Abstract
Respiratory viral infections cause mild to severe diseases, such as common cold, bronchiolitis and pneumonia and are associated with substantial burden for society. To test new molecules for shortening, alleviating the diseases or to develop new therapies, relevant human in vitro models are mandatory. MucilAir™, a human standardized air-liquid interface 3D airway epithelial culture holds in vitro specific mechanisms to counter invaders comparable to the in vivo situation, such as mucus production, mucociliary clearance, and secretion of defensive molecules. The objective of this study was to test the relevance of such a model for the discovery and validation of antiviral drugs. Fully differentiated 3D nasal epithelium cultures were inoculated with picornaviruses, a coronavirus and influenza A viruses in the absence or in the presence of reference antiviral drugs. Results showed that, rupintrivir efficiently inhibits the replication of respiratory picornaviruses in a dose dependent manner and prevents the impairment of the mucociliary clearance. Similarly, oseltamivir reduced the replication of influenza A viruses in a dose dependent manner and prevented the impairment of the epithelial barrier function and cytotoxicity until 4 days of infection. In addition we found that Rhinovirus B14, C15 and influenza A(H1N1) induce significant increase of β Defensins 2 and Cathelicidin release with different time course. These results reveal that a large panel of epithelial functions is modified upon viral infection and validate MucilAir™ as a pertinent tool for pre-clinical antiviral drug testing.
Keywords: Antiviral screening, Respiratory infection, Human in vitro airway model, Innate immune response
Highlights
-
•
Reference antivirals inhibit in a dose-dependent manner the respiratory virus production in MucilAir™.
-
•
Respiratory viruses induce specific antimicrobial peptide expression and functional changes in MucilAir™.
-
•
Antivirals prevent virus-induced dysfunctions, the disruption of epithelial barrier and the decrease of mucociliary clearance.
-
•
MucilAir™ is a suitable model to produce clinical respiratory virus isolates and to perform antiviral drugs screening.
1. Introduction
Human reconstituted airway epithelial 3D models are highly relevant and useful tools for pharmacology, toxicology and biology in general. Several standardized models are commercially available, stratified along the respiratory tract, as nasal, tracheal and bronchial MucilAir™, and recently SmallAir™ originated from small airways (Huang et al., 2017). MucilAir™ is a well-validated human in vitro model containing ciliated, goblet and basal cells, representing the upper airway epithelia (Huang et al., 2011). As the airway epithelium is the first line of defence against microbes, MucilAir™ provides a robust and convenient platform for studying the viral or bacterial infections in vitro, allowing dissecting the molecular and cellular mechanisms of host-pathogen interactions. It constitutes also a platform for medium throughput testing of therapeutic compounds.
The upper respiratory infection also known as common cold is mostly caused by viruses. While the majority of these infections are self-limiting, however severe complications, as bronchiolitis and pneumonia, may occur particularly in children or elderly patients. Moreover, upper respiratory infections are widely implicated in acute exacerbation of cystic fibrosis, asthma, and chronic obstructive pulmonary disease (Jackson and Johnston, 2010; Seemungal et al., 2000; Wat et al., 2008). Among the respiratory pathogen viruses, rhinoviruses, respiratory syncytial viruses and influenza viruses have high prevalence.
Human Rhinoviruses (RVs) and human Enteroviruses (EVs) belong to the Picornaviridae family which are non-enveloped, positive sense single stranded RNA viruses. Human Rhinoviruses are divided into RV-A, RV-B and RV-C species. Influenza viruses belong to the Orthomyxoviridae family, and are enveloped, negative sense single stranded, segmented RNA viruses. Human coronaviruses (HCoV) belong to Coronaviridae family are enveloped positive sense single stranded RNA viruses.
Seasonal respiratory illnesses present a substantial burden for healthcare worldwide. Currently the treatment of most viral respiratory infections is supportive, but in some cases the use of antiviral drugs is necessary, for instance infants at high-risk, immunocompromised patients or in chronic respiratory diseases. Thus, oseltamivir, zanamivir and peramivir are recommended for influenza, while ribavirin, and palivizumab are used to treat and prevent, respectively, respiratory syncytial virus infections (https://www.cdc.gov/flu/professionals/antivirals/summary-clinicians.htm); (Jha et al., 2016). However, no Food and Drug Administration approved antivirals are available to rhinoviruses and enteroviruses or coronaviruses causing Severe Acute Respiratory and Middle East Respiratory Syndromes. Clearly there is an unmet need for this antiviral drugs. Moreover, testing of antiviral agents is, at least in part, hampered by a limited number of relevant animal models for these viruses. In this context, the standardized nasal human 3D epithelial MucilAir™ model having functional characteristics similar to in vivo situation, such as mucus production, mucociliary clearance, and secretion of cytokines and chemokines, appears as a suitable model to support the development of new antivirals.
The aim of this study was to evaluate and validate MucilAir™ as a potential in vitro platform for developing antiviral compounds against respiratory viruses. For this purpose, reference antiviral molecules were tested on fully differentiated 3D nasal epithelium cultures inoculated with targeted respiratory viruses. In addition, to emphasize the accuracy of MucilAir™ to mimic the in vivo situation, we also characterised the antimicrobial response of the model by measuring respiratory virus induced specific release of antimicrobial peptides.
In this study, one representative of each rhinovirus species, RV-A16, RV-B14 and RV-C15, as well as EV-D68 as representative of respiratory enteroviruses were chosen for the inoculation of the MucilAir™ cultures. Rupintrivir is an irreversible inhibitor of human rhinovirus 3C protease and has a broad antirhinoviral activity (Zalman et al., 2000). Despite not having passed Phase II clinical trials in natural infections because of poor pharmacokinetic properties, rupintrivir has consistently shown antiviral activity both in vitro and in humans with experimentally induced rhinovirus colds (Hayden et al., 2003; Patick et al., 1999), and was therefore chosen as reference drug in this study. From the Influenza A genus, H1N1 and H3N2 subtypes were used in this study. As antiviral, we follow the WHO recommendation, which advise the use of oseltamivir, a neuraminidase inhibitor, to treat and prevent influenza infections. Finally, from human coronaviruses, HCoV-OC43 was used for MucilAir™ infection. No reference antiviral is currently available for this strain.
2. Materials and methods
2.1. MucilAir™ culturing
Airway cells were obtained from patients undergoing surgical polypectomy. All experimental procedures were explained in full, and all subjects provided informed consent. The study was conducted according to the declaration of Helsinki on biomedical research (Hong Kong amendment, 1989), and received approval from local ethics commission. Human airway epithelial cells were isolated and expanded by two passages to preserve the physiological characteristics of the cells. Airway epithelia then can be reconstituted from individual donors or, to lessen differences between donors, from a mixture of human airway cells from different donors (MucilAir™-Pool). For viral infections, MucilAir™-Pool airway epithelia were reconstituted with a mixture of human nasal cells isolated from 14 different donors, and cultured at the air–liquid interface (ALI) in MucilAir™ culture medium (EP04MM), ready-to-use, chemically defined, serum-free (Epithelix Sàrl, Geneva, Switzerland), in 24-well plates with 6.5-mm Transwell® inserts (cat #3470, Corning Incorporated, Tweksbury, USA). The MucilAir™ models are stable for months and generate reproducible data in several domains, such as toxicology, therapeutic drug study or disease model (Baxter et al., 2015), (Balogh Sivars et al., 2017), (Rocca et al., 2016), (Sonneville et al., 2017).
2.2. Viruses and inoculation
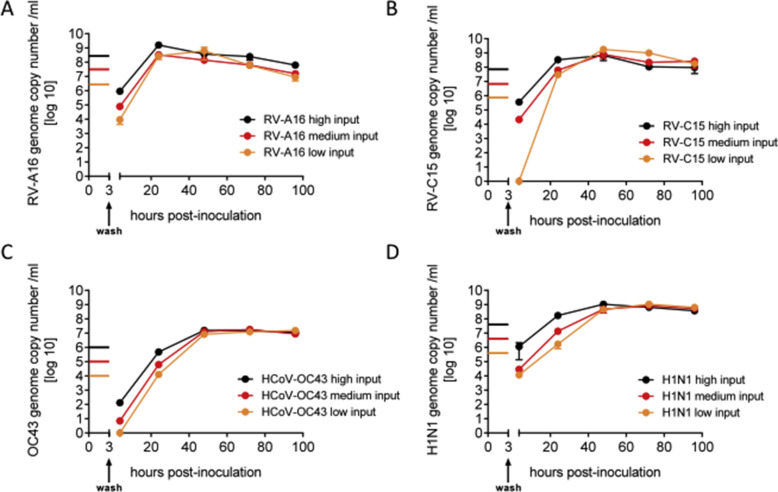
Rhinovirus (RV) A16 and B14 were purchased from American Type Culture Collection (Manassas, USA), while Rhinovirus C15, EV-D68, HCoV-OC43, influenza A(H1N1) and influenza A(H3N2) were isolated directly on MucilAir™ from clinical specimen as previously described (Essaidi-Laziosi et al., 2017; Tapparel et al., 2013). Influenza lineage was determined by PCR, hemagglutination inhibition assay and partial sequencing for A/Switzerland/7717739/2013(H1N1), which is very close to the reference strain A/California/07/09 and for A/Switzerland/8004462/2013(H3N2). Viral stocks for the experiments were produced on MucilAir™, collecting apical washes with culture medium. Production of several days were pooled and quantified by Taqman qPCR, aliquoted and stored at −80 °C. Prior to infection, the apical side of the MucilAir™ cultures were washed once with culture medium and transferred to a new plate containing EP04MM culture medium. Inoculations were performed with 100 μl of culture medium containing different concentrations of virus applied to the apical side of the cultures for 3 h at 34 °C. Three concentrations of inoculation were tested in preliminary experiments in MucilAir™ (Supplementary Fig. 1). Non-infected controls were exposed also to 100 μl of culture medium on the apical side for 3 h. The age of the cultures varied between 50 and 70 days of ALI. Viruses were eliminated after the incubation period by three rapid washing steps. Remaining viruses after the washing were verified by a 20 min apical wash and quantified by qPCR.
2.3. Antiviral drugs
Rupintrivir was purchased from Santa Cruz Biotechnology (Dallas, US). The neuraminidase inhibitor oseltamivir carboxylate (oseltamivir) was obtained from Carbosynth (Compton, UK). Drugs were diluted in DMSO, (final concentration was 0.25%) and used at 0.05 μM, 0.5 μM, 5μΜ and 0.01, 0.1, 1, 10 μM respectively. Antivirals were added concomitantly to viral inoculation in the basal media and renewed all along the infection. Culture media was changed every day.
2.4. Virus genome copy number
Apical washes with 200 μl of culture medium were performed for 20 min in the incubator at 3 h, day 1, 2, 3, and 4. RNA extracted from the apical washes with the QIAamp® Viral RNA kit (Qiagen) or from MucilAir™ tissue with RNeasy kit (Qiagen) was quantified by quantitative RT-PCR (qPCR) (QuantiTect Probe RT-PCR, Qiagen) in a Taqman ABI 7000 from Applied Biosystems (ThermoFisher Scientific, Waltham, US). Primers and Taqman probes as well as the amplification parameters were described previously (Essaidi-Laziosi et al., 2017; Schibler et al., 2012). Briefly, 5′ UTR region was amplified with 5′ AGC CTG CGT GCC KGC C 3′ forward and 5′ GAA ACA CGG ACA CCC AAA GTA GT 3′ reverse primers for rhinoviruses; 5′ GCT GCG YTG GCG GCC 3′ forward and 5′ GAA ACA CGG ACA CCC AAA GTA GT 3′ reverse primers for enterovirus. For influenza A(H1N1) virus, MP gene was amplified with 5′ GAC CRA TCC TGT CAC CTC TGA C 3′ forward and 5′ AGG GCA TTY TGG ACA AAK CGT CTA 3 reverse primers; for influenza A(H3N2), NS gene was used with 5′ TCC TCA AYT CAC TCT TCG AGC G 3′ forward and 5′ CGG TGC TCT TGA CCA AAT TGG 3′ reverse primers; for coronavirus, N2 gene was amplified with 5′ STC GAT CGG GAC CCA AGT AG 3′ forward and 5′ CCT TCC TGA GCC TTC AAT ATA GTA ACC 3 reverse primers.
2.5. Cell culture-based titration
MDCK (Madin-Darby canine kidney) cells were obtained from the American Type Culture Collection (Manassas, US) and cultured in Dulbecco's Modified Eagle Medium (DMEM) and GlutaMAX (31966021, ThermoFisher Scientific) supplemented with 10% (v/v) fetal bovine serum (FBS) (P40-37500, Pan Biotech) and 100 μg/mL of penicillin and streptomycin (15140-122, Gibco). HeLa cells were obtained from the American Type Culture Collection (Manassas, US) and cultured in Minimum Essential Medium Eagle (MEM) (M4655, Sigma) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% l-glutamine (25030-024, Gibco), 0.5% fungizone (15290-026, Gibco), 1% gentamycine (15710-049, Gibco), 0.2% vancomycine (A18390250, AppliChem). Cultures were maintained at 37 °C in a 5% CO2 atmosphere. The infection medium of MDCK was similar but with 2.5% FBS, whereas for the HeLa was the McCoy's 5A (26600, Gibco) supplemented with 2% FBS, 0.4% fungizone, 0.2% vancomycine, 0.5% gentamycine, 3% MgCl2 1M (6-5232-3, Hanselar AG, Switzerland).
Viral titers of the RV-A16 and influenza A(H1N1) samples were determined as log10TCID50/ml by the endpoint method of Reed and Muench in HeLa and MDCK cells respectively (Flint, 2015). Confluent monolayers were inoculated in quadruplicates with 10-fold serial dilutions of the viral samples and were placed at 34 °C. 16 h post infection, the cells were washed with phosphate-buffered saline (PBS) and fixed with cold methanol and acetone (1:1) for 1 min and permeabilised with PBS-Triton 0.1% (Fluka). Infected cells were immunostained using the mouse J2 anti-dsRNA antibody (Scicons, diluted 1:250) or the mouse anti-Influenza A antibody (#5001, Light Diagnostics, diluted 1:100) and the anti-mouse HRP linked secondary antibody (# 7076, Cell Signaling Technology, diluted 1:1000). Cells were incubated for 15 min with the active substrate (D4293-50SET, Sigma) diluted in deionized water and subsequently washed with phosphate-buffered saline (PBS). Positive wells were scored under a light microscope.
2.6. Trans-epithelial electrical resistance (TEER)
After addition of 200 μl of culture medium to the apical compartment of the tissue cultures, resistance was measured across cultures with an EVOMX volt-ohm-meter (World Precision Instruments, Sarasota, US). Resistance values (Ω) were converted to TEER (Ω.cm2) by using the following formula: TEER (Ω.cm2) = (resistance value (Ω) - 100(Ω)) x 0.33 (cm2), where 100 Ω is the resistance of the membrane and 0.33 cm2 is the total surface of the epithelium. Virus induced changes are presented as percentage of the non-infected control.
2.7. Cytotoxicity measure
For the lactate dehydrogenase assay, 100 μl from the basal medium was incubated with the reaction mixture of the Cytotoxicity Detection KitPLUS, following manufacturer's instructions (Sigma, Roche; ST Louis, USA). To determine the percentage of cytotoxicity, the following equation was used (A = absorbance values): Cytotoxicity (%) = (A (exp value)-A (low control)/A (high control)-A (low control))*100. The high control value corresponds to a 10% Triton X-100 treatment applied to the culture for 24 h. A threshold limit of 5% of the toxicity index reflects the physiological cell turnover in MucilAir™ cultures.
2.8. Mucociliary clearance (MCC)
Polystyrene microbeads of 30 μm diameter (Sigma, 84135) were added to the apical surface of the airway cultures. The movements of the microbeads were video tracked and recorded at 2 frames per second for 30 images, at 34 °C. Video acquisition setup was composed of a Sony XCD-U100CR camera connected to an Olympus BX51 microscope with a 5× objective and a heated microscope plate. Three movies were taken per insert. Average beads velocity (μm/sec) was calculated with the ImageProPlus 6.0 software.
2.9. ELISA
Human β-defensins 2 (BD-2) and Cathelicidin antimicrobial peptide (CAMP) were quantified in both basal medium and apical wash according to manufacturer's instructions (Cusabio, Wuhan, CN). Samples were diluted to 1:100 for ΒD− 2 analysis, but used pure for CAMP measurements.
2.10. Statistics
Data are presented as mean ± standard error of the mean. For statistical comparison one-way or two-way analysis of variance were performed with multiple comparison tests using GraphPad Prims software (version 6.01, La Jolla, USA) (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
3. Results
3.1. Reference antivirals inhibit virus replication in a dose dependent manner in MucilAir™
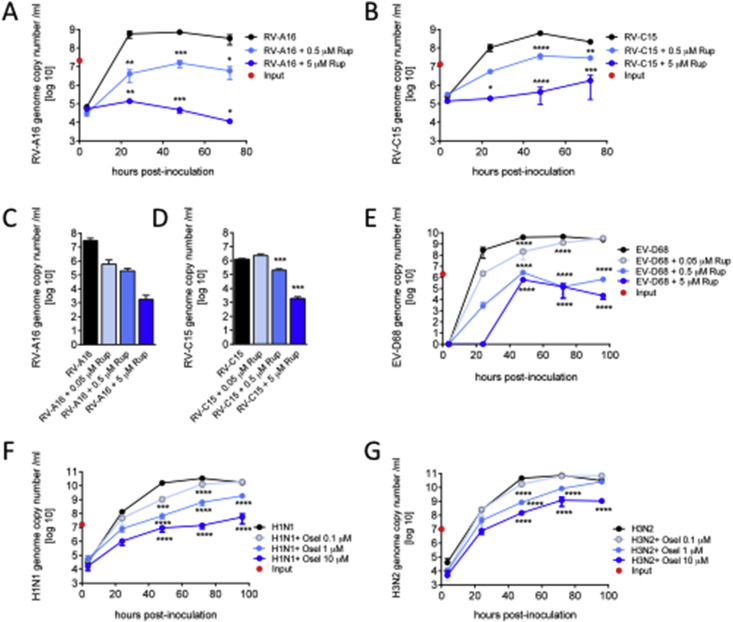
Preliminary tests indicated that rupintrivir was more efficient to inhibit viral production when added to the basal medium than on the apical side. Indeed, rupintrivir administered basally with concentrations ranging from 0.05 to 5 μM showed a dose-dependent inhibition of the RV-A16 (Fig. 1 A) and C15 viral production (Fig. 1B) as reported by virus quantification performed from apical wash at 24, 48 and 72 h post-inoculation (n = 2 for all conditions). Rupintrivir inhibition was 4 orders of magnitude for RV-A16 and 2 orders of magnitude for RV-C15 at 72 h compared to the control. Virus quantification performed at 24 h in the MucilAir™ tissue confirmed that rupintrivir inhibited viral replication during RV-A16 (Fig. 1C, n = 2) and RV-C15 infections (Fig. 1D, n = 3). Similarly, rupintrivir inhibited EV-D68 production in a dose dependent manner. At 96 h, the amplitude of the inhibition reached 4.5 logs for the 5 μM rupintrivir condition (Fig. 1E, n = 3).
Fig. 1.
Reference antivirals inhibit viral production in a dose dependent manner in MucilAir™. A–B: Rupintrivir of 0.5 and 5 μM were applied in the basal culture medium concomitantly with Rhinovirus A16 and C15 (n = 2). RNA was quantified from apical samples collection by qPCR and expressed as genome copy number/ml in time post-inoculation. C–D: in MucilAir™ tissue viral contents were quantified by qPCR at 24 h in the presence of 0.05, 0.5 and 5 μM of rupintrivir following Rhinovirus A16 and C15 inoculations (n = 2 and 3). E: Time course of the apical viral load after Enterovirus D68 inoculation in the presence of 0.5 and 5 μM of rupintrivir (n = 3). F–G: Time course of the apical viral load after influenza A(H1N1) and influenza A(H3N2) inoculation in the presence of 0.1, 1 and 10 μM of oseltamivir (n = 3 and 3). One way (C–D) or two-way ANOVA tests were performed with multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
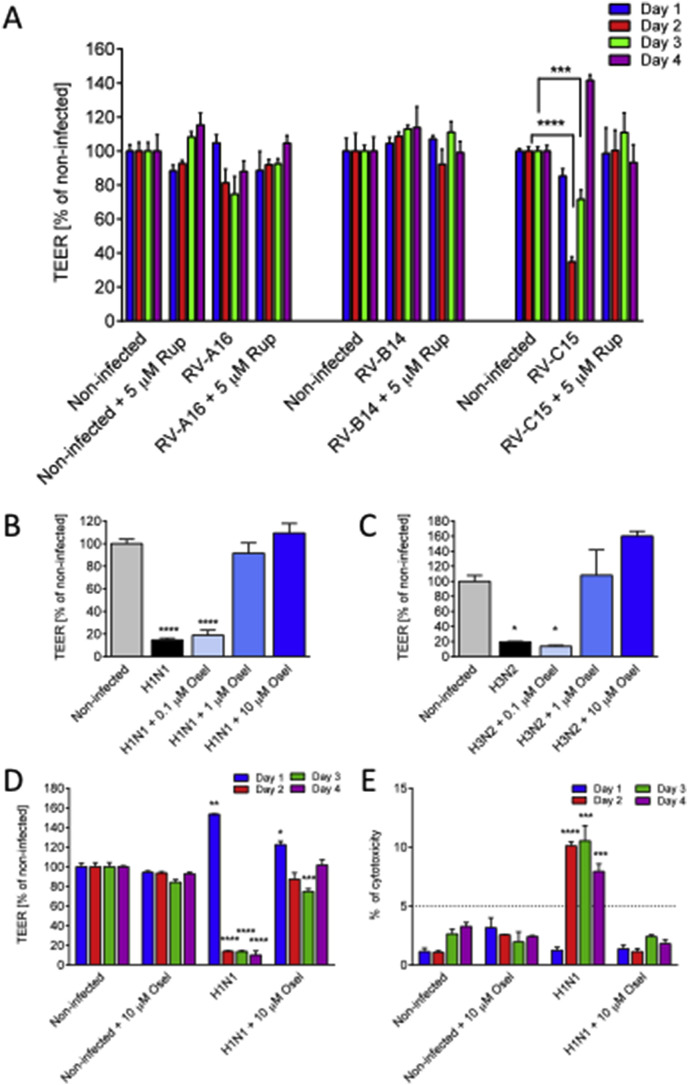
Regarding reference anti-influenza compound, oseltamivir was administered to the basal medium of MucilAir™ at concentrations ranging from 0.1 to 10 μM to mimic human therapeutic condition, where a mean plasma concentration of 300 μg/L (1 μM) was reported after orally administered oseltamivir (Davies, 2010). We found that oseltamivir treatment reduced H1N1 (Fig. 1F, n = 3) and H3N2 (Fig. 1G, n = 3) influenza virus replication in a dose-dependent manner. The amplitudes of reduction resulting from 10 μM oseltamivir treatment were 2.5 and 2 logs respectively at 96 h. Importantly, the most efficient doses of antivirals (rupintrivir 5 μM and oseltamivir 10 μM) did not show any toxic effects on MucilAir™ cultures, as documented by the analysis of physiological functions in the non-infected but drug treated condition, i.e TEER (Fig. 2 A, D) and mucociliary clearance (Fig. 3 A, C).
Fig. 2.
Antivirals prevent virus-induced disruption of MucilAir™ epithelial tissue. A: Transepithelial electrical resistance (TEER) expressed in percentage of non-infected control following Rhinovirus A16, B14 and C15 inoculation at 24, 48, 72 and 96 h (n = 6, 3 and 6) in the presence or absence of rupintrivir of 5 μM. B–C: TEER percentage in the presence of different concentrations of oseltamivir with influenza A(H1N1) and influenza A(H3N2) infections at 96 h (n = 3). D-E. Time course of the influenza A(H1N1) induced tissue integrity changes (n = 3), represented by TEER (D) and cytotoxicity percentage (E) (threshold limit value of 5% cytotoxicity corresponds to a physiological cell turnover in MucilAir™ long term culture). One-way ANOVA tests were performed daily with multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
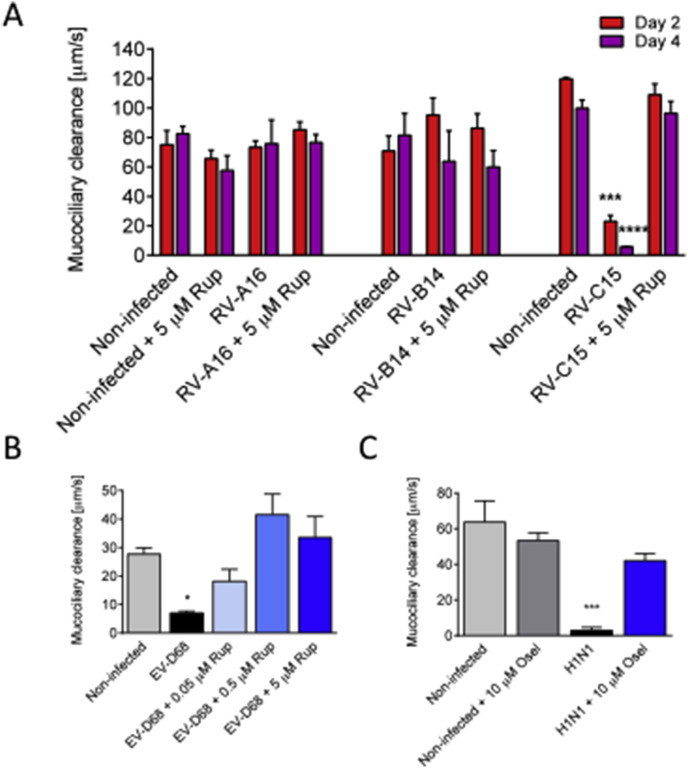
Fig. 3.
Antivirals prevent the diminution of the MucilAir™ mucociliary clearance (MCC) induced by Rhinovirus C15, Enterovirus D68 and influenza A(H1N1) viruses. A: MCC measured at 48 and 96 h at 34 °C following Rhinovirus A16, B14 and C15 inoculation in the absence or presence of antiviral rupintrivir of 5 μM (n = 5, 3 and 3). B: MCC measured at 96 h at room temperature following Enterovirus D68 inoculation in the absence or presence of antiviral rupintrivir of 5 μM (n = 3). C: MCC measured at 96 h at 34 °C following influenza A(H1N1) inoculation in the absence or presence of antiviral oseltamivir of 10 μM (n = 3). One-way ANOVA tests were performed daily with multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
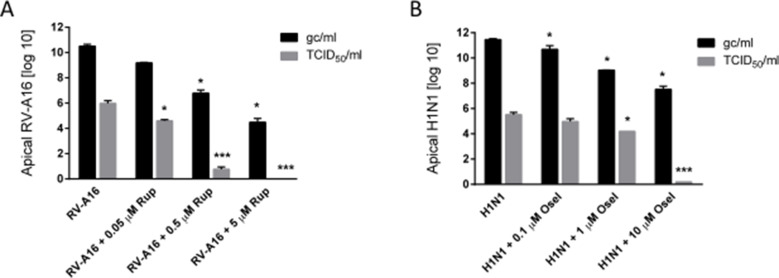
Because the genome copy number by itself does not give information about infectivity of the viral yields, a cell culture-based virus titration was performed. To this end, the 50% tissue infectivity dose (TCID50) was determined on common cell lines. An independent series of experiments was carried out with one representative virus from rhinovirus, RV-A16 treated with 3 concentrations of rupintrivir and with one representative virus from influenza A, H1N1 treated with 3 concentrations of oseltamivir. Based on the genome copy number results, infectious titer was assessed on apical washes collected 48 h post infection of MucilAir™ tissues by inoculation of these samples in Hela and MDCK cell lines. Comparison of genome copy number and infectious titers are presented in Supplementary Fig. 2. These new TCID50 results confirmed the antiviral efficacy of rupintrivir and oseltamivir in MucilAir™, which was first observed by genome copy number quantification. Results showed that the general pattern of antiviral induced dose-dependent inhibition is quite similar between the two methods. However the decay of viral particles at the highest antivirals doses seems more marked when estimated by the cell culture-based method, rather than the molecular method. In addition, rough estimation of rupintrivir IC50 for RV-A16 gave 6.5 μM based on genome copy number quantification and 0.8 μM by using the TCID50 method. Similarly, IC50 of oseltamivir for influenza A (H1N1) was estimated to 9 nM by genome copy number and 1 μM with TCID50. Although the limited number of antiviral dilutions does not permit to establish confident IC50 values, these results suggest differences in sensitivity between the two methods.
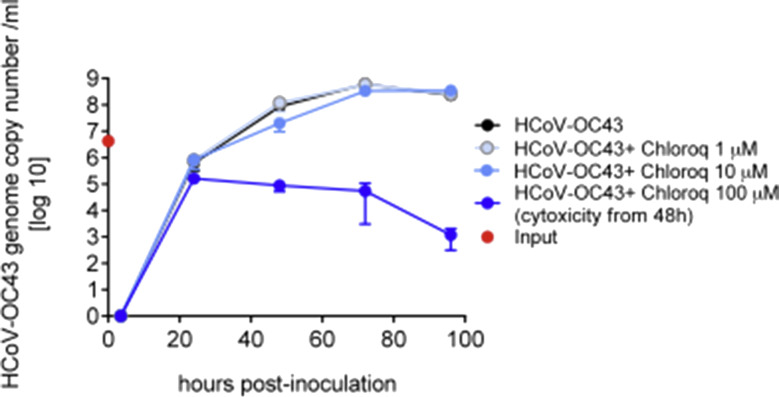
To inhibit HCoV-OC43 amplification on MucilAir™, several antivirals (among them chloroquine) were tested but none of these was efficient in our model. Chloroquine was applied at 1, 10 and 100 μM concentrations in the basal medium of MucilAir™, and showed cytotoxicity for its highest concentration as reported by microscopic observation and LDH assay from 48 h (Supplementary Fig. 3).
3.2. Antivirals prevent virus-induced disruption of barrier function in MucilAir™
The measurement of transepithelial electrical resistance (TEER) is a rapid, non-invasive method to assess tissue integrity. In addition, TEER is a highly dynamic parameter, which is sensitive to various stimuli. TEER values measured from a MucilAir™ culture typically fell between 200 and 600 Ω cm2. TEER values can show substantial reversible or irreversible modifications. A TEER value between 100 and 200 Ω cm2 is considered as very low, but the change may be yet reversible. A loss of tissue integrity corresponds to a TEER <100 Ω cm2. When an epithelium is seriously damaged, the decrease of TEER is associated with an increase of LDH release (cytotoxicity assay) or a decrease of the cell viability. MucilAir™, being long term cultures, have a physiological cell turnover showing maximum 5% cytotoxicity or cell replacement. Significant increase above of 5% cytotoxicity is associated with the decrease of TEER. Therefore, both TEER and cytotoxicity should be measured simultaneously. To compare antiviral efficacy during different virus infections, TEER values were presented as percentage of non-infected control cultures. Among the tested rhinoviruses, only RV-C15 induced a TEER diminution below 100 Ω cm2, showing 71.6 ± 5.5% of control at 48 h (n = 6, p < 0.0001), and 34.9 ± 2.8% of control at 72 h (p < 0.001) (Fig. 2A) without any increase of LDH release (data not shown). Interestingly, TEER was fully restored at 96 h with a value even higher than the control (141.6 ± 3.2%), suggesting that the epithelium was able to recover from the damage induced by RV-C15 within a short period of time. All TEER changes were prevented by a rupintrivir treatment (n = 5). RV-A16 produced a moderate decrease in TEER, which did not reach the level of significance. In contrast, both H1N1 and H3N2 influenza viruses caused a decrease of TEER below 100 Ω cm2 at 96 h, reaching 14.7 ± 1.6% of control (n = 3, p < 0.0001) and 19.4 ± 0.6% of control respectively (Fig. 2B, C, n = 3, p < 0.05). Again, TEER diminution was prevented in a dose dependent manner by the antiviral oseltamivir (n = 3). The H1N1 induced tissue disruption started from 48 h, resulting in 13.8 ± 0.7 TEER % of control (n = 3, p < 0.0001) and was associated with significant cytotoxicity, as demonstrated by an increase in LDH release, reaching 10.1 ± 0.3% toxicity index (p < 0.0001, compared to the non-infected control) (Fig. 2D and E). These TEER and LDH values remained affected in time, at 72 (13.7 ± 0.9 TEER %, p < 0.0001; 10.5 ± 1.3 LDH %, p < 0.001) and 96 h (10 ± 4.1 TEER %, p < 0.0001; 7.9 ± 0.6 LDH %, p < 0.001), suggesting no spontaneous repair in the case of influenza A(H1N1) infection in the observed time window (four days post-inoculation).
Of note, 5 μM rupintrivir and 10 μM oseltamivir treatments did not induce any significant changes in TEER (Fig. 2A, D) or LDH release (Fig. 2E), pointing out the lack of toxicity of these two drugs on the MucilAir™ cultures.
3.3. Antivirals prevent the decrease of mucociliary clearance following rhinovirus C15, enterovirus D68 and influenza A(H1N1) infections
The mucociliary clearance (MCC) is the primary defence mechanism to move away pathogens and particles from the respiratory airways. This function is fine-tuned by the quantity and quality of mucus production as well as the number of cilia and their synchronized beating. To measure MCC in the infected MucilAir™, microbeads were added on the apical surface of the epithelia and videos were taken in order to measure velocities of their movement. As mentioned, MCC is determined by various parameters and is highly dependent on temperature. At 34 °C, mean MCC ranged from 65 to 120 μm/s in non-infected MucilAir™ cultures. No significant mucociliary clearance changes were observed following RV-A16 and RV-B14 infection (n = 5 and n = 3, respectively). In contrast, RV-C15 infection caused a strong decrease of MCC at 48 h (23.1 ± 3.9 compared to 120 ± 1 μm/s in the non-infected control, n = 3, p < 0.001), and a complete arrest at 96 h (5.8 ± 0.3 vs. 100 ± 5.5 μm/s in the control, p < 0.0001) (Fig. 3A). This reduction was not accompanied by any cytotoxicity (data not shown). Of note, the addition of 5 μM rupintrivir to the culture media efficiently prevented the dramatic effect of RV-C15 on MCC (Fig. 3A). A preventive effect of rupintrivir was also observed during EV-D68 infection, where it counteracted in a dose dependent manner the decrease of MCC (7.1 ± 0.6 vs. 27.7 ± 2.3 μm/s in the control, n = 3, p < 0.05) measured at 96 h at room temperature (Fig. 3B). In a similar way, we found that oseltamivir was efficient to prevent the MCC arrest (2.8 ± 1.7 vs. 63.7 ± 12 μm/s in the control, n = 3) measured at 96 h at 34 °C after H1N1 infection (Fig. 3C). Rupintrivir at 5 μM and oseltamivir at 10 μM alone did not modify significantly the MCC of the MucilAir™ (Fig. 3A, C).
3.4. Rhinoviruses and influenza A(H1N1) induce an increase of antimicrobial peptides
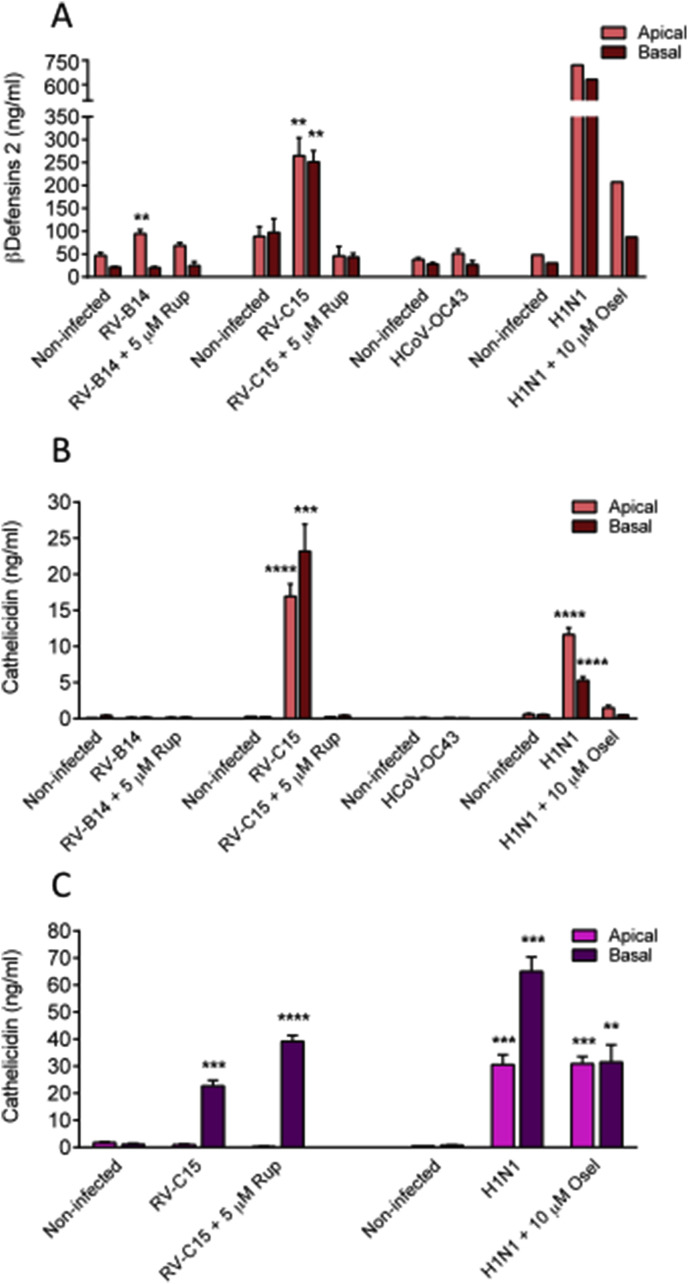
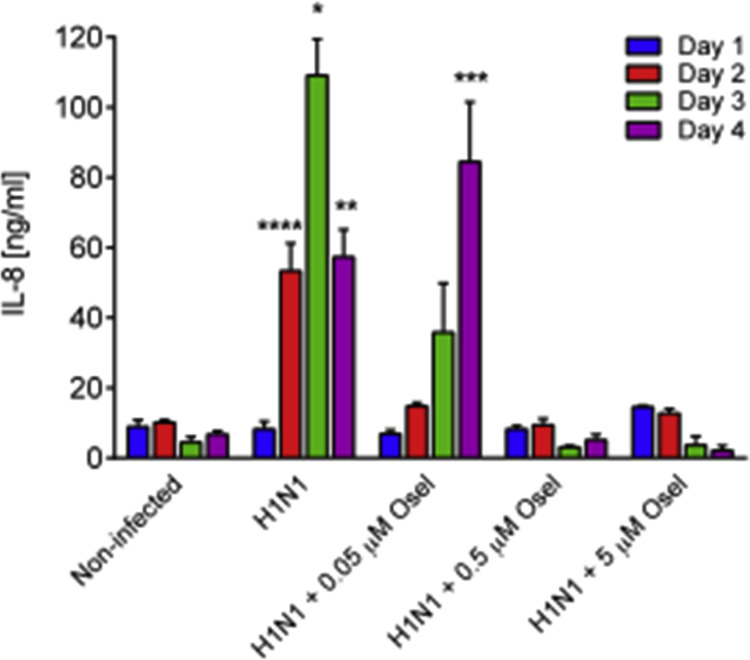
It has been recently shown by Essaidi-Laziosi et al., that following viral infection, MucilAir™ epithelial cells, as their in vivo counterparts, release a multitude of chemokines and cytokines. This local inflammation reaction is efficiently prevented by antivirals, in a dose dependent manner (Supplementary Fig. 4). However, another important primary innate host defence mechanism in the airway epithelial cells is the production of antimicrobial peptides (AMPs). Indeed, epithelial cells synthesize and secrete AMPs either constitutively or inducibly upon viral and bacterial infections (Hariri and Cohen, 2016). Here we measured by ELISA assays the apical and basal secretion of defensins and cathelicidin, which represent the two main respiratory epithelium AMP classes, during different viral infections. Non-infected cultures constitutively secreted 20–100 ng/ml β−defensins2 (BD-2) measured either basally or apically (Fig. 4 A). The quantity of BD-2 was significantly increased after RV-B14 infection (94.5 ± 8.6 vs. 46.6 ± 6.2 ng/ml, in non-infected, p < 0.05) at the apical side and following RV-C15 infection at both sides of MucilAir™ cultures, at 48 h post-inoculation (apical: 264.8 ± 39.2 vs. non-infected 88.5 ± 21.1, p < 0.05; basal: 251.3 ± 24.6 vs. 97.3 ± 29.8 ng/ml in non-infected, p < 0.05). Similarly, influenza A(H1N1) induced a huge increase in BD-2 secretion at both apical and basal sides of the cultures (apical: 721 vs. 48 ng/ml in control; basal: 631 vs. 30 ng/ml in control) (Fig. 4A). In contrast, HCoV-OC43 did not upregulate BD-2 secretion. Overall, rupintrivir and oseltamivir antivirals inhibited the production of BD-2 induced by viral infections.
Fig. 4.
Rhinoviruses and influenza A(H1N1) induce an increase of antimicrobial peptides. A. β−defensins 2 concentration in apical and basal sides of MucilAir™ measured at 48 h following Rhinovirus B14, C15, OC43 coronavirus and influenza A(H1N1) virus inoculation in the absence or presence of antivirals (n = 3, 4, 3 and 1). B: Cathelicidin concentration in apical and basal sides of MucilAir™ measured at 48 h following Rhinovirus B14, C15, OC43 coronavirus and influenza A(H1N1) virus inoculation in the absence or presence of antivirals (n = 3, 3, 3 and 3). C: Cathelicidin concentration in apical and basal sides of MucilAir™ measured at 96 h following Rhinovirus C15 and influenza A(H1N1) virus inoculation in the absence or presence of antivirals (n = 3 and 3). One-way ANOVA tests were performed daily with multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Regarding cathelicidin (CAMP), its secretion was significantly induced at both sides of MucilAir™ cultures after 48 h of infection with RV-C15 and influenza A(H1N1), but not with RV-B14 or HCoV-OC43 (Fig. 4B). No significant quantity of CAMP was detected in non-infected control cultures. The CAMP apical concentration was increased to 16.9 ± 1.7 ng/ml after RV-C15 infection (p < 0.0001) and to 11.6 ± 0.9 ng/ml following influenza A(H1N1) infection (n = 3, p < 0.0001) and the basal secretion was increased to 23.5 ± 3.7 ng/ml with RV-C15 (p < 0.001) and 5.3 ± 0.5 ng/ml with influenza A(H1N1) (n = 3, p < 0.0001). At this time point (48 h), both rupintrivir and oseltamivir antivirals prevented the production of CAMP in treated cultures. However, CAMP secretion profile changed with time. After 96 h of infection, no more CAMP was detected in the apical side of the RV-C15 inoculated cultures, while levels are still high (22.7 ± 2.1 ng/ml, n = 3, p < 0.001) in the basal side (Fig. 4C). High level of CAMP was also detected in the basal side of the RV-C15 + rupintrivir cultures (39.1 ± 2.2 ng/ml), contrasting with the protective effect previously observed with the rupintrivir at 48 h. For the influenza A(H1N1) condition, at 96 h we observed a strong induction of CAMP both in the apical and in the basal side of the cultures (30.6 ± 3.6 ng/ml and 65.1 ± 5.3 ng/ml, respectively). Similarly to rupintrivir, oseltamivir did not prevent any more CAMP secretion, as reported by the high concentrations measured in all compartments of the influenza A(H1N1) + oseltamivir cultures (apical: 30.9 ± 2.7 ng/ml, basal: 31.5 ± 6.4 ng/ml).
4. Discussion
Disabilities resulting from upper respiratory infections, which are mostly caused by viruses such as common cold, bronchiolitis and pneumonia represent an important cost for the society. To test new molecules intended to shorten or alleviate these pathologies, or to develop new antiviral approaches, relevant human models are clearly mandatory, due to species as well as cellular tropism of each virus. To study respiratory infections, traditionally cell lines and undifferentiated monolayer epithelial cells in submerged cultures were used, which couldn't replicate faithfully the real mechanisms of viral infections in human respiratory tract. Therefore, increasing number of studies turned recently toward ALI models, because of its high physiological and functional relevance, to investigate certain aspects of the respiratory viral infections (Berman et al., 2016; Farsani et al., 2015; Griggs et al., 2017; Jones et al., 2016). In this study, we undertook a rather holistic analysis of the physiological modifications happening in a human 3D airway ALI culture upon viral infection and revealed the importance of an innate immune response which is virus strain dependent. Reference antivirals efficiently inhibited viral replications in MucilAir™ cultures as assessed by two independent virus quantification methods, genome copy number determination and infectious virus titer obtained by TCID50. Moreover, we established that MucilAir™ cultures allow to screen and rank antivirals.
Overall, our data support the use of the standardized MucilAir™ airway as a promising preclinical model for the design and the test of novel antiviral strategies. In comparison with other air-liquid interface 3D approaches (Berman et al., 2016; Farsani et al., 2015; Griggs et al., 2017; Jones et al., 2016), MucilAir™ is made of low passage human airway epithelial cells ensuring a maximum similarity with physiological condition and the reconstruction is standardized with quality control using functional tests. Some important assets of MucilAir™ for pre-clinical testing have already been addressed. An absorption study demonstrated the capacity of the model to predict respiratory transport in human (Reus et al., 2014). Xenobiotic metabolism gene profiling demonstrated strong correspondences with normal human lung and activity of CYP1A1/1B1 and CYP2A6/2A13 were confirmed in MucilAir™ (Baxter et al., 2015), suggesting again high functional similarities with the in vivo situation.
4.1. Virus-host (MucilAir™) interactions
During this study we found that the levels of different respiratory virus productions in MucilAir™ are comparable to human infection conditions (Chiu et al., 2005; Esposito et al., 2014; Kennedy et al., 2014; Ngaosuwankul et al., 2010), but with differences in virus shedding kinetics. Indeed, after an early rapid phase of virus replication we noticed a steady state virus production in MucilAir™ cultures. Such a long term and persistent virus production may result from the combination of two factors: a limited number of infected cells in MucilAir™, as it has been previously demonstrated by immunocytochemistry (Essaidi-Laziosi et al., 2017); as well as the absence of immune cells in the model. A possible explanation for the limited virus spreading observed in MucilAir™ is the upregulation of specific antimicrobial peptides reported here, as well as the induction and release of specific cytokines and chemokines (Essaidi-Laziosi et al., 2017). Of note, these in vitro results are in frame with human in situ localization studies which showed that only a small proportion of the epithelial cells are infected by rhinoviruses in the upper and lower airways (Arruda et al., 1995; Mosser et al., 2002).
4.2. MucilAir™ epithelial barrier and mucociliary clearance functions are selectively affected by different virus strains
The nasal epithelial cells composing MucilAir™ are connected together via cell junctions known as desmosomes, adherens and tight junctions to form a physical barrier against inhaled pathogens, irritant or allergens, thus mimicking the in vivo situation. Here we found that RV-C15 leads to the loss of the barrier function of the epithelial tissue at an earlier time point of the infection. This result is in agreement with other data reported in vitro with the 16HBE14o-human bronchial epithelial cell line or in vivo in mice, where RV infection reduced TEER by dissociating zona occludens-1 from tight junction complex and E-cadherin from adherens junctions (Sajjan et al., 2008; Yeo and Jang, 2010). Interestingly, we noticed that this loss of integrity was transient since TEER was restored with time, due to a spontaneous repair of the epithelium. However, all these barrier dysfunctions were fully prevented by rupintrivir treatment, which therefore might be useful to limit the exacerbation of the infection. Conversely, we found that influenza viruses induced tissue disruption was not reversible in MucilAir™, certainly because of the high cytotoxicity caused by these viruses, as revealed by high LDH release. Indeed, an overactive innate immune response and an excessive production of reactive oxygen species have been documented for H5N1 influenza and other conditions causing acute lung injury (Imai et al., 2008).
Mucociliary clearance (MCC) primary function is to eliminate microbes and airborne particles. Ciliated cells generate a synchronized movement in the periciliary fluid, which is not viscous. Above that a viscous mucus gel layer lies down, composed of secreted glycosylated mucin proteins. The sticky mucus traps pathogens and the cilia move them away. The majority of virus strains tested in this study (RV-C15, EV-D68 and influenza A(H1N1)) caused MCC impairment, with the exception of RV-A16 and RV-B14. MCC disruption is the consequence of perturbation of a highly complex, multi-component system: such as cilia blockage or desynchronization, modification of the periciliary fluid layer and change of the viscoelasticity of the mucus, etc. (Gizurarson, 2015). While the precise reason of MCC dysfunction caused by each type of virus is not known, it is clearly related to the cell tropism of the different respiratory viruses used in this study (Dijkman et al., 2013; Essaidi-Laziosi et al., 2017). Importantly, in the absence of antiviral treatment, no spontaneous restoration of the MCC was detected, even 4 days after the initial inoculation. The decrease of the MCC is particularly favourable for a secondary bacterial infection (Pittet et al., 2010). To relieve the symptoms caused by viral infections, it would be beneficial to restore the mucociliary clearance.
4.3. Virus strain-dependent induction of innate immune responses in MucilAir™
Epithelial cells also generate and secrete antimicrobial peptides, which directly interact with pathogens. Two major classes of AMPs are present in human respiratory airways, defensins and cathelicidins. These compounds have a wide range of antiviral effects, including directly attacking viruses, inhibiting viral entry and preventing viral replication (Tripathi et al., 2013; Wilson et al., 2013). AMPs have multiple functions related to their ability to disrupt membranes and they can also function also as potent immune regulators (Lai and Gallo, 2009).
Our study provides for the first time, to the best of our knowledge, a detailed characterization of human AMPs upregulation, measured in a human 3D nasal epithelium model, upon various respiratory virus infections. First, we found that BD-2 is upregulated in MucilAir™ by Rhinovirus B14, C15 and influenza A(H1N1) measured 2 days post-inoculation. These results are in perfect agreement with the BD-2 level measured during an in vivo experimental RV-A16 infection in healthy adults. (Proud et al., 2004). In this study, BD-2 expression was reported significantly elevated on day 2 and related to symptom scores. In MucilAir™ cultures, we observed that in the presence of antivirals, the level of BD-2 induction is lower or no induction occurs at all. Further investigations are needed to determine if the modification of AMP levels by antivirals will have positive or negative effects on the course of the infection.
Second, we found that CAMP upregulation time course is dependent on virus strains in MucilAir™. RV-C15 induced CAMP concentration reached a maximum two days post-inoculation at the apical side, while the basal CAMP concentration remained stable over time. Influenza A(H1N1) induced secretion of CAMP showed a slower kinetics with a maximum concentration reached four days post-infection, both apically and basally. However, as with RV-C15, the secretion of CAMP was more important in the basal compartment than in the apical side. A possible explanation is that the accumulation of AMPs at the basal site resulted from the transient or persistent disruption of the epithelium barrier. On the other hand, we can speculate that the different time course of apical and basal release may reflect the dual function of CAMP in innate immunity: an early direct action on the microbes at the apical side, and a later immune regulatory action via recruitment and activation of immune cells at the basal side. Taken together, these results underscore the importance and the complexity of the host innate immune responses at work in MucilAir™.
5. Conclusion
In summary, these results demonstrate that human nasal airway epithelium reconstituted in vitro is an efficient tool for antiviral drug development. Our model could be useful for single or multiple viral infections, viral and bacterial co-infections. Further developments to evaluate antiviral response using diseased epithelia based on donor stratification (asthma, COPD) can be envisioned.
Acknowledgments
We thank C. Mas (OncoTheis) for discussion and comments on the manuscript. We are grateful to M. Caul-Futy (Epithelix) for technical assistance and M. Essaidi-Laziosi, L. Royston and I. Piuz (Laboratory of Virology, Prof. C. Tapparel) for technical advice and assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.antiviral.2018.06.007.
Appendix A. Supplementary data
The following is/are the supplementary data related to this article:
Supplementary Fig. 1.
Human rhinoviruses, Coronavirus OC43 and Influenza A H1N1 virus show efficient growth in MucilAir™. Apical virus production measured by qPCR and expressed as genome copy number/ml in time post-inoculation. Three different concentrations, high, medium and low, were used for an apical inoculation of 3 h for A: Rhinovirus A16, (n = 2), B: Rhinovirus C15 (n = 2), C: OC43 coronavirus (n = 3) and D: influenza A(H1N1) virus (n = 2).
Supplementary Fig. 2.
Comparison of the antiviral induced inhibition of viral production in MucilAir™ using genome copy number determination and TCID50 titration in cell lines. A: Rupintrivir of 0.05, 0.5 and 5 mM were applied in the basal culture medium concomitantly with Rhinovirus A16 apical inoculation (n = 2 for each condition). RV-A16 was quantified from apical samples collection at 48 h post-inoculation by qPCR, expressed as genome copy number/ml, and was inoculated in Hela cells for TCID50/ml determination after 16 h post-inoculation. B: Three concentrations of oseltamivir (0.1, 1 and 10 mM) were applied to the basal culture medium concomitantly with influenza A(H1N1) apical inoculation of the MucilAir™ tissues (n = 2 for each condition). (n = 2 for each condition). H1N1 was quantified from apical samples collection at 48 h post-inoculation by qPCR, expressed as genome copy number/ml, and was inoculated in MDCK cells for TCID50/ml determination after 16 h post-inoculation. One way ANOVA tests were performed with multiple comparisons using virus alone condition for both methods (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Supplementary Fig. 3.
Chloroquine antiviral has no effect on human coronavirus OC43 production in MucilAir™. Apical virus production measured by qPCR and expressed in genome copy number/ml as a function of time post-inoculation. Three concentrations of chloroquine (1, 10 and 100 mM) were applied in the basal media concomitantly with HCoV-OC43 apical inoculation of the MucilAir™ tissues (n = 2 for each condition). Note the cytotoxicity for the highest concentration of chloroquine from 48 h, assessed by microscopic observation and LDH assay.
Supplementary Fig. 4.
Interleukin 8 concentration in basal sides of MucilAir™ measured at 24, 48, 72 and 96 h following influenza A(H1N1) virus inoculation in the absence or presence of antivirals at different concentrations (n = 3).One way ANOVA tests were performed daily with multiple comparisons (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
References
- Arruda E., Boyle T.R., Winther B., Pevear D.C., Gwaltney J.M., Jr., Hayden F.G. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J. Infect. Dis. 1995;171:1329–1333. doi: 10.1093/infdis/171.5.1329. [DOI] [PubMed] [Google Scholar]
- Balogh Sivars K., Sivars U., Hornberg E., Zhang H., Branden L., Bonfante R., Huang S., Constant S., Robinson I., Betts C.J., Aberg P. A 3D human airway model enables prediction of respiratory toxicity of inhaled drugs in vitro. Toxicol. Sci. Off. J. Soc. Toxicol. 2018;162(1):301–308. doi: 10.1093/toxsci/kfx255. [DOI] [PubMed] [Google Scholar]
- Baxter A., Thain S., Banerjee A., Haswell L., Parmar A., Phillips G., Minet E. Targeted omics analyses, and metabolic enzyme activity assays demonstrate maintenance of key mucociliary characteristics in long term cultures of reconstituted human airway epithelia. Toxicol. Vitro Inter. J. Publ. Assoc. BIBRA. 2015;29:864–875. doi: 10.1016/j.tiv.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Berman R., Jiang D., Wu Q., Chu H.W. alpha1-Antitrypsin reduces rhinovirus infection in primary human airway epithelial cells exposed to cigarette smoke. Int. J. Chronic Obstr. Pulm. Dis. 2016;11:1279–1286. doi: 10.2147/COPD.S105717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.S., Chan K.H., Chu K.W., Kwan S.W., Guan Y., Poon L.L., Peiris J.S. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B.E. Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J. Antimicrob. Chemother. 2010;65(Suppl. 2):ii5–ii10. doi: 10.1093/jac/dkq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Koekkoek S.M., Deijs M., Jonsdottir H.R., Molenkamp R., Ieven M., Goossens H., Thiel V., van der Hoek L. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 2013;87:6081–6090. doi: 10.1128/JVI.03368-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Daleno C., Scala A., Castellazzi L., Terranova L., Sferrazza Papa S., Longo M.R., Pelucchi C., Principi N. Impact of rhinovirus nasopharyngeal viral load and viremia on severity of respiratory infections in children. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2014;33:41–48. doi: 10.1007/s10096-013-1926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi-Laziosi M., Brito F., Benaoudia S., Royston L., Cagno V., Fernandes-Rocha M., Piuz I., Zdobnov E., Huang S., Constant S., Boldi M.O., Kaiser L., Tapparel C. Propagation of respiratory viruses in human airway epithelia reveals persistent virus-specific signatures. J. Allergy Clin. Immunol. 2018;141(6):2074–2084. doi: 10.1016/j.jaci.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsani S.M., Deijs M., Dijkman R., Molenkamp R., Jeeninga R.E., Ieven M., Goossens H., van der Hoek L. Culturing of respiratory viruses in well-differentiated pseudostratified human airway epithelium as a tool to detect unknown viruses. Influenza Other Respir. Viruses. 2015;9:51–57. doi: 10.1111/irv.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S.J.R., V.R., Rall G.F., Skalka A.M., Enquist L.W. fourth ed. ASM Press; Washington, DC: 2015. Principles of Virology. [Google Scholar]
- Gizurarson S. The effect of cilia and the mucociliary clearance on successful drug delivery. Biol. Pharm. Bull. 2015;38:497–506. doi: 10.1248/bpb.b14-00398. [DOI] [PubMed] [Google Scholar]
- Griggs T.F., Bochkov Y.A., Basnet S., Pasic T.R., Brockman-Schneider R.A., Palmenberg A.C., Gern J.E. Rhinovirus C targets ciliated airway epithelial cells. Respir. Res. 2017;18:84. doi: 10.1186/s12931-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri B.M., Cohen N.A. New insights into upper airway innate immunity. Am. J. Rhinol. Allergy. 2016;30:319–323. doi: 10.2500/ajra.2016.30.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F.G., Turner R.B., Gwaltney J.M., Chi-Burris K., Gersten M., Hsyu P., Patick A.K., Smith G.J., 3rd, Zalman L.S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Boda B., Vernaz J., Ferreira E., Wiszniewski L., Constant S. Establishment and characterization of an in vitro human small airway model (SmallAir) Eur. J. Pharm. Biopharm. Off. J. Arbeitsgem. Pharm. Verfahrenstech. E.V. 2017;118:68–72. doi: 10.1016/j.ejpb.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Huang S., Wiszniewski L., Constant S. The use of in vitro 3D cell models in drug development for respiratory diseases. Drug Discov. Develop. Present Futur. 2011;8 [Google Scholar]
- Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., Ermolaeva M., Veldhuizen R., Leung Y.H., Wang H., Liu H., Sun Y., Pasparakis M., Kopf M., Mech C., Bavari S., Peiris J.S., Slutsky A.S., Akira S., Hultqvist M., Holmdahl R., Nicholls J., Jiang C., Binder C.J., Penninger J.M. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.J., Johnston S.L. The role of viruses in acute exacerbations of asthma. J. Allergy Clin. Immunol. 2010;125:1178–1187. doi: 10.1016/j.jaci.2010.04.021. quiz 1188–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A., Jarvis H., Fraser C., Openshaw P.J.M. Respiratory syncytial virus. In: Hui D.S., Rossi G.A., Johnston S.L., editors. SARS, MERS and Other Viral Lung Infections, Sheffield UK. 2016. [Google Scholar]
- Jones J.C., Marathe B.M., Lerner C., Kreis L., Gasser R., Pascua P.N., Najera I., Govorkova E.A. A novel endonuclease inhibitor exhibits broad-spectrum anti-influenza virus activity in vitro. Antimicrob. Agents Chemother. 2016;60:5504–5514. doi: 10.1128/AAC.00888-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy J.L., Shaker M., McMeen V., Gern J., Carper H., Murphy D., Lee W.M., Bochkov Y.A., Vrtis R.F., Platts-Mills T., Patrie J., Borish L., Steinke J.W., Woods W.A., Heymann P.W. Comparison of viral load in individuals with and without asthma during infections with rhinovirus. Am. J. Respir. Crit. Care Med. 2014;189:532–539. doi: 10.1164/rccm.201310-1767OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Gallo R.L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser A.G., Brockman-Schneider R., Amineva S., Burchell L., Sedgwick J.B., Busse W.W., Gern J.E. Similar frequency of rhinovirus-infectible cells in upper and lower airway epithelium. J. infect. dis. 2002;185:734–743. doi: 10.1086/339339. [DOI] [PubMed] [Google Scholar]
- Ngaosuwankul N., Noisumdaeng P., Komolsiri P., Pooruk P., Chokephaibulkit K., Chotpitayasunondh T., Sangsajja C., Chuchottaworn C., Farrar J., Puthavathana P. Influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses. Virol. J. 2010;7:75. doi: 10.1186/1743-422X-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patick A.K., Binford S.L., Brothers M.A., Jackson R.L., Ford C.E., Diem M.D., Maldonado F., Dragovich P.S., Zhou R., Prins T.J., Fuhrman S.A., Meador J.W., Zalman L.S., Matthews D.A., Worland S.T. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999;43:2444–2450. doi: 10.1128/aac.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet L.A., Hall-Stoodley L., Rutkowski M.R., Harmsen A.G. Influenza virus infection decreases tracheal mucociliary velocity and clearance of Streptococcus pneumoniae. Am. J. Respir. Cell Mol. Biol. 2010;42:450–460. doi: 10.1165/rcmb.2007-0417OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud D., Sanders S.P., Wiehler S. Human rhinovirus infection induces airway epithelial cell production of human beta-defensin 2 both in vitro and in vivo. J. Immunol. 2004;172:4637–4645. doi: 10.4049/jimmunol.172.7.4637. [DOI] [PubMed] [Google Scholar]
- Reus A.A., Maas W.J., Jansen H.T., Constant S., Staal Y.C., van Triel J.J., Kuper C.F. Feasibility of a 3D human airway epithelial model to study respiratory absorption. Toxicol. Vitro Inter. J. Publ. Assoc. BIBRA. 2014;28:258–264. doi: 10.1016/j.tiv.2013.10.025. [DOI] [PubMed] [Google Scholar]
- Rocca J., Manin S., Hulin A., Aissat A., Verbecq-Morlot W., Pruliere-Escabasse V., Wohlhuter-Haddad A., Epaud R., Fanen P., Tarze A. New use for an old drug: COX-independent anti-inflammatory effects of sulindac in models of cystic fibrosis. Br. J. Pharmacol. 2016;173:1728–1741. doi: 10.1111/bph.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan U., Wang Q., Zhao Y., Gruenert D.C., Hershenson M.B. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am. J. Respir. Crit. Care Med. 2008;178:1271–1281. doi: 10.1164/rccm.200801-136OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler M., Yerly S., Vieille G., Docquier M., Turin L., Kaiser L., Tapparel C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J. Clin. Microbiol. 2012;50:2868–2872. doi: 10.1128/JCM.06752-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemungal T.A., Harper-Owen R., Bhowmik A., Jeffries D.J., Wedzicha J.A. Detection of rhinovirus in induced sputum at exacerbation of chronic obstructive pulmonary disease. Eur. Respir. J. 2000;16:677–683. doi: 10.1034/j.1399-3003.2000.16d19.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville F., Ruffin M., Coraux C., Rousselet N., Le Rouzic P., Blouquit-Laye S., Corvol H., Tabary O. MicroRNA-9 downregulates the ANO1 chloride channel and contributes to cystic fibrosis lung pathology. Nat. Commun. 2017;8:710. doi: 10.1038/s41467-017-00813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C., Sobo K., Constant S., Huang S., Van Belle S., Kaiser L. Growth and characterization of different human rhinovirus C types in three-dimensional human airway epithelia reconstituted in vitro. Virology. 2013;446:1–8. doi: 10.1016/j.virol.2013.06.031. [DOI] [PubMed] [Google Scholar]
- Tripathi S., Tecle T., Verma A., Crouch E., White M., Hartshorn K.L. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J. Gen. Virol. 2013;94:40–49. doi: 10.1099/vir.0.045013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wat D., Gelder C., Hibbitts S., Cafferty F., Bowler I., Pierrepoint M., Evans R., Doull I. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros. Off. J. Euro. Cystic Fibros. Soc. 2008;7:320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.S., Wiens M.E., Smith J.G. Antiviral mechanisms of human defensins. J. Mol. Biol. 2013;425:4965–4980. doi: 10.1016/j.jmb.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo N.K., Jang Y.J. Rhinovirus infection-induced alteration of tight junction and adherens junction components in human nasal epithelial cells. Laryngoscope. 2010;120:346–352. doi: 10.1002/lary.20764. [DOI] [PubMed] [Google Scholar]
- Zalman L.S., Brothers M.A., Dragovich P.S., Zhou R., Prins T.J., Worland S.T., Patick A.K. Inhibition of human rhinovirus-induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 2000;44:1236–1241. doi: 10.1128/aac.44.5.1236-1241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]