell transformation and oncogenesis. Among the strategies developed by viruses to escape the host immune response, mechanisms involving viral non-coding RNAs have recently been discovered. Viruses can express microRNAs that directly target cellular transcripts, while others produce long non-coding RNAs acting as microRNA sponges or epigenetic modulators. These strategies, based on ribonucleic acids, control the cell's fate without eliciting immune responses due to the absence of viral proteins. Virally encoded miRNAs were first identified in DNA viruses. Most of them are generated from endonucleolytic cleavage of long viral transcripts. However, retroviruses were initially assumed not to encode miRNAs because of the potential self-cleavage of their RNA genome (Gillet et al., 2016).
To date, four retroviruses -- bovine leukemia virus (BLV), bovine foamy virus (BFV), avian leucosis virus (ALV-J) and simian foamy virus (SFV) – have been shown to express viral miRNAs in infected cells. BLV naturally infects bovine species to cause a benign lymphocytosis, but approximately 5% of infections lead to B-cell leukemia/lymphoma. Remarkably, BLV persists and replicates in vivo in the absence of significant levels of viral mRNA transcription. In contrast, BLV miRNAs are highly expressed in infected cells, representing up to 40% of all cellular miRNAs. Consequently, because ribonucleic acids likely are less immunogenic than viral protein antigens, BLV miRNAs could alter the cell's fate by escaping immune recognition (Gillet et al., 2016).
6. The network in 2016
As with the 2014 appearance of CHIKV in the Western Hemisphere, the sudden emergence and dissemination of ZIKV have driven similar cooperative responses at the GVN, including the formation of a second task force of experts to exchange information and share resources; the organization of a webinar for business leaders to inform them about the virus and prospects for therapies and vaccines (http://www.btsmeetings.com/zika); providing expert information to the news media to ensure accurate reporting; and acquiring a grant from the Allergan Foundation to establish a bank of acute and convalescent sera from confirmed ZIKV patients to aid in developing better diagnostics.
Among other accomplishments, Shyamasundran Kottilil and colleagues at the Institute of Human Virology in Baltimore began a project with the GVN, and funded by the Gilead Foundation, to develop a training model for physicians using DAA treatment for HCV patients in India. Members of the HTLV-1 task force published a commentary on the need for HTLV-1 screening prior to organ transplant, and an agenda for future research (Gallo et al., 2016, Willems et al., 2017). Other GVN investigators published new research on West Nile virus and rabies virus (Kobayashi et al., 2016, Phongphaew et al., 2017, Anindita et al., 2016).
During the past year, the Network expanded the number of cooperating research centers by adding the University of Miami, Emory University and Tulane University School of Medicine in the USA and the International Vaccine Institute in South Korea (http://gvn.org/coe/). These centers are expected to strengthen cooperative research on vaccines, antiviral drugs, and emerging viruses such as ZIKV and Ebola virus.
7. Plans for 2017
Among other projects, the GVN staff in Baltimore expects to assist with a pilot study of hepatitis B in Arunachal Pradesh, India. Funded by the John Martin Foundation, the project will screen 30,000 people, provide vaccinations for those who are not infected, and develop a longitudinal cohort to treat patients with chronic hepatitis B infections.
Members of the GVN task force on HTLV-1 expect to publish additional commentaries and reviews during 2017. Similarly, a GVN task force report on ZIKV emergence and research priorities is in preparation. GVN staff also will continue to seek additional patient serum samples for the ZIKV serum bank based at the University of Texas Medical Branch in Galveston, Texas. Additionally, a project to sequence and conduct phylogenetic analyses of collected Zika virus isolates has begun in cooperation with the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), which is funded by the National Institute of Allergy and Infectious Diseases (USA).
The GVN will host its fourth annual Short Course on Medical Virology for young investigators. The Short Course attracts an international class of 15–20 postdoctoral students and fellows for five days of meetings and lectures with experts in medical virology research and clinical practice. Information about the course and registration is available online at http://www.gvn.org.
The 2017 international GVN meeting will be in Melbourne, Australia during September 25–27. It will be co-hosted by the GVN Centers of Excellence, the Institut Pasteur (Paris) and the Peter Doherty Institute for Infection and Immunity (Melbourne). The Scientific Organizing Committee includes Sharon Lewin, Director, The Peter Doherty Institute for Infection and Immunity, Australia; Robert Gallo, Co-Founder & Scientific Director, GVN, Institute of Human Virology at the University of Maryland School of Medicine; William Hall, Co-Founder, GVN, University College Dublin; Dr. Marc Lecuit, Director of the Biology of Infection Unit at Institut Pasteur, France; Damian Purcell, Viral Infectious Diseases, Doherty Institute, Australia; Peter Revill, Senior Medical Scientist, Victorian Infectious Diseases Reference Laboratory, Australia; and Didier Fontenille, Director, Institut Pasteur in Cambodia. Among the major topics of the 2017 meeting will be HTLV-1 infections among indigenous Australian populations; emerging viruses in Australia and Southeast Asia; and viruses causing hemorrhagic syndromes and persistent infections.
Acknowledgements
We thank the participants who presented their data at the 2016 International GVN Meeting. We thank the Japanese Society of Virology and Hokkaido University organizers for their outstanding management of the meeting and related events, and the Japanese hosts: Hokkaido University's Research Center for Zoonosis Control, the Global Institution for Collaborative Research and Education (GI-CoRE) Global Station for Zoonosis Control (GSZ) and the National Institute of Infectious Diseases (NIID). Support also was provided by numerous sponsors who are noted at http://gvn.org/sapporo_2016.
References
- Anindita P.D., Sasaki M., Nobori H., Sato A., Carr M., Ito N. Generation of recombinant rabies viruses encoding NanoLuc luciferase for antiviral activity assays. Virus Res. 2016;215:121–128. doi: 10.1016/j.virusres.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Annunziata C., Buonaguro L., Buonaguro F.M., Tornesello M.L. Characterization of the human papillomavirus (HPV) integration sites into genital cancers. Pathol. Oncol. Res. 2012;18(4):803–808. doi: 10.1007/s12253-012-9507-y. [DOI] [PubMed] [Google Scholar]
- de Ávila A.I., Gallego I., Soria M.E., Gregori J., Quer J., Esteban J.I. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS One. 2016;11:e0164691. doi: 10.1371/journal.pone.0164691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporale M., Di Gialleonorado L., Janowicz A., Wilkie G., Shaw A., Savini G. Virus and host factors affecting the clinical outcome of bluetongue virus infection. J. Virol. 2014;88(18):10399–10411. doi: 10.1128/JVI.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagrèverie H.M., Delaugerre C., Lewin S.R., Deeks S.G., Li J.Z. Ongoing clinical trials of human immunodeficiency virus latency-reversing and immunomodulatory agents. Open Forum Infect. Dis. 2016;7(4) doi: 10.1093/ofid/ofw189. ofw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D. Is virology dead? mBio. 2014;5 doi: 10.1128/mBio.01003-14. 2e01003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts T.R., Bagley K., Prado I.J., Bobb K.L., Schwartz J.A., Xu R. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc. Natl. Acad. Sci. U. S. A. 2015;112(9):E992–E999. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama W., Marzi A., Carmody A.B., Maruyama J., Kuroda M., Miyamoto H. Fcγ-receptor IIa-mediated Src signaling pathway is essential for the antibody-dependent enhancement of Ebola virus infection. PLoS Pathog. 2016;12(12):e1006139. doi: 10.1371/journal.ppat.1006139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R.C. History of the discoveries of the first human retroviruses: HTLV-1 and HTLV-2. Oncogene. 2005;5:5926–5930. doi: 10.1038/sj.onc.1208980. Review. [DOI] [PubMed] [Google Scholar]
- Gallo R.C., Willems L., Hasegawa H. Global virus Network's task force on HTLV-1. Blood. 2016;128(26):3029–3031. doi: 10.1182/blood-2016-09-739433. Screening transplant donors for HTLV-1 and -2. [DOI] [PubMed] [Google Scholar]
- Gérardin P., Couderc T., Bintner M., Tournebize P., Renouil M., Lémant J. Chikungunya virus-associated encephalitis: a cohort study on La Réunion Island, 2005-2009. Neurology. 2016;86(1):94–102. doi: 10.1212/WNL.0000000000002234. [DOI] [PubMed] [Google Scholar]
- Gillet N.A., Hamaidia M., de Brogniez A., Gutiérrez G., Renotte N., Reichert M. Bovine leukemia virus small noncoding RNAs are functional elements that regulate replication and contribute to oncogenesis in vivo. PLoS Pathog. 2016;12(4):e1005588. doi: 10.1371/journal.ppat.1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.E. Alphavirus encephalomyelitis: mechanisms and approaches to prevention of neuronal damage. Neurotherapeutics. 2016;13(3):455–460. doi: 10.1007/s13311-016-0434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haredy A.M., Takenaka N., Yamada H., Sakoda Y., Okamatsu M., Yamamoto N. An MDCK cell culture-derived formalin-inactivated influenza virus whole-virion vaccine from an influenza virus library confers cross-protective immunity by intranasal administration in mice. Clin. Vaccine Immunol. 2013;20(7):998–1007. doi: 10.1128/CVI.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski A.B., Muir A.J. Direct-acting antiviral medications for chronic hepatitis C virus infection. Gastroenterology Hepatology. 2011;7(3):154–162. [PMC free article] [PubMed] [Google Scholar]
- Karlas A., Berre S., Couderc T., Varjak M., Braun P., Meyer M. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat. Commun. 2016;12:11320. doi: 10.1038/ncomms11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Suzuki T., Kawaguchi A., Phongphaew W., Yoshii K., Iwano T. Rab8b regulates transport of West Nile virus particles from recycling endosomes. J. Biol. Chem. 2016;291(12):6559–6568. doi: 10.1074/jbc.M115.712760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman M.M., Cannon P.M., Currier J.S., June C.H., Kiem H.P., Kuritzkes D.R. A cure for HIV infection: “not in my lifetime” or “just around the corner”? Pathogens Immun. 2016;1(1):154–164. doi: 10.20411/pai.v1i1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A. New organization pledges scientific expertise for viral outbreaks. Nat. Med. 2011;17:394. doi: 10.1038/nm0411-394a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSweegan E., Weaver S.C., Lecuit M., Frieman M., Morrison T.E., Hrynkow S. The global virus network: challenging chikungunya. Antivir. Res. 2015;120:147–152. doi: 10.1016/j.antiviral.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzi E., Caporale M., Rocchi M., Martín V., Gamino V., di Provvido A. Follicular dendritic cell disruption as a novel mechanism of virus-induced immunosuppression. Proc. Natl. Acad. Sci. U. S. A. 2016;113(41):E6238–E6247. doi: 10.1073/pnas.1610012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire C.E., Geisbert J.B., Agans K.N., Thi E.P., Lee A.C., Fenton K.A. Passive immunotherapy: assessment of convalescent serum against Ebola virus makona infection in nonhuman primates. J. Infect. Dis. 2016;214(suppl 3):S367–S374. doi: 10.1093/infdis/jiw333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S., Arii J., Koyanagi N., Kato A., Kawaguchi Y. The interaction between herpes simplex virus 1 tegument proteins UL51 and UL14 and its role in virion morphogenesis. J. Virol. 2016;90(19):8754–8767. doi: 10.1128/JVI.01258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phongphaew W., Kobayashi S., Sasaki M., Carr M., Hall W.W., Orba Y. Valosin-containing protein (VCP/p97) plays a role in the replication of West Nile virus. Virus Res. 2017;15(228):114–123. doi: 10.1016/j.virusres.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X.C., Shi M., Tian J.H., Lin X.D., Gao D.Y., He J.R. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. U. S. A. 2014;111(18):6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin A., Ashkenazy H., Smelyanski L., Kaplan G., Penn O., Weiss-Ottolenghi Y. Deep panning: steps towards probing the IgOme. PLoS One. 2012;7(8):e41469. doi: 10.1371/journal.pone.0041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E.O., Aman M.J. Feverish quest for Ebola immunotherapy: straight or cocktail? Trends Microbiol. 2016;24(9):684–686. doi: 10.1016/j.tim.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Saphire E.O., Dye J.M., Kobinger G.P., Zeitlin L., Chandran K., Garry R.F. How to turn competitors into collaborators. Nature. 2017;541(7637):283–285. doi: 10.1038/541283a. [DOI] [PubMed] [Google Scholar]
- Sun G., Larsen C.N., Baumgarth N., Klem E.B., Scheuermann R.H. Comprehensive annotation of mature peptides and genotypes for Zika virus. PLoS One. 2017;12(1):e0170462. doi: 10.1371/journal.pone.0170462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Kawaguchi A., Ainai A., Tamura S., Ito R., Multihartina P. Relationship of the quaternary structure of human secretory IgA to neutralization of influenza virus. Proc. Natl. Acad. Sci. U. S. A. 2015;112(25):7809–7814. doi: 10.1073/pnas.1503885112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornesello M.L., Annunziata C., Buonaguro L., Losito S., Greggi S., Buonaguro F.M. TP53 and PIK3CA gene mutations in adenocarcinoma, squamous cell carcinoma and high-grade intraepithelial neoplasia of the cervix. J. Transl. Med. 2014;16(12):255. doi: 10.1186/s12967-014-0255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.E., Podolsky K.A., Bartesaghi A., Kuybeda O., Grandinetti G., Wohlbold T.J. Cryo-electron microscopy structures of chimeric hemagglutinin displayed on a Universal influenza vaccine candidate. MBio. 2016;7(2):e00257. doi: 10.1128/mBio.00257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Costa F., Garcia-Blanco M.A., Ko A.I., Ribeiro G.S., Saade G. Zika virus: history, emergence, biology, and prospects for control. Antivir. Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Ottolenghi Y., Gershoni J.M. Profiling the IgOme: meeting the challenge. FEBS Lett. 2014;588(2):318–325. doi: 10.1016/j.febslet.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Hasegawa H., Accolla R., Bangham C., Bazarbachi A., Bertazzoni U. Reducing the global burden of HTLV-1 infection: an agenda for research and action. Antivir. Res. 2017;37:41–48. doi: 10.1016/j.antiviral.2016.10.015. [DOI] [PubMed] [Google Scholar]